The Honey Bee Body Surface as a Microbial Hub: Connectivity Shaped by Monoculture vs. Polyculture Farming
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Treatments
2.2. Sample Collection
2.3. DNA Extraction and Bioinformatic Analysis
2.4. Statistical Analysis
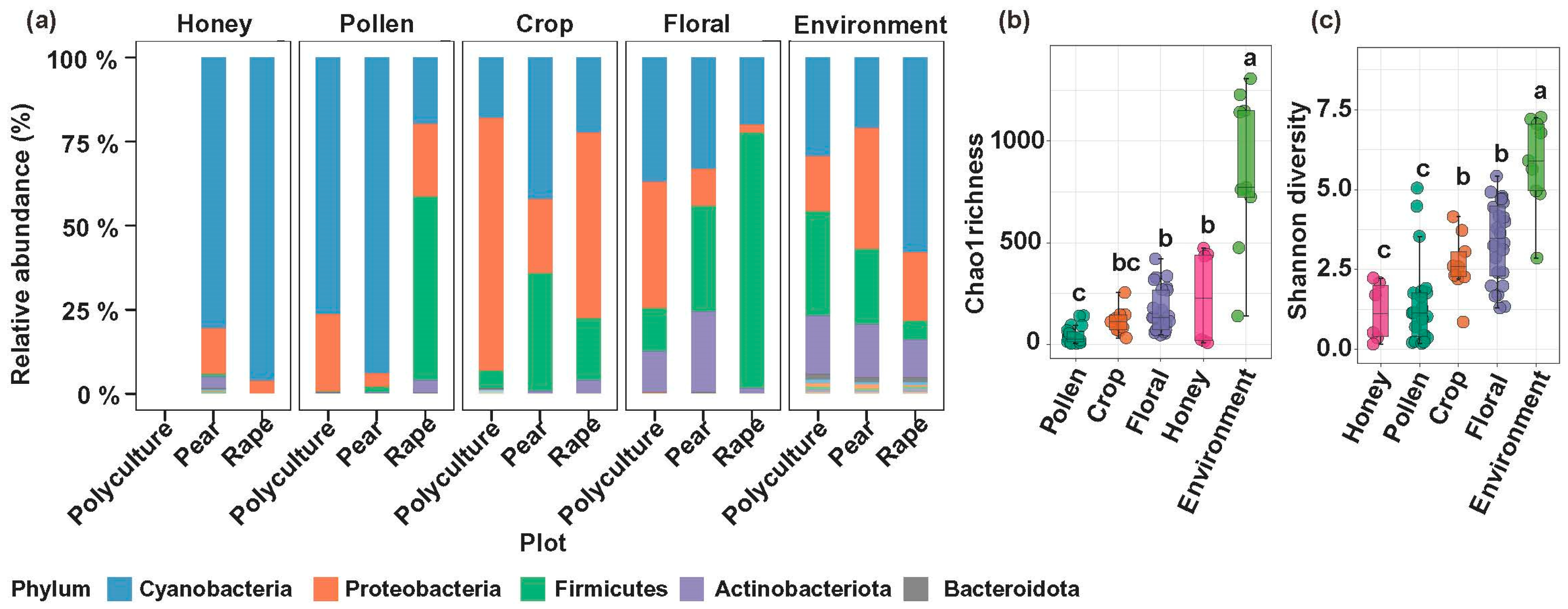
3. Results
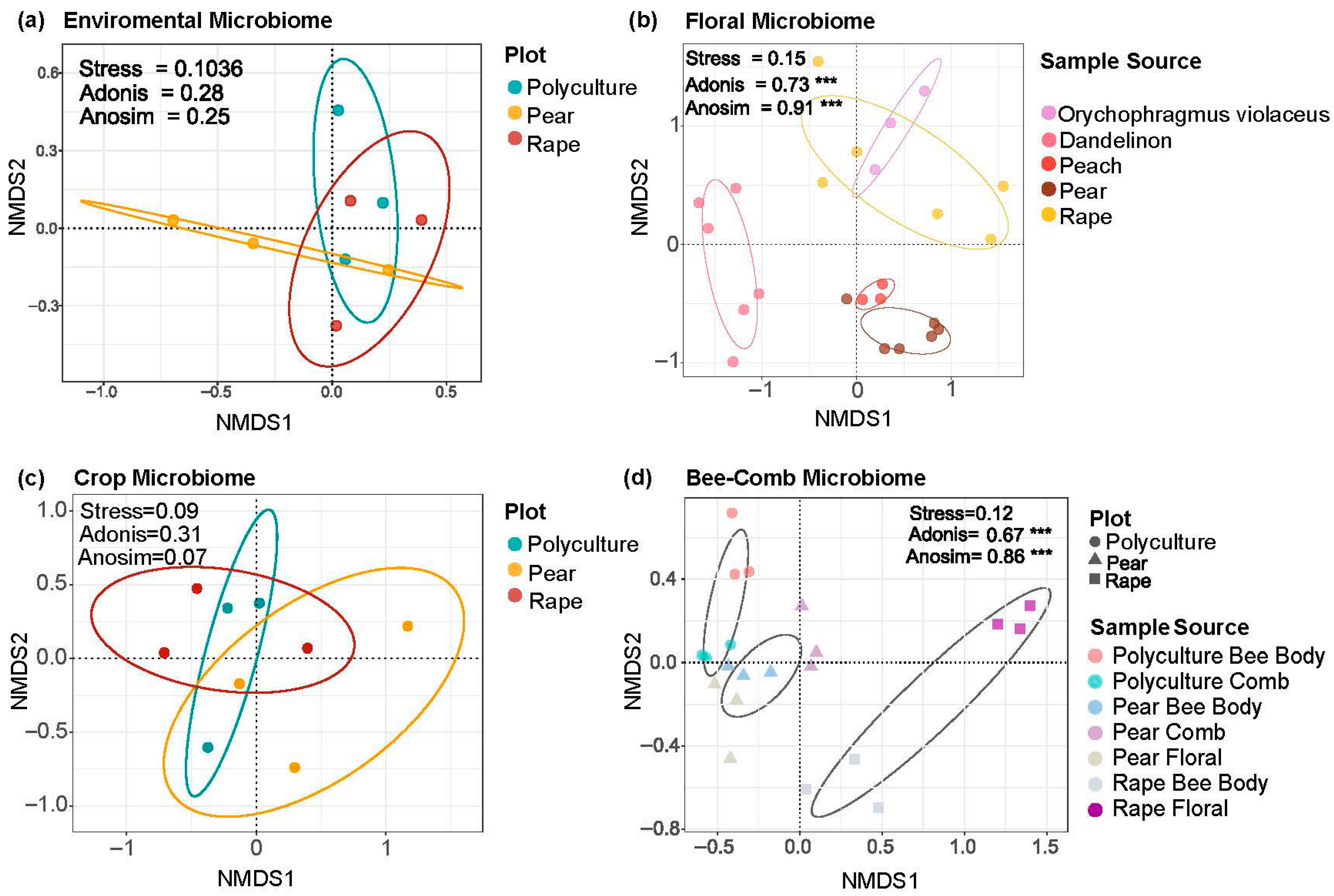
3.1. Plant-Driven Differentiation of Floral and Bee-Associated Microbiomes
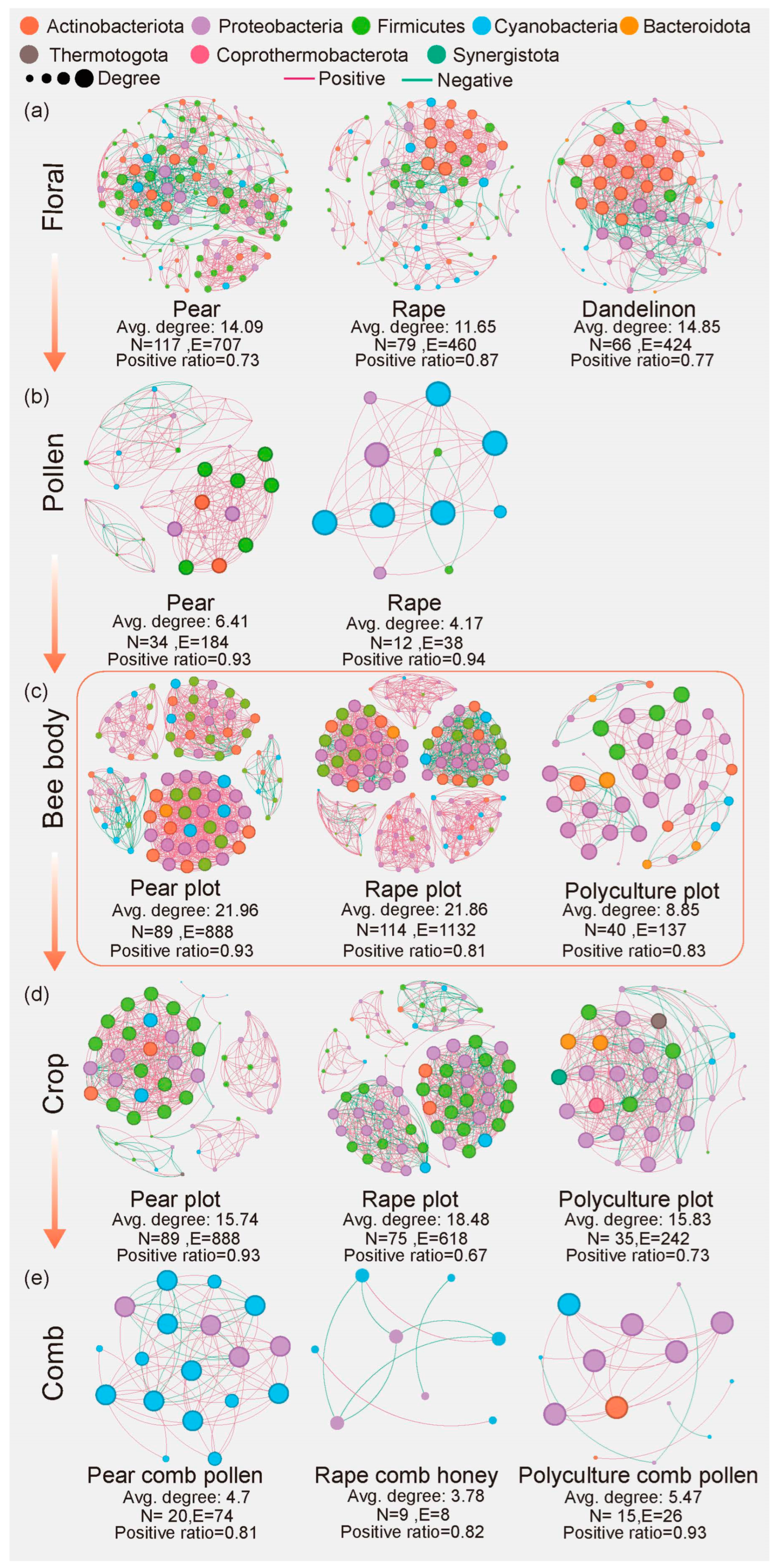
3.2. Bee Body as a Microbial Hub with Distinct Responses to Cultivation Plots
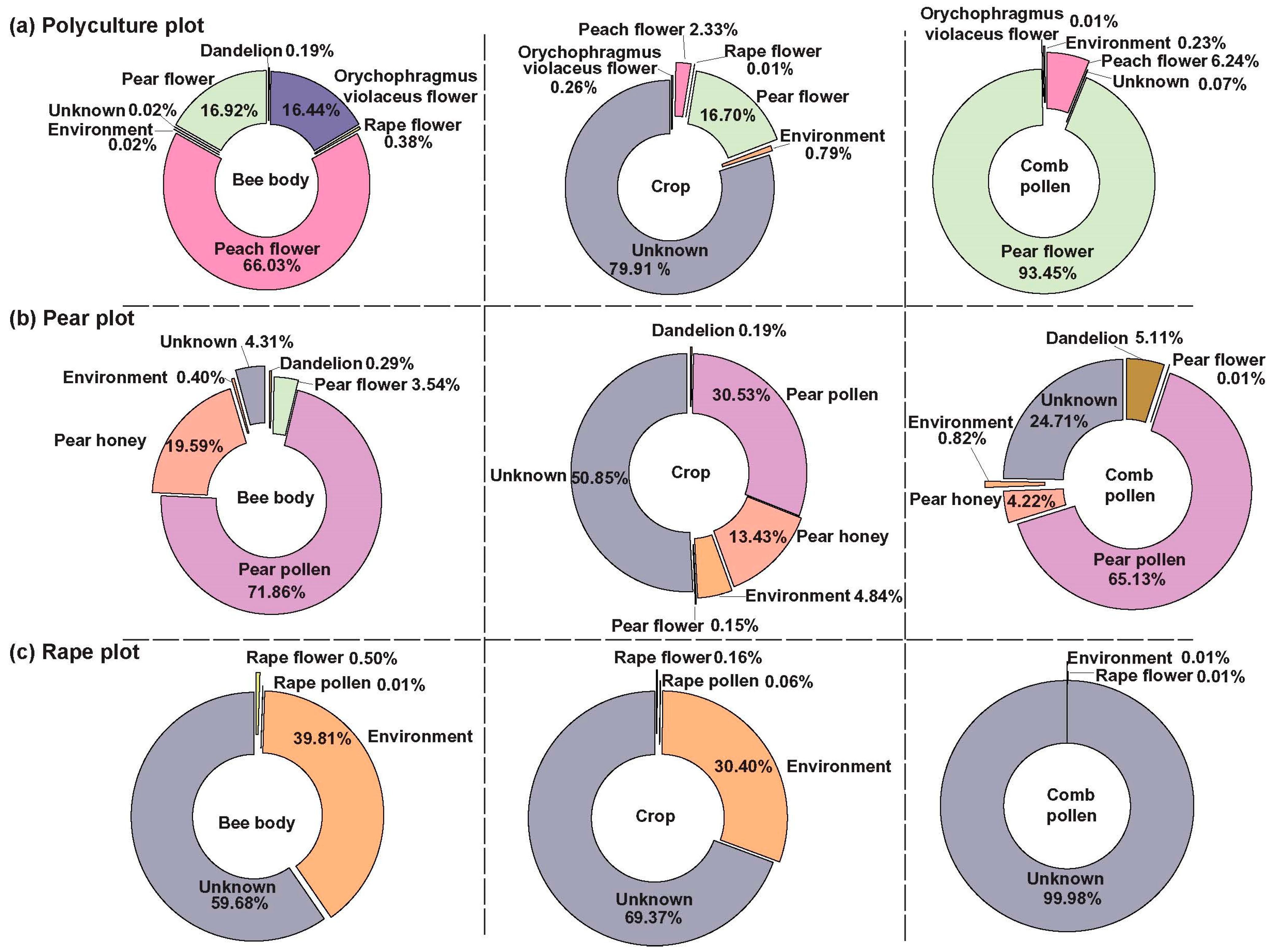
3.3. Bees Carry the Microbial Signature of the Cropping System
4. Discussion
4.1. Floral Microbiomes of Different Nectar Plants Remain Relatively Stable
4.2. Bees as a Keystone in Microbial Communication Within Agricultural Systems
4.3. Variability in Microbial Sources and the Unique Composition of Crop Microbiomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding Pollinators and Their Values to Human Well-Being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef]
- Klein, A.-M.; Boreux, V.; Fornoff, F.; Mupepele, A.-C.; Pufal, G. Relevance of Wild and Managed Bees for Human Well-Being. Curr. Opin. Insect Sci. 2018, 26, 82–88. [Google Scholar] [CrossRef]
- Resci, I.; Cilia, G. The Use of Honey Bee (Apis mellifera L.) as Biological Monitors for Pathogenic Bacteria and Antimicrobial Resistance: A Systematic Review. Environ. Pollut. 2023, 333, 122120. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee Declines Driven by Combined Stress from Parasites, Pesticides, and Lack of Flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Winfree, R.; Aguilar, R.; Vázquez, D.P.; LeBuhn, G.; Aizen, M.A. A Meta-Analysis of Bees’ Responses to Anthropogenic Disturbance. Ecology 2009, 90, 2068–2076. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The Worldwide Importance of Honey Bees as Pollinators in Natural Habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef]
- Olhnuud, A.; Wen, J.; Yu, J.; Lyu, F.; Zhang, Q. Responses of Insect Pollinators to Habitat Fragmentation: A Global Meta-Analysis. J. Appl. Ecol. 2025, 62, 2502–2514. [Google Scholar] [CrossRef]
- Mazel, F.; Prasad, A.; Engel, P. Host Specificity of Gut Microbiota Associated with Social Bees: Patterns and Processes. Microbiol. Mol. Biol. Rev. 2025, 89, e00080-23. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Moran, N.A. The Honeybee Microbiota and Its Impact on Health and Disease. Nat. Rev. Microbiol. 2024, 22, 122–137. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Engel, P. Genomic Diversity Landscape of the Honey Bee Gut Microbiota. Nat. Commun. 2019, 10, 446. [Google Scholar] [CrossRef]
- Navarro-Escalante, L.; Ashraf, A.H.M.Z.; Leonard, S.P.; Barrick, J.E. Protecting Honey Bees through Microbiome Engineering. Curr. Opin. Insect Sci. 2025, 72, 101416. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut Microbial Communities of Social Bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Ricigliano, V.A.; Fine, J.D.; Nicklisch, S.C.T. Harnessing Biotechnology for Bee Pollinator Health. Trends Biotechnol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of Labor in Honey Bee Gut Microbiota for Plant Polysaccharide Digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The Role of the Gut Microbiome in Health and Disease of Adult Honey Bee Workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Mihrete, T.B.; Mihretu, F.B. Crop Diversification for Ensuring Sustainable Agriculture, Risk Management and Food Security. Glob. Chall. 2025, 9, 2400267. [Google Scholar] [CrossRef]
- Schaeffer, R.N.; Crowder, D.W.; Illán, J.G.; Beck, J.J.; Fukami, T.; Williams, N.M.; Vannette, R.L. Disease Management during Bloom Affects the Floral Microbiome but Not Pollination in a Mass-Flowering Crop. J. Appl. Ecol. 2023, 60, 64–76. [Google Scholar] [CrossRef]
- Rering, C.C.; Vannette, R.L.; Schaeffer, R.N.; Beck, J.J. Microbial Co-Occurrence in Floral Nectar Affects Metabolites and Attractiveness to a Generalist Pollinator. J. Chem. Ecol. 2020, 46, 659–667. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; Hara, B.; Henry, M.; Stevens, H. The Vegan Package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar] [CrossRef]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe’er, I.; Halperin, E. FEAST: Fast Expectation-Maximization for Microbial Source Tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Shi, Y.; Zhu, J.; Zhao, C.; Wang, J.; Liu, Z.; Fu, X.; Liu, X.; Yan, J.; Yuan, M.; et al. The Spatial Variation of Soil Bacterial Community Assembly Processes Affects the Accuracy of Source Tracking in Ten Major Chinese Cities. Sci. China Life Sci. 2021, 64, 1546–1559. [Google Scholar] [CrossRef]
- Warren, M.L.; Tsuji, K.; Decker, L.E.; Kishi, M.; Yang, J.; Howe, A.C.; Fukami, T. Bacteria in Honeybee Crops Are Decoupled from Those in Floral Nectar and Bee Mouths. Microb. Ecol. 2025, 88, 46. [Google Scholar] [CrossRef]
- Tiusanen, M.; Becker-Scarpitta, A.; Wirta, H. Distinct Communities and Differing Dispersal Routes in Bacteria and Fungi of Honey Bees, Honey, and Flowers. Microb. Ecol. 2024, 87, 100. [Google Scholar] [CrossRef]
- Junker, R.R.; Keller, A. Microhabitat Heterogeneity across Leaves and Flower Organs Promotes Bacterial Diversity. FEMS Microbiol. Ecol. 2015, 91, fiv097. [Google Scholar] [CrossRef]
- Zemenick, A.T.; Vannette, R.L.; Rosenheim, J.A. Linked Networks Reveal Dual Roles of Insect Dispersal and Species Sorting for Bacterial Communities in Flowers. Oikos 2021, 130, 697–707. [Google Scholar] [CrossRef]
- Temmermans, J.; Legein, M.; Checchia, I.; Felis, G.E.; Smets, W.; Karise, R.; Lebeer, S. Agricultural Practices and Pollinators Modulate the Anthosphere Microbiome. ISME Commun. 2025, 5, ycaf026. [Google Scholar] [CrossRef]
- Vannette, R.L.; Fukami, T. Nectar Microbes Can Reduce Secondary Metabolites in Nectar and Alter Effects on Nectar Consumption by Pollinators. Ecology 2016, 97, 1410–1419. [Google Scholar] [CrossRef]
- Ushio, M.; Yamasaki, E.; Takasu, H.; Nagano, A.J.; Fujinaga, S.; Honjo, M.N.; Ikemoto, M.; Sakai, S.; Kudoh, H. Microbial Communities on Flower Surfaces Act as Signatures of Pollinator Visitation. Sci. Rep. 2015, 5, 8695. [Google Scholar] [CrossRef]
- de Vega, C.; Álvarez-Pérez, S.; Albaladejo, R.G.; Steenhuisen, S.-L.; Lachance, M.-A.; Johnson, S.D.; Herrera, C.M. The Role of Plant–Pollinator Interactions in Structuring Nectar Microbial Communities. J. Ecol. 2021, 109, 3379–3395. [Google Scholar] [CrossRef]
- Tourbez, C.; Gómez-Martínez, C.; González-Estévez, M.Á.; Lázaro, A. Pollen Analysis Reveals the Effects of Uncovered Interactions, Pollen-Carrying Structures, and Pollinator Sex on the Structure of Wild Bee-Plant Networks. Insect Sci. 2023, 31, 971–988. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Rehan, S.M. Environmental Effects on Bee Microbiota. Microb. Ecol. 2023, 86, 1487–1498. [Google Scholar] [CrossRef]
- Keller, A.; McFrederick, Q.S.; Dharampal, P.; Steffan, S.; Danforth, B.N.; Leonhardt, S.D. (More than) Hitchhikers through the Network: The Shared Microbiome of Bees and Flowers. Curr. Opin. Insect Sci. 2021, 44, 8–15. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; de Miranda, J.R.; Doublet, V.; et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. Mbio 2016, 7, e02164-15. [Google Scholar] [CrossRef]
- Evans, J.D.; Lopez, D.L. Bacterial Probiotics Induce an Immune Response in the Honey Bee (Hymenoptera: Apidae). J. Econ. Entomol. 2004, 97, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Walderdorff, L.; Laval-Gilly, P.; Bonnefoy, A.; Falla-Angel, J. Imidacloprid Intensifies Its Impact on Honeybee and Bumblebee Cellular Immune Response When Challenged with LPS (Lippopolysacharide) of Escherichia coli. J. Insect Physiol. 2018, 108, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.N.; Samad-zada, F.; Chau, K.D.; Rehan, S.M. Microbiome and Floral Associations of a Wild Bee Using Biodiversity Survey Collections. Environ. Microbiol. 2024, 26, e16657. [Google Scholar] [CrossRef]
- Martin, V.N.; Schaeffer, R.N.; Fukami, T. Potential Effects of Nectar Microbes on Pollinator Health. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210155. [Google Scholar] [CrossRef]
- Quevedo-Caraballo, S.; de Vega, C.; Lievens, B.; Fukami, T.; Álvarez-Pérez, S. Tiny but Mighty? Overview of a Decade of Research on Nectar Bacteria. New Phytol. 2025, 245, 1897–1910. [Google Scholar] [CrossRef] [PubMed]
- Al-Sherif, A.A.; Mazeed, A.M.; Ewis, M.A.; Nafea, E.A.; Hagag, E.-S.E.; Kamel, A.A. Activity of Salivary Glands in Secreting Honey-Elaborating Enzymes in Two Subspecies of Honeybee (Apis mellifera L.). Physiol. Entomol. 2017, 42, 397–403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Guo, B.; Yi, X.; Sun, Q.; Sun, K.; Guo, L.; Guo, Y. The Honey Bee Body Surface as a Microbial Hub: Connectivity Shaped by Monoculture vs. Polyculture Farming. Insects 2026, 17, 53. https://doi.org/10.3390/insects17010053
Guo B, Yi X, Sun Q, Sun K, Guo L, Guo Y. The Honey Bee Body Surface as a Microbial Hub: Connectivity Shaped by Monoculture vs. Polyculture Farming. Insects. 2026; 17(1):53. https://doi.org/10.3390/insects17010053
Chicago/Turabian StyleGuo, Baobei, Xueyan Yi, Qihang Sun, Ke Sun, Lina Guo, and Yuan Guo. 2026. "The Honey Bee Body Surface as a Microbial Hub: Connectivity Shaped by Monoculture vs. Polyculture Farming" Insects 17, no. 1: 53. https://doi.org/10.3390/insects17010053
APA StyleGuo, B., Yi, X., Sun, Q., Sun, K., Guo, L., & Guo, Y. (2026). The Honey Bee Body Surface as a Microbial Hub: Connectivity Shaped by Monoculture vs. Polyculture Farming. Insects, 17(1), 53. https://doi.org/10.3390/insects17010053






