Alternative Characterizations of Methyl Lucidone-Responsive Differentially Expressed Genes in Drosophila melanogaster Using DEG-by-Index Ratio Transformation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila melanogaster Sample Acquisition and Preparation for RNA-Seq
2.2. Data Processing of 18 D. melanogaster RNA-Seqs
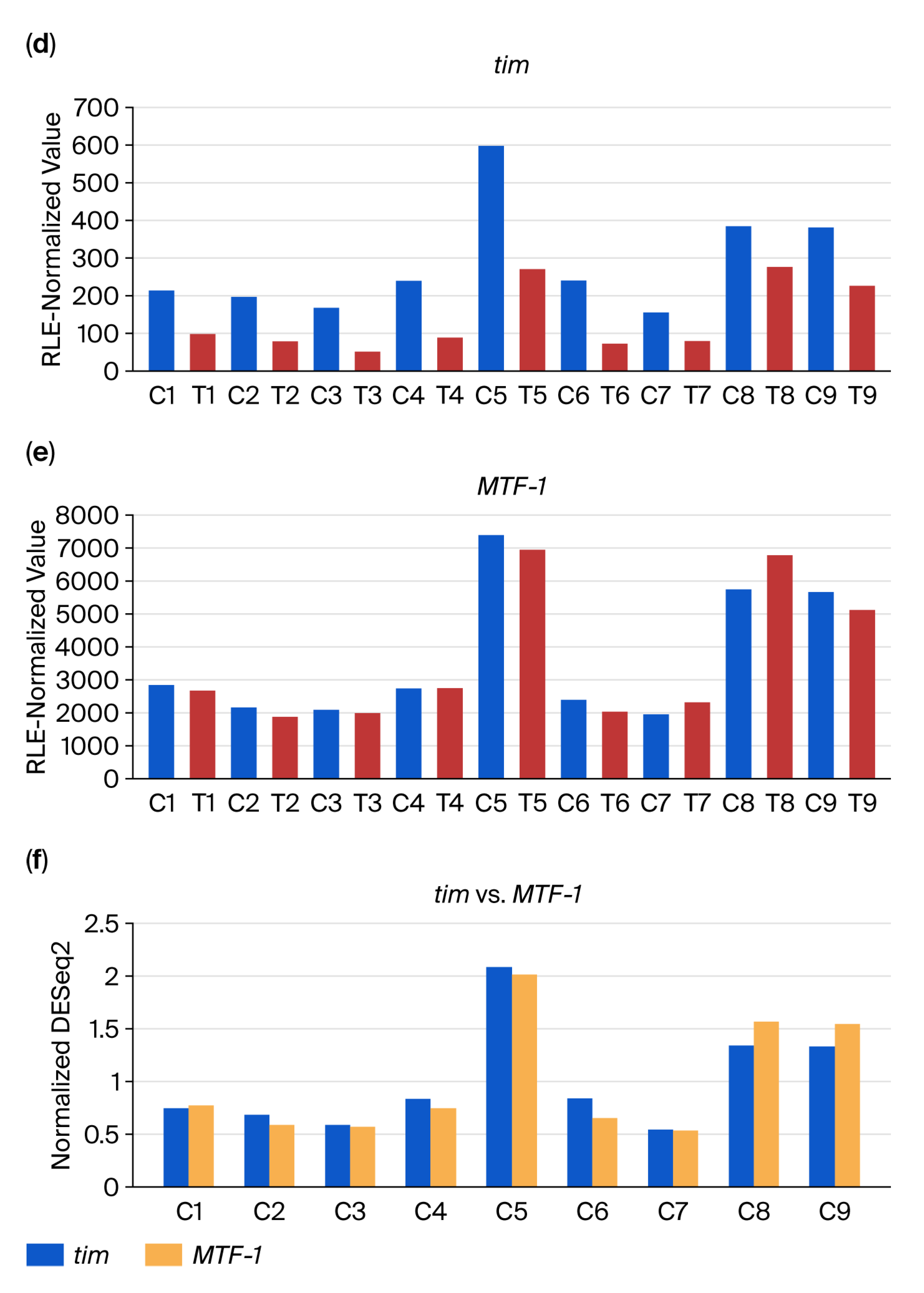
2.3. DiRT Analysis Workflow
2.4. DESeq2/edgeR Analyses
2.5. Heatmaps
2.6. Methods for p-Value Calculation and Adjustment
2.7. KEGG Pathway Analysis of DiRT-Derived DEGs
3. Results
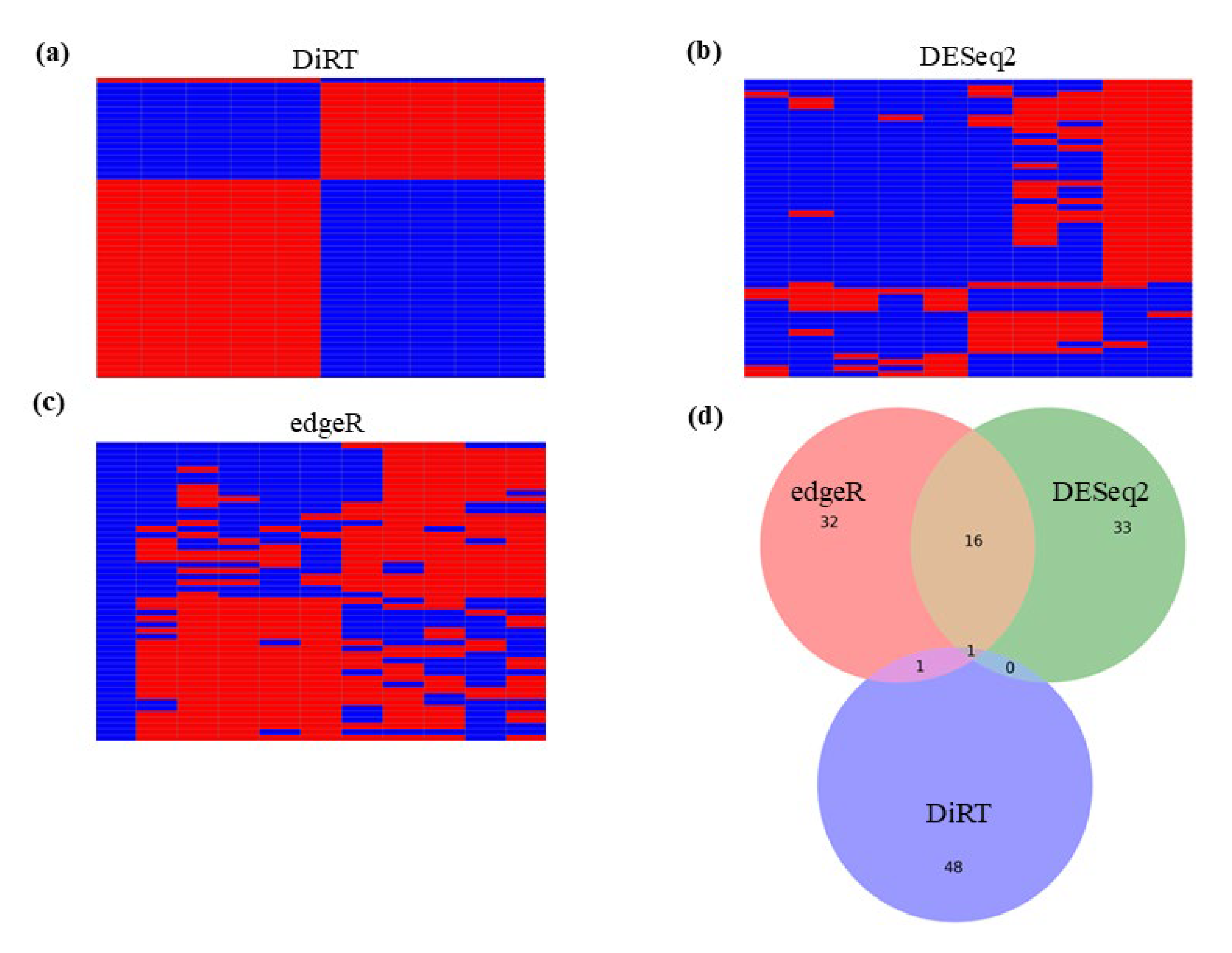
3.1. Analayes of Methyl-Lucidone-Treated D. melanogaster RNA-Seq Using DESeq2 and edgeR
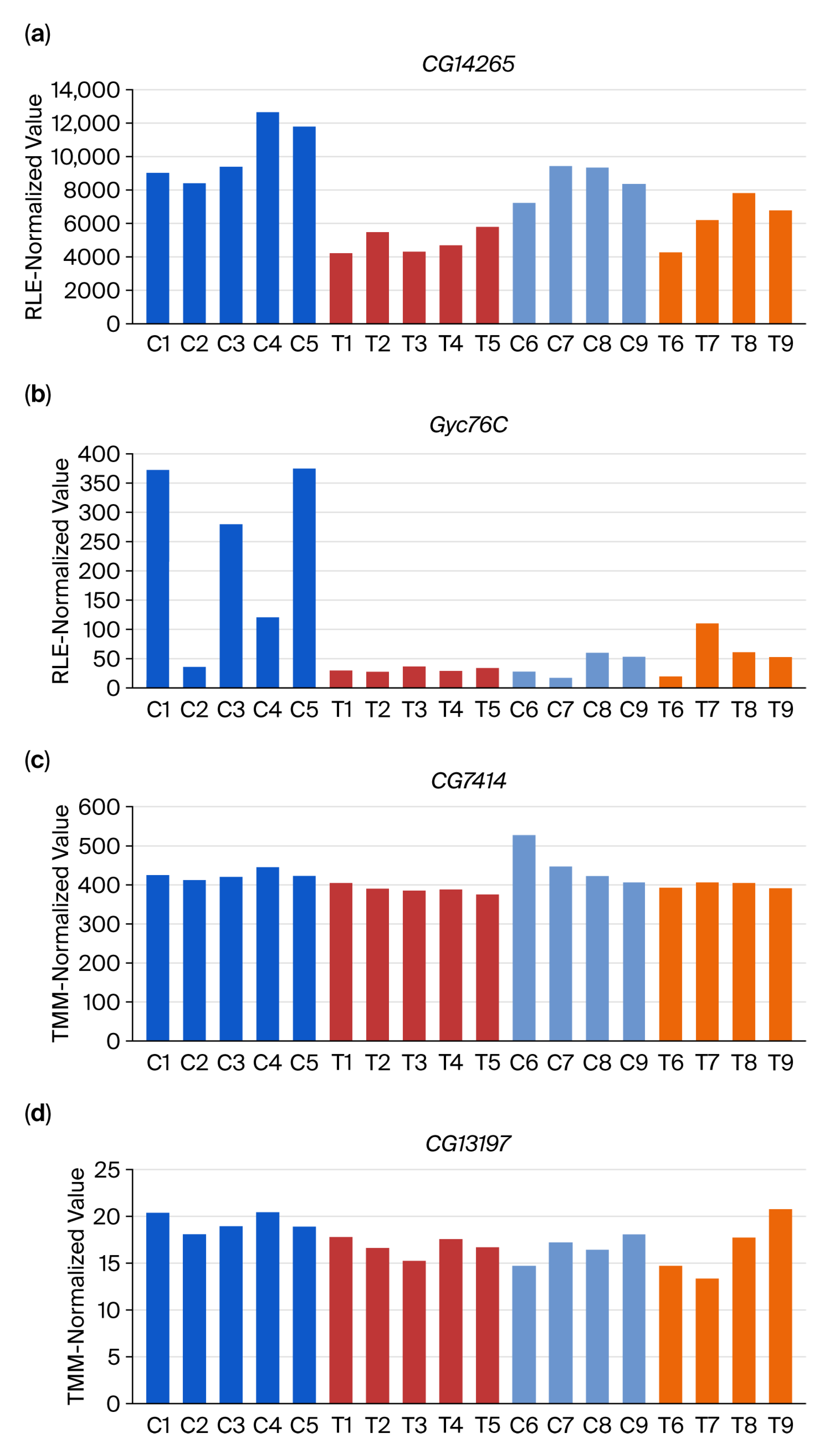
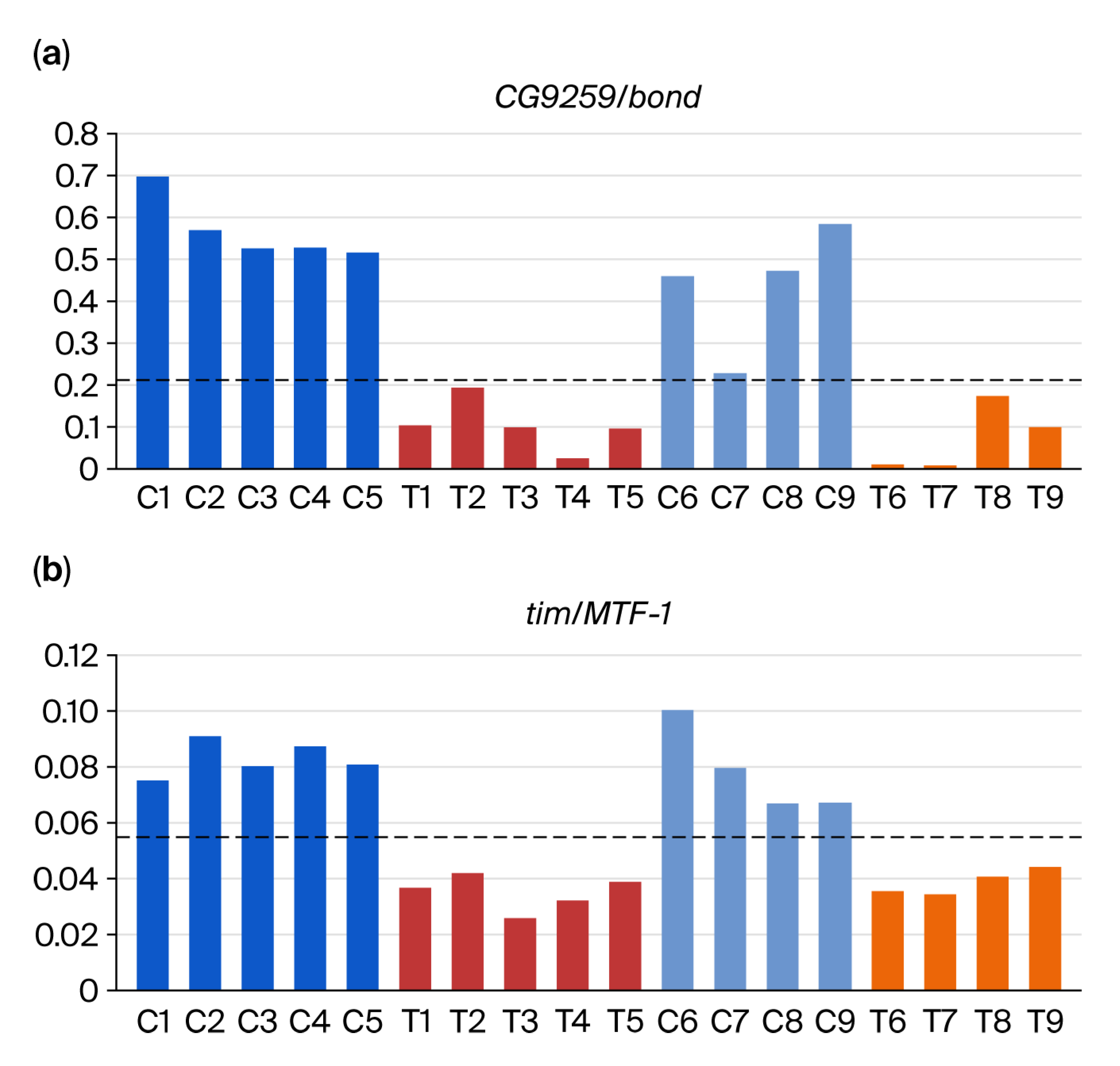
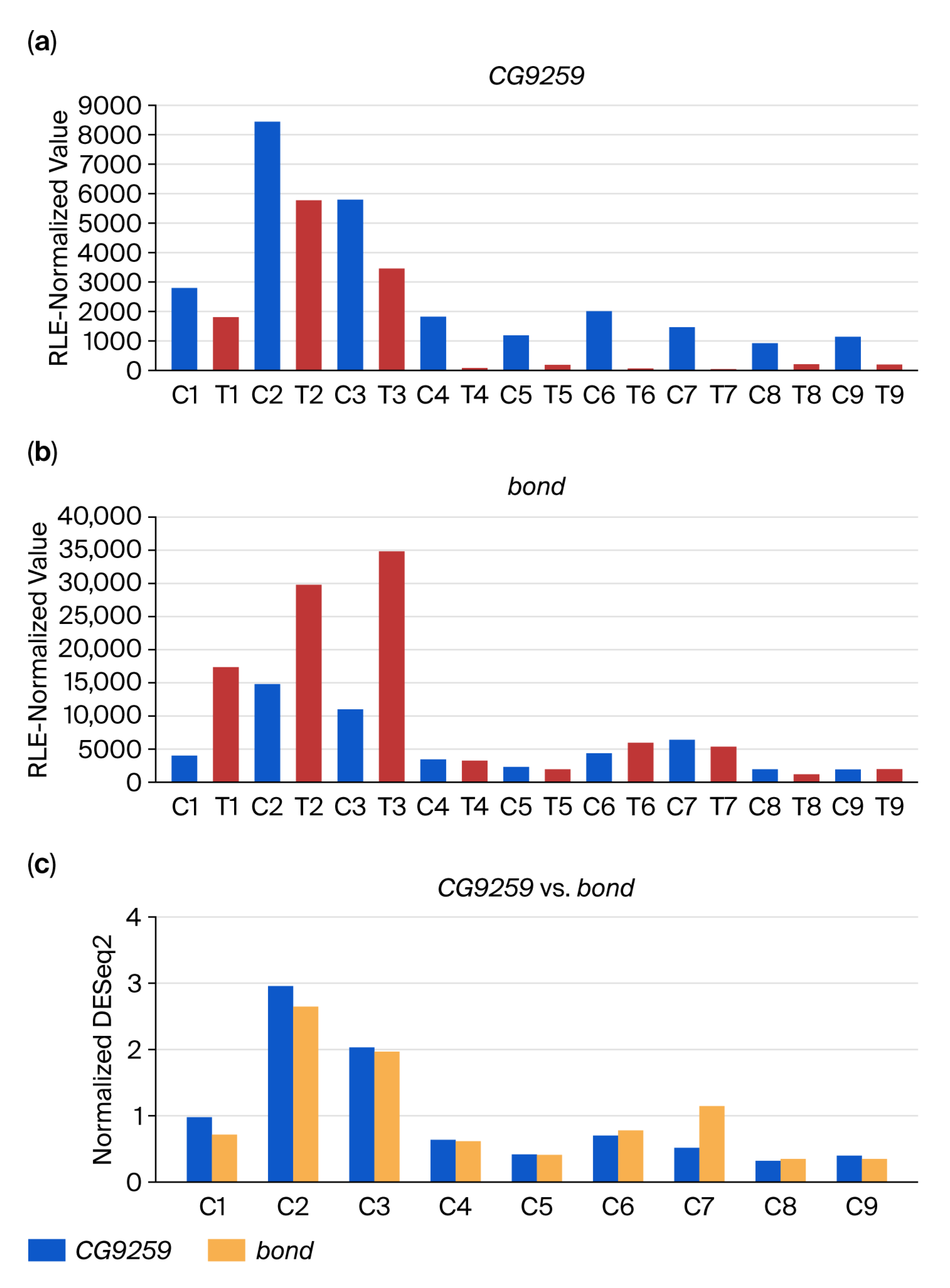
3.2. DEG-by-Index Ratio Transformation (DiRT) Normalization Identifies Reproducible DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEG | differentially expressed gene |
| RLE | relative log expression |
| TMM | trimmed mean of M-values |
| DiRT | DEG-by-index ratio transformation |
| JH | juvenile hormone |
| RNA-Seq | RNA sequencing |
| CDA | compositional data analysis |
| NSD | normalized standard deviation |
References
- Lee, S.H.; Oh, H.W.; Fang, Y.; An, S.B.; Park, D.S.; Song, H.H.; Oh, S.R.; Kim, S.Y.; Kim, S.; Kim, N.; et al. Identification of plant compounds that disrupt the insect juvenile hormone receptor complex. Proc. Natl. Acad. Sci. USA 2015, 112, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.W.; Jeon, J.H.; Jeong, S.A.; Kim, J.-A.; Park, D.-S.; Shin, Y.; Oh, H.-W. A plant diterpene counteracts juvenile hormone-mediated gene regulation during Drosophila melanogaster larval development. PLoS ONE 2018, 13, e0200706. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-W.; Jeon, J.-H.; Kim, J.-A.; Park, D.-S.; Shin, Y.-J.; Oh, H.-W. Inducible expression of several Drosophila melanogaster genes encoding juvenile hormone binding proteins by a plant diterpene secondary metabolite, methyl lucidone. Insects 2022, 13, 420. [Google Scholar] [CrossRef] [PubMed]
- Riddiford, L.M. How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocrinol. 2012, 179, 477–484. [Google Scholar] [CrossRef]
- Jindra, M.; Bellés, X.; Shinoda, T. Molecular basis of juvenile hormone signaling. Curr. Opin. Insect Sci. 2015, 11, 39–46. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Aitchison, J. The statistical analysis of compositional data. J. R. Stat. Soc. Ser. B Methodol. 1982, 44, 139–177. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Vu, M.; Fernandes, A.D. Compositional uncertainty should not be ignored in high-throughput sequencing data analysis. Austrian J. Stat. 2016, 45, 73–87. [Google Scholar] [CrossRef]
- Quinn, T.P.; Erb, I.; Gloor, G.; Notredame, C.; Richardson, M.F.; Crowley, T.M. A field guide for the compositional analysis of any-omics data. Gigascience 2019, 8, giz107. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, J.M.; Wilson, S.K.; Kasa, M.; Rise, J.S.; Topalidou, I.; Ailion, M. The SEK-1 p38 MAP kinase pathway modulates Gq signaling in Caenorhabditis elegans. G3 2017, 7, 2979–2989. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, M.; Viken, K.; Wang, Q.; Jatap, P.; Bitterman, P.; Ingbar, D.; Wendt, C. Bronchoalveolar lavage fluid protein expression in acute respiratory distress syndrome provides insights into pathways activated in subjects with different outcomes. Sci. Rep. 2017, 7, 7464. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- dos Santos, G.; Schroeder, A.J.; Goodman, J.L.; Strelets, V.B.; Crosby, M.A.; Thurmond, J.; Emmert, D.B.; Gelbart, W.M.; Flybase Consortium. FlyBase: Introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015, 43, D690–D697. [Google Scholar] [CrossRef]
- Sirén, J.; Välimäki, N.; Mäkinen, V. Indexing graphs for path queries with applications in genome research. IEEE ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.; Liu, S. Juvenile Hormone Studies in Drosophila melanogaster. Front. Physiol. 2022, 12, 785320. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shin, S.W.; Zou, Z.; Saha, T.T.; Raikhel, A.S. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. USA 2012, 109, 16576–16581. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pandey, A.; Motro, U.; Bloch, G. Juvenile Hormone Affects the Development and Strength of Circadian Rhythms in Young Bumble Bee (Bombus terrestris) Workers. Neurobiol. Sleep Circadian Rhythm. 2020, 9, 100056. [Google Scholar] [CrossRef]
- Panda, S.; Hogenesch, J.B.; Kay, S.A. Circadian rhythms from flies to human. Nature 2002, 417, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J. Transcriptomic and proteomic effects of gene deletion are not evolutionarily conserved. Genome Res. 2025, 35, 512–521. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Sample ID | Reads Assigned to dm6-Annotated Genes | NCBI Accession # | Condition |
|---|---|---|---|
| C1 | 67,399,437 | SRR22894279 | EtOH-treated |
| C2 | 94,044,531 | SRR22891343 | EtOH-treated |
| C3 | 92,115,653 | SRR22891368 | EtOH-treated |
| C4 | 68,365,272 | SRR22891367 | EtOH-treated |
| C5 | 74,929,527 | SRR22891357 | EtOH-treated |
| T1 | 50,864,597 | SRR22891355 | Methyl lucidone-treated |
| T2 | 91,480,523 | SRR22891354 | Methyl lucidone-treated |
| T3 | 92,471,264 | SRR22891353 | Methyl lucidone-treated |
| T4 | 67,774,651 | SRR22891366 | Methyl lucidone-treated |
| T5 | 76,983,129 | SRR22891362 | Methyl lucidone-treated |
| C6 | 65,937,883 | SRR22891360 | EtOH-treated |
| C7 | 62,985,695 | SRR22891359 | EtOH-treated |
| C8 | 63,522,838 | SRR22891358 | EtOH-treated |
| C9 | 69,304,669 | SRR22891356 | EtOH-treated |
| T6 | 66,503,607 | SRR22891364 | Methyl lucidone-treated |
| T7 | 66,182,329 | SRR22891363 | Methyl lucidone-treated |
| T8 | 69,941,736 | SRR22891361 | Methyl lucidone-treated |
| T9 | 65,816,863 | SRR22891365 | Methyl lucidone-treated |
| KEGG Pathways | Gene Count | p-Value | Benjamini |
|---|---|---|---|
| Proteasome | 8 | 2.8 × 10−6 | 0.00015 |
| Drug metabolism—cytochrome P450 | 8 | 0.000015 | 0.00033 |
| Metabolism of xenobiotics via cytochrome P450 | 8 | 0.000019 | 0.00033 |
| Drug metabolism—other enzymes | 7 | 0.001 | 0.014 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.W.; Kim, J.A.; Jeon, J.H.; Park, K.; Lee, S.; Oh, H.-W. Alternative Characterizations of Methyl Lucidone-Responsive Differentially Expressed Genes in Drosophila melanogaster Using DEG-by-Index Ratio Transformation. Insects 2025, 16, 898. https://doi.org/10.3390/insects16090898
Shin SW, Kim JA, Jeon JH, Park K, Lee S, Oh H-W. Alternative Characterizations of Methyl Lucidone-Responsive Differentially Expressed Genes in Drosophila melanogaster Using DEG-by-Index Ratio Transformation. Insects. 2025; 16(9):898. https://doi.org/10.3390/insects16090898
Chicago/Turabian StyleShin, Sang Woon, Ji Ae Kim, Jun Hyoung Jeon, Kunhyang Park, SooJin Lee, and Hyun-Woo Oh. 2025. "Alternative Characterizations of Methyl Lucidone-Responsive Differentially Expressed Genes in Drosophila melanogaster Using DEG-by-Index Ratio Transformation" Insects 16, no. 9: 898. https://doi.org/10.3390/insects16090898
APA StyleShin, S. W., Kim, J. A., Jeon, J. H., Park, K., Lee, S., & Oh, H.-W. (2025). Alternative Characterizations of Methyl Lucidone-Responsive Differentially Expressed Genes in Drosophila melanogaster Using DEG-by-Index Ratio Transformation. Insects, 16(9), 898. https://doi.org/10.3390/insects16090898







