Ionotropic Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Identification
2.2. Orthologous Analysis of the IRs
2.3. Phylogenetic Analysis of the IRs
2.4. Tests of Positive Selection
3. Results
3.1. Identification and Phylogenetic Analysis of IR Genes
3.2. Analysis of Orthologous IR Genes
3.3. Selective Pressures on IR Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berg, C.C. Flora Malesiana Precursor for the Treatment of Moraceae 1: The Main Subdivision of Ficus: The Subgenera. Blumea 2003, 48, 166–177. [Google Scholar] [CrossRef]
- Galil, J.; Eisikowitch, D. Flowering Cycles and Fruit Types of Ficus Sycomorus in Israel. New Phytol. 1968, 67, 745–758. [Google Scholar] [CrossRef]
- Hossaert-McKey, M.; Soler, C.; Schatz, B.; Proffit, M. Floral Scents: Their Roles in Nursery Pollination Mutualisms. Chemoecology 2010, 20, 75–88. [Google Scholar] [CrossRef]
- Moe, A.M.; Weiblen, G.D. Pollinator-Mediated Reproductive Isolation among Dioecious Fig Species (Ficus, Moraceae). Evolution 2012, 66, 3710–3721. [Google Scholar] [CrossRef] [PubMed]
- Conchou, L.; Cabioch, L.; Rodriguez, L.J.; Kjellberg, F. Daily Rhythm of Mutualistic Pollinator Activity and Scent Emission in Ficus Septica: Ecological Differentiation between Co-Occurring Pollinators and Potential Consequences for Chemical Communication and Facilitation of Host Speciation. PLoS ONE 2014, 9, e103581. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, X.; Herre, E.A.; McKey, D.; Machado, C.A.; Yu, W.B.; Cannon, C.H.; Arnold, M.L.; Pereira, R.A.S.; Ming, R. Genomic Evidence of Prevalent Hybridization throughout the Evolutionary History of the Fig-Wasp Pollination Mutualism. Nat. Commun. 2021, 12, 718. [Google Scholar] [CrossRef]
- Ware, A.B.; Compton, S.G. Repeated Evolution of Elongate Multiporous Plate Sensilla in Female Fig Wasps (Hymenoptera: Agaonidae: Agaoninae). Proc. K. Ned. Akad. Wet. 1992, 95, 275–292. [Google Scholar]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Walker, W.B.; Dong, S.; Wang, G. Identification and Functional Characterization of Sex Pheromone Receptors in Beet Armyworm Spodoptera Exigua (Hübner). Insect Biochem. Mol. Biol. 2013, 43, 747–754. [Google Scholar] [CrossRef]
- Ning, C.; Yang, K.; Xu, M.; Huang, L.-Q.; Wang, C.-Z. Functional Validation of the Carbon Dioxide Receptor in Labial Palps of Helicoverpa Armigera Moths. Insect Biochem. Mol. Biol. 2016, 73, 12–19. [Google Scholar] [CrossRef]
- Chang, H.; Liu, Y.; Ai, D.; Jiang, X.; Dong, S.; Wang, G. A Pheromone Antagonist Regulates Optimal Mating Time in the Moth Helicoverpa Armigera. Curr. Biol. 2017, 27, 1610–1615.e3. [Google Scholar] [CrossRef]
- Abuin, L.; Bargeton, B.; Ulbrich, M.H.; Isacoff, E.Y.; Kellenberger, S.; Benton, R. Functional Architecture of Olfactory Ionotropic Glutamate Receptors. Neuron 2011, 69, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Ai, M.; Blais, S.; Park, J.-Y.; Min, S.; Neubert, T.A.; Suh, G.S. Ionotropic Glutamate Receptors IR64a and IR8a Form a Functional Odorant Receptor Complex in Vivo in Drosophila. J. Neurosci. 2013, 33, 10741–10749. [Google Scholar] [CrossRef] [PubMed]
- Rytz, R.; Croset, V.; Benton, R. Ionotropic Receptors (IRs): Chemosensory Ionotropic Glutamate Receptors in Drosophila and Beyond. Insect Biochem. Mol. Biol. 2013, 43, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Koh, T.-W.; He, Z.; Gorur-Shandilya, S.; Menuz, K.; Larter, N.K.; Stewart, S.; Carlson, J.R. The Drosophila IR20a Clade of Ionotropic Receptors Are Candidate Taste and Pheromone Receptors. Neuron 2014, 83, 850–865. [Google Scholar] [CrossRef]
- Ni, L.; Klein, M.; Svec, K.V.; Budelli, G.; Chang, E.C.; Ferrer, A.J.; Benton, R.; Samuel, A.D.; Garrity, P.A. The Ionotropic Receptors IR21a and IR25a Mediate Cool Sensing in Drosophila. Elife 2016, 5, e13254. [Google Scholar] [CrossRef]
- Ganguly, A.; Pang, L.; Duong, V.-K.; Lee, A.; Schoniger, H.; Varady, E.; Dahanukar, A. A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Rep. 2017, 18, 737–750. [Google Scholar] [CrossRef]
- Xiao, J.-H.; Yue, Z.; Jia, L.-Y.; Yang, X.-H.; Niu, L.-H.; Wang, Z.; Zhang, P.; Sun, B.-F.; He, S.-M.; Li, Z. Obligate Mutualism within a Host Drives the Extreme Specialization of a Fig Wasp Genome. Genome Biol. 2013, 14, 1–18. [Google Scholar] [CrossRef]
- Chen, L.; Segar, S.T.; Chantarasuwan, B.; Wong, D.-M.; Wang, R.; Chen, X.; Yu, H. Adaptation of Fig Wasps (Agaodinae) to Their Host Revealed by Large-Scale Transcriptomic Data. Insects 2021, 12, 815. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; Zhang, S.; Chen, S.; Wang, Y.; Wen, P.; Ma, X.; Shi, Y.; Qi, R.; Yang, Y. Genomes of the Banyan Tree and Pollinator Wasp Provide Insights into Fig-Wasp Coevolution. Cell 2020, 183, 875–889.e17. [Google Scholar] [CrossRef]
- Chen, L.; Feng, C.; Wang, R.; Nong, X.; Deng, X.; Chen, X.; Yu, H. A Chromosome-Level Genome Assembly of the Pollinating Fig Wasp Valisia Javana. DNA Res. 2022, 29, dsac014. [Google Scholar] [CrossRef]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef]
- Mayer, M.L.; Ghosal, A.; Dolman, N.P.; Jane, D.E. Crystal Structures of the Kainate Receptor GluR5 Ligand Binding Core Dimer with Novel GluR5-Selective Antagonists. J. Neurosci. 2006, 26, 2852–2861. [Google Scholar] [CrossRef] [PubMed]
- Croset, V.; Schleyer, M.; Arguello, J.R.; Gerber, B.; Benton, R. A Molecular and Neuronal Basis for Amino Acid Sensing in the Drosophila Larva. Sci. Rep. 2016, 6, 34871. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Zhang, M.; Üçpunar, H.K.; Svensson, T.; Quillery, E.; Gompel, N.; Ignell, R.; Grunwald Kadow, I.C. Ionotropic Chemosensory Receptors Mediate the Taste and Smell of Polyamines. PLoS Biol. 2016, 14, e1002454. [Google Scholar] [CrossRef]
- Silbering, A.F.; Rytz, R.; Grosjean, Y.; Abuin, L.; Ramdya, P.; Jefferis, G.S.; Benton, R. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. J. Neurosci. 2011, 31, 13357–13375. [Google Scholar] [CrossRef] [PubMed]
- Knecht, Z.A.; Silbering, A.F.; Ni, L.; Klein, M.; Budelli, G.; Bell, R.; Abuin, L.; Ferrer, A.J.; Samuel, A.D.; Benton, R. Distinct Combinations of Variant Ionotropic Glutamate Receptors Mediate Thermosensation and Hygrosensation in Drosophila. Elife 2016, 5, e17879. [Google Scholar] [CrossRef]
- Guo, M.; Krieger, J.; Große-Wilde, E.; Mißbach, C.; Zhang, L.; Breer, H. Variant Ionotropic Receptors Are Expressed in Olfactory Sensory Neurons of Coeloconic Sensilla on the Antenna of the Desert Locust (Schistocerca gregaria). Int. J. Biol. Sci. 2013, 10, 1. [Google Scholar] [CrossRef]
- Pitts, R.J.; Derryberry, S.L.; Zhang, Z.; Zwiebel, L.J. Variant Ionotropic Receptors in the Malaria Vector Mosquito Anopheles Gambiae Tuned to Amines and Carboxylic Acids. Sci. Rep. 2017, 7, 40297. [Google Scholar] [CrossRef]
- Tang, R.; Jiang, N.-J.; Ning, C.; Li, G.-C.; Huang, L.-Q.; Wang, C.-Z. The Olfactory Reception of Acetic Acid and Ionotropic Receptors in the Oriental Armyworm, Mythimna Separata Walker. Insect Biochem. Mol. Biol. 2020, 118, 103312. [Google Scholar] [CrossRef]
- De Bruyne, M.; Foster, K.; Carlson, J.R. Odor Coding in the Drosophila Antenna. Neuron 2001, 30, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Hallem, E.A.; Carlson, J.R. Coding of Odors by a Receptor Repertoire. Cell 2006, 125, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Nong, X.; Fan, S.; Bhanumas, C.; Deng, X.; Wang, R.; Chen, X.; Compton, S.G. Olfactory and Gustatory Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies. Gene 2023, 850, 146953. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Li, W.; Schuler, M.A.; Berenbaum, M.R. Diversification of Furanocoumarin-Metabolizing Cytochrome P450 Monooxygenases in Two Papilionids: Specificity and Substrate Encounter Rate. Proc. Natl. Acad. Sci. USA 2003, 100, 14593–14598. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Su, Z.-H.; Sasaki, A.; Kusumi, J.; Chou, P.-A.; Tzeng, H.-Y.; Li, H.-Q.; Yu, H. Pollinator Sharing, Copollination, and Speciation by Host Shifting among Six Closely Related Dioecious Fig Species. Commun. Biol. 2022, 5, 284. [Google Scholar] [CrossRef]
- Yu, H.; Tian, E.; Zheng, L.; Deng, X.; Cheng, Y.; Chen, L.; Wu, W.; Tanming, W.; Zhang, D.; Compton, S.G. Multiple Parapatric Pollinators Have Radiated across a Continental Fig Tree Displaying Clinal Genetic Variation. Mol. Ecol. 2019, 28, 2391–2405. [Google Scholar] [CrossRef]
- Wang, R.; Yang, Y.; Jing, Y.; Segar, S.T.; Zhang, Y.; Wang, G.; Chen, J.; Liu, Q.-F.; Chen, S.; Chen, Y. Molecular Mechanisms of Mutualistic and Antagonistic Interactions in a Plant–Pollinator Association. Nat. Ecol. Evol. 2021, 5, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Karpe, S.D.; Dhingra, S.; Brockmann, A.; Sowdhamini, R. Computational Genome-Wide Survey of Odorant Receptors from Two Solitary Bees Dufourea Novaeangliae (Hymenoptera: Halictidae) and Habropoda Laboriosa (Hymenoptera: Apidae). Sci. Rep. 2017, 7, 10823. [Google Scholar] [CrossRef] [PubMed]
- Cruaud, A.; Rønsted, N.; Chantarasuwan, B.; Chou, L.S.; Clement, W.L.; Couloux, A.; Cousins, B.; Genson, G.; Harrison, R.D.; Hanson, P.E. An Extreme Case of Plant–Insect Codiversification: Figs and Fig-Pollinating Wasps. Syst. Biol. 2012, 61, 1029–1047. [Google Scholar] [CrossRef]
- Yu, H.; Liao, Y.; Cheng, Y.; Jia, Y.; Compton, S.G. More Examples of Breakdown the 1: 1 Partner Specificity between Figs and Fig Wasps. Bot. Stud. 2021, 62, 1–12. [Google Scholar] [CrossRef]
- Getahun, M.N.; Wicher, D.; Hansson, B.S.; Olsson, S.B. Temporal Response Dynamics of Drosophila Olfactory Sensory Neurons Depends on Receptor Type and Response Polarity. Front. Cell. Neurosci. 2012, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.A.; Ignell, R.; Carlson, J.R. Chemosensory Coding by Neurons in the Coeloconic Sensilla of the Drosophila antenna. J. Neurosci. 2005, 25, 8359–8367. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Godino, L.L.; Rytz, R.; Bargeton, B.; Abuin, L.; Arguello, J.R.; Peraro, M.D.; Benton, R. Olfactory Receptor Pseudo-Pseudogenes. Nature 2016, 539, 93–97. [Google Scholar] [CrossRef]
- Ai, M.; Min, S.; Grosjean, Y.; Leblanc, C.; Bell, R.; Benton, R.; Suh, G.S. Acid Sensing by the Drosophila Olfactory System. Nature 2010, 468, 691–695. [Google Scholar] [CrossRef]
- Zhang, Y.V.; Ni, J.; Montell, C. The Molecular Basis for Attractive Salt-Taste Coding in Drosophila. Science 2013, 340, 1334–1338. [Google Scholar] [CrossRef]
- Knecht, Z.A.; Silbering, A.F.; Cruz, J.; Yang, L.; Croset, V.; Benton, R.; Garrity, P.A. Ionotropic Receptor-Dependent Moist and Dry Cells Control Hygrosensation in Drosophila. Elife 2017, 6, e26654. [Google Scholar] [CrossRef]
- Gupta, K.; Dhawan, R.; Kajla, M.; Misra, T.; Kumar, S.; Gupta, L. The evolutionary divergence of STAT transcription factor in different Anopheles species. Gene 2017, 596, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Garg, S.; Gupta, L.; Gupta, K.; Diagne, C.T.; Misse, D.; Pompon, J.; Kumar, S.; Saxena, V. Delineating the role of Aedes aegypti ABC transporter gene family during mosquito development and arboviral infection via transcriptome analyses. Pathogens 2021, 10, 1127. [Google Scholar] [CrossRef] [PubMed]

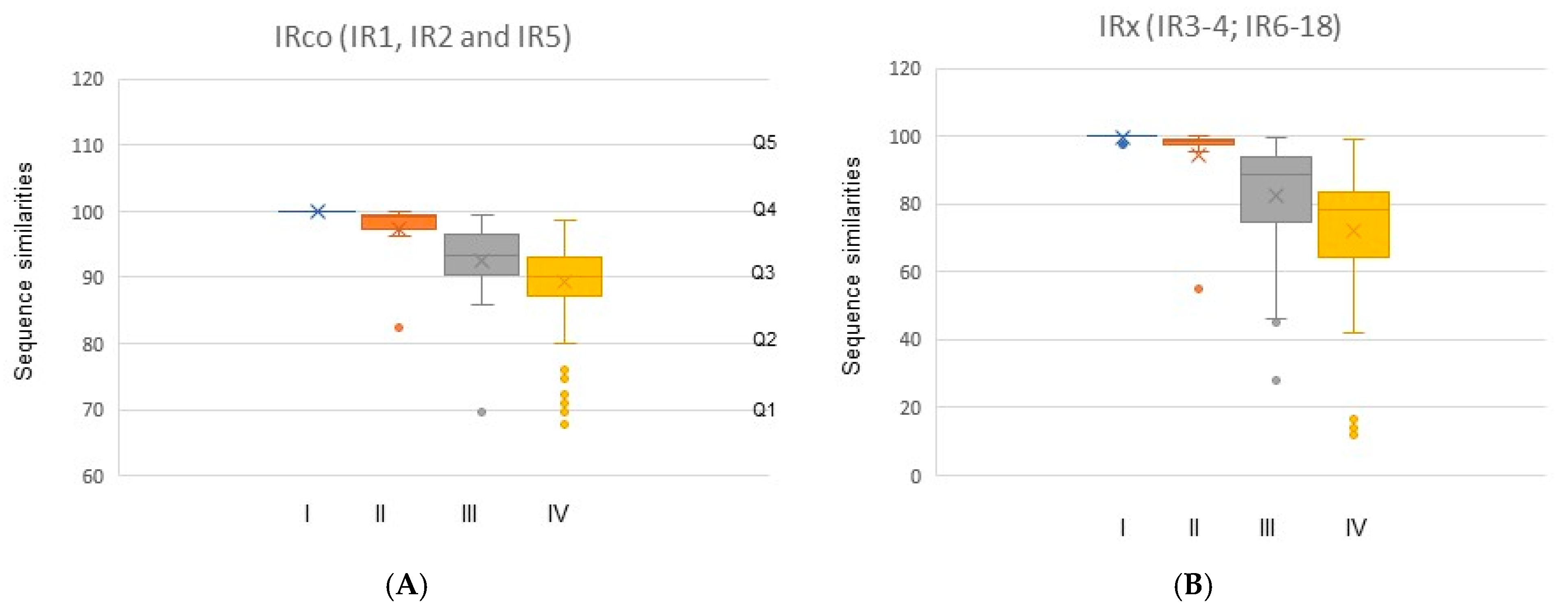
| Fig Wasp Taxa | Host Ficus | Number of IRs in Each Orthologous Group (IR 1–18) | T OG | T IR (1) | T IR (2) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||||
| Blastophaga 1 | F. abeli | 1 | 1 | 1 | - | 1 | 1 | 1 | - | - | - | - | - | - | - | - | - | - | - | 6 | 6 | 6 |
| Blastophaga 2 | F. erecta var. beecheyana | - | 1 | 1 | - | 1 | 1 | - | - | 1 | - | 1 | 1 | 1 | - | - | 1 | - | - | 9 | 9 | 9 |
| Blastophaga 3 | F. variolosa | - | 1 | 2 | - | 1 | - | - | - | - | - | - | 2 | - | - | - | - | - | - | 4 | 6 | 6 |
| Blastophaga 4 | F. pyriformis | - | 1 | 1 | - | 1 | 1 | 1 | - | 1 | 1 | 1 | - | - | - | 1 | - | 1 | - | 10 | 10 | 10 |
| Blastophaga 5 | F. formosa | 1 | 2 | - | - | - | 1 | 1 | - | 1 | - | - | - | 1 | - | - | - | - | - | 6 | 7 | 7 |
| Ceratosolen appendiculatus | F. variegata | 1 | - | 1 | - | 1 | 1 | - | - | 1 | 1 | 1 | - | - | - | 1 | 1 | - | - | 9 | 9 | 10 * |
| C. constrictus | F. fistulosa | 1 | 1 | - | - | - | 1 | - | 1 | - | - | - | - | - | - | - | 1 | 1 | 1 | 7 | 7 | 8 * |
| C. fusciceps | F. racemosa | 1 | 1 | 1 | 1 | - | 1 | - | - | - | - | - | - | 1 | 2 | - | - | - | - | 7 | 8 | 10 * |
| C. gravelyi | F. semicordata | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | - | - | - | - | 1 | - | - | 9 | 9 | 9 |
| C. solmsi | F. hispida | 1 | 2 | 1 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | - | - | - | - | - | 8 | 9 | 9 |
| Eupristina altissima | F. altissima | 1 | - | 2 | 1 | - | - | - | - | - | - | - | - | - | 2 | 1 | - | - | - | 5 | 7 | 7 |
| Kradibia tentacularis | F. montana | 1 | - | 1 | 1 | 1 | 1 | 1 | - | - | - | - | - | - | - | 1 | - | 1 | - | 8 | 8 | 8 |
| Platyscapa sp. 1 | F. concinna | 1 | - | - | 1 | - | - | - | 1 | - | - | 1 | - | - | - | - | - | - | 1 | 5 | 5 | 7 |
| P. quadraticeps | F. religiosa | 1 | 1 | - | - | - | - | - | 1 | - | - | 1 | 1 | - | 1 | - | - | - | 1 | 7 | 7 | 7 |
| Platyscapa sp. 2 | F. rumphii | - | - | - | 1 | 1 | - | - | - | - | - | 1 | - | - | - | - | - | - | - | 3 | 3 | 4 * |
| Valisia esquirolianae | F. triloba | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | - | 1 | - | 1 | - | - | - | - | - | - | 9 | 9 | 9 |
| V. cf. filippina | F. ruficaulis var. antaoensis | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0 | 0 | 7 * |
| V. malayana | F. grossularioides | - | 3 | - | 1 | - | 1 | - | - | 1 | - | - | - | - | - | - | - | - | - | 4 | 6 | 6 |
| Valisia sp. 1 | F. langkokensis | 1 | 1 | 2 | 2 | 1 | 1 | - | - | - | - | - | - | - | - | - | 1 | 1 | - | 8 | 10 | 10 |
| V. medusa | F. chartacea | - | 1 | - | 1 | - | - | 2 | - | 1 | 1 | - | 1 | 1 | 1 | - | - | - | - | 8 | 9 | 9 |
| V. compacta | F. fulva | 2 | - | 1 | 1 | - | 1 | - | 1 | - | 1 | 1 | - | 1 | - | - | - | - | - | 8 | 9 | 9 |
| V. javana sp. 1 | F. hirta | 1 | 1 | 1 | 1 | 1 | - | 1 | - | 1 | - | - | - | - | - | - | - | 1 | - | 8 | 8 | 8 |
| V. javana sp. 2 | F. hirta | 1 | 1 | 1 | 1 | 2 | - | 1 | 1 | - | - | - | - | - | - | - | - | - | - | 7 | 8 | 9 |
| V. javana sp. 7 | F. hirta | 1 | 1 | 2 | 1 | 1 | - | 1 | 1 | - | 1 | - | - | - | 1 | 2 | - | - | - | 10 | 12 | 12 |
| V. javana sp. 8 | F. hirta | 1 | 1 | 1 | - | - | - | 1 | 1 | 1 | 1 | - | - | 1 | - | - | - | - | - | 8 | 8 | 9 * |
| Total number of IRs in each orthologous group | 19 | 21 | 21 | 16 | 14 | 13 | 12 | 10 | 9 | 8 | 7 | 7 | 6 | 7 | 6 | 5 | 5 | 3 | ||||
| Number of taxa where present | 18 | 17 | 17 | 15 | 13 | 13 | 11 | 10 | 9 | 8 | 7 | 6 | 6 | 5 | 5 | 5 | 5 | 3 | 189 | 205 | ||
| Mean | 6.9 | 7.6 | 8.2 | |||||||||||||||||||
| SE | 2.4 | 2.4 | 1.7 | |||||||||||||||||||
| Sequence Identities of IRs (%) | Number of IRs in Each Orthologous Group (IR 1–18) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| In same Blastophaga sp. | 100 | 100 | 100 | - | 100 | 100 | 100 | - | 97.4 | - | - | 100 | 100 | - | - | - | - | - |
| In Blastophaga wasps | - | 99.4 | 99.1–99.6 | - | 82.3–100 | 99.5–99.6 | 97.4–98.7 | - | 97.4–99.6 | - | 99.2 | - | - | - | - | - | - | - |
| In Ceratosolen wasps | 88.8–94 | 69.8–87.4 | 44.9–82.6 | 83.4–87 | 93.3 | 58.9–73.5 | - | 91.4–93.4 | 88.1 | 89 | - | - | - | - | - | 80.5–85.4 | - | - |
| In Platyscapa wasps | 96.4 | - | - | 88.6 | - | - | - | 92.6 | - | - | 54.3–69.5 | - | - | - | - | - | - | 94.1 |
| In V. javana wasps | 98.4–99.6 | 96.2–99.2 | 55–98.6 | 96–98.9 | 97.9–99.5 | - | 98–100 | 95.6–97 | 98.3 | 97.4 | - | - | - | - | - | - | - | - |
| In Valisia wasps | 91.2–96.7 | 85.8–98.7 | 49.7–99.1 | 80.9–99.1 | 92.5–99.3 | 79.1–95.6 | 91.7–99.4 | 89.9–97 | 86.7–95.3 | 93.3–98.7 | - | 47.1 | 28.8–85.9 | 54.2 | - | 69.6–85.4 | 85.2 | - |
| Between genera in each group | 70.9–98.7 | 63.5–83.8 | 41.7–90.2 | 72.6–89.7 | 67.9–96.8 | 59.1–81.3 | 68.1–86.6 | 75.4–92.6 | 64.4–89.2 | 83.1–88.1 | 41.7–99.2 | 44.5–77.1 | 12.8–85.8 | 45.3–81.7 | - | 69.6–84.6 | 11.7–85.2 | 14.5–14.9 |
| In each group | 70.9–100 | 63.5–100 | 41.7–100 | 72.6–99.1 | 67.9–100 | 58.5–100 | 68.1–100 | 75.4–97 | 64.4–99.6 | 83.1–98.7 | 41.7–99.2 | 44.5–100 | 12.8–100 | 45.3–81.7 | 20.7–72.3 | 69.6–85.4 | 11.7–85.2 | 14.5–94.1 |
| Gene | Model | lnL H0 Versus H1 | Mates of Parameter dN/dS (ω) | df | 2ΔlnL | p-Value |
|---|---|---|---|---|---|---|
| IR1 | Free-ratio | −1022.584 | Variable ω | 33 | 33.568 | 0.220 |
| One-ratio | −1039.368 | ω = 0.035 | ||||
| IR2 | Free-ratio | −1244.001 | Variable ω | 31 | 27.050 | 0.335 |
| One-ratio | −1257.526 | ω = 0.136 | ||||
| IR3 | Free-ratio | −2814.628 | Variable ω | 31 | 57.032 | 0.002 * |
| One-ratio | −2843.144 | ω = 0.057 | ||||
| IR4 | Free-ratio | −978.290 | Variable ω | 27 | 26.566 | 0.244 |
| One-ratio | −991.573 | ω = 0.045 | ||||
| IR5 | Free-ratio | −726.358 | Variable ω | 23 | 12.932 | 0.477 |
| One-ratio | −732.824 | ω = 0.022 | ||||
| IR6 | Free-ratio | −1934.165 | Variable ω | 23 | 42.834 | 0.004 * |
| One-ratio | −1955.582 | ω = 0.140 | ||||
| IR7 | Free-ratio | −2010.821 | Variable ω | 19 | 16.640 | 0.307 |
| One-ratio | −2019.141 | ω = 0.079 | ||||
| IR8 | Free-ratio | −2159.311 | Variable ω | 17 | 34.518 | 0.004 * |
| One-ratio | −2176.570 | ω = 0.106 | ||||
| IR9 | Free-ratio | −792.710 | Variable ω | 15 | 8.348 | 0.455 |
| One-ratio | −796.884 | ω = 0.059 | ||||
| IR10 | Free-ratio | −1823.814 | Variable ω | 13 | 33.150 | 0.001 * |
| One-ratio | −1840.389 | ω = 0.065 | ||||
| IR11 | Free-ratio | −185.188 | Variable ω | 11 | 13.518 | 0.130 |
| One-ratio | −191.947 | ω = 0.168 | ||||
| IR12 | Free-ratio | −1686.123 | Variable ω | 9 | 22.448 | 0.004 * |
| One-ratio | −1697.347 | ω = 0.113 | ||||
| IR13 | Free-ratio | −105.870 | Variable ω | 9 | 2.156 | 0.494 |
| One-ratio | −106.948 | ω = 0.019 | ||||
| IR14 | Free-ratio | −738.105 | Variable ω | 7 | 3.584 | 0.413 |
| One-ratio | 739.897 | ω = 0.109 | ||||
| IR15 | Free-ratio | −239.273 | Variable ω | 7 | 12.978 | 0.036 * |
| One-ratio | −245.762 | ω = 0.060 | ||||
| IR16 | Free-ratio | −737.614 | Variable ω | 7 | 9.294 | 0.116 |
| One-ratio | −742.261 | ω = 0.120 | ||||
| IR17 | Free-ratio | −247.287 | Variable ω | 7 | 6.666 | 0.232 |
| One-ratio | −250.620 | ω = 0.049 |
| Gene | Clade | Model | lnL | Estimates of Parameters dN/dS | df | 2ΔlnL | p-Value | Positively Selected Sites |
|---|---|---|---|---|---|---|---|---|
| IR4 | 2 | Model A | −9787.966 | ω0 = 0.065, ω1 = 1, ω2 = 999 | 1 | 61.022 | 0.000 | 46A * |
| Null A | −9818.477 | ω0 = 0.064, ω1 = 1, ω2 = 1 | ||||||
| Vjb | Model A | −9815.376 | ω0 = 0.063, ω1 = 1, ω2 = 1 | 1 | 0 | 0.500 | 317I * | |
| Null A | −9815.376 | ω0 = 0.063, ω1 = 1, ω2 = 1 | ||||||
| IR9 | Bta | Model A | −4218.911 | ω0 = 0.070, ω1 = 1, ω2 = 999 | 1 | 30.338 | 0.000 | 51I ** |
| Null A | −4234.08 | ω0 = 0.070, ω1 = 1, ω2 = 1 | ||||||
| IR11 | Plq | Model A | −6009.32 | ω0 = 0.129, ω1 = 1, ω2 = 4.622 | 1 | 6.672 | 0.005 | 330V * |
| Null A | −6012.656 | ω0 = 0.125, ω1 = 1, ω2 = 1 | ||||||
| IR15 | Vjc | Model A | −3440.001 | ω0 = 0.056, ω1 = 1, ω2 = 1.973 | 1 | 2.126 | 0.072 | 245F *, 249T *, 251T *, 310E **, 314Y **, 315R **, 361S **, 363A ** |
| Null A | −3441.06421 | ω0 = 0.055, ω1 = 1, ω2 = 1 | ||||||
| Krt | Model A | −3454.803 | ω0 = 0.064, ω1 = 1, ω2 = 1 | 1 | 0 | 0.500 | 366W * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Nong, X.; Huang, W.; Yang, L.; Bhanumas, C.; Xiong, Y.; Dai, S. Ionotropic Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies. Insects 2025, 16, 679. https://doi.org/10.3390/insects16070679
Yu H, Nong X, Huang W, Yang L, Bhanumas C, Xiong Y, Dai S. Ionotropic Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies. Insects. 2025; 16(7):679. https://doi.org/10.3390/insects16070679
Chicago/Turabian StyleYu, Hui, Xiaojue Nong, Weicheng Huang, Ling Yang, Chantarasuwan Bhanumas, Yongmei Xiong, and Seping Dai. 2025. "Ionotropic Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies" Insects 16, no. 7: 679. https://doi.org/10.3390/insects16070679
APA StyleYu, H., Nong, X., Huang, W., Yang, L., Bhanumas, C., Xiong, Y., & Dai, S. (2025). Ionotropic Receptor Genes in Fig Wasps: Evolutionary Insights from Comparative Studies. Insects, 16(7), 679. https://doi.org/10.3390/insects16070679






