Chromosomal Inversions in Chromosome U of Drosophila subobscura: A Story from Population Studies to Molecular Level
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Strains
2.2. DNA Extraction and Sequencing
2.3. Bioinformatics of Sequencing Data
2.4. PCR Validation and Sanger Sequencing
3. Results
3.1. Inversion U6
3.2. Inversion U8
3.3. Description of “Ziga-Zaga”, a Fold-Back Element in the Subobscura Group
4. Discussion
4.1. Limitations of the Methods Used to Molecularly Characterize Inversions and Their Breakpoints
4.2. Origin of the Inversions U6 and U8
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sturtevant, A.H. Genetic Factors Affecting the Strength of Linkage in Drosophila. Proc. Natl. Acad. Sci. USA 1917, 3, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Painter, T.S. A New Method for the Study of Chromosome Rearrangements and the Plotting of Chromosome Maps. Science 1933, 78, 585–586. [Google Scholar] [CrossRef]
- Sharakhov, I.V.; Sharakhova, M.V. Chromosomal Inversions and Their Impact on Insect Evolution. Curr. Opin. Insect Sci. 2024, 66, 101280. [Google Scholar] [CrossRef]
- Krimbas, C.B.; Powell, J.R. Drosophila Inversion Polymorphism; Krimbas, C.B., Powell, J.R., Eds.; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Krimbas, C.B.; Loukas, M. The Inversion Polymorphism of Drosophila subobscura. In Evolutionary Biology; Hecht, H., Steere, W., Wallace, B., Eds.; Plenum Press: New York, NY, USA; London, UK, 1980; pp. 163–234. [Google Scholar]
- Krimbas, C.B.; Loukas, M. Evolution of the Obscura Group Drosophila Species. I. Salivary Chromosomes and Quantitative Characters in D. Subobscura and Two Closely Related Species. Heredity 1984, 53, 469–482. [Google Scholar] [CrossRef]
- Solé, E.; Balanyà, J.; Sperlich, D.; Serra, L. Long-Term Changes in the Chromosomal Inversion Polymorphism of Drosophila subobscura. I. Mediterranean Populations from Southwestern Europe. Evolution 2002, 56, 830–835. [Google Scholar]
- Kunze-Mühl, E.; Müller, E. Weitere Untersuchungen Uber Die Chromosomale Struktur Und Die Natürlichen Strukturtypen von Drosophila subobscura Coll. Chromosoma 1958, 9, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Prevosti, A.; Ribó, G.; Serra, L.; Aguadé, M.; Balanyà, J.; Monclús, M.; Mestres, F. Colonization of America by Drosophila subobscura: Experiment in Natural Populations That Supports the Adaptive Role of Chromosomal-Inversion Polymorphism. Proc. Natl. Acad. Sci. USA 1988, 85, 5597–5600. [Google Scholar] [CrossRef] [PubMed]
- Orengo, D.J.; Puerma, E.; Aguadé, M. Monitoring Chromosomal Polymorphism in Drosophila Subobscura over 40 Years. Entomol. Sci. 2016, 19, 215–221. [Google Scholar] [CrossRef]
- Orengo, D.J.; Prevosti, A. Temporal Changes in Chromosomal Polymorphism of Drosophila subobscura Related to Climatic Changes. Evolution 1996, 50, 1346–1350. [Google Scholar] [CrossRef]
- Sturtevant, A.H.; Beadle, G.W. The Relations of Inversions in the X Chromosome of Drosophila melanogaster to Crossing over and Disjunction. Genetics 1936, 21, 554–604. [Google Scholar] [CrossRef]
- Dobzhansky, T. Genetics of Natural Populations. XIX. Origin of Heterosis through Natural Selection in Populations of Drosophila pseudoobscura. Genetics 1950, 35, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Papaceit, M.; Segarra, C.; Aguadé, M. Structure and Population Genetics of the Breakpoints of a Polymorphic Inversion in Drosophila subobscura. Evolution 2013, 67, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Puerma, E.; Orengo, D.J.; Salguero, D.; Papaceit, M.; Segarra, C.; Aguadé, M. Characterization of the Breakpoints of a Polymorphic Inversion Complex Detects Strict and Broad Breakpoint Reuse at the Molecular Level. Mol. Biol. Evol. 2014, 31, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Orengo, D.J.; Puerma, E.; Papaceit, M.; Segarra, C.; Aguadé, M. A Molecular Perspective on a Complex Polymorphic Inversion System with Cytological Evidence of Multiply Reused Breakpoints. Heredity 2015, 114, 610–618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Puerma, E.; Orengo, D.J.; Aguadé, M. Multiple and Diverse Structural Changes Affect the Breakpoint Regions of Polymorphic Inversions across the Drosophila Genus. Sci. Rep. 2016, 6, 36248. [Google Scholar] [CrossRef]
- Puerma, E.; Orengo, D.J.; Aguadé, M. The Origin of Chromosomal Inversions as a Source of Segmental Duplications in the Sophophora Subgenus of Drosophila. Sci. Rep. 2016, 6, 30715. [Google Scholar] [CrossRef]
- Puerma, E.; Orengo, D.J.; Aguadé, M. Inversion Evolutionary Rates Might Limit the Experimental Identification of Inversion Breakpoints in Non-Model Species. Sci. Rep. 2017, 7, 17281. [Google Scholar] [CrossRef]
- Puerma, E.; Orengo, D.J.; Cruz, F.; Gómez-Garrido, J.; Librado, P.; Salguero, D.; Papaceit, M.; Gut, M.; Segarra, C.; Alioto, T.S.; et al. The High-Quality Genome Sequence of the Oceanic Island Endemic Species Drosophila Guanche Reveals Signals of Adaptive Evolution in Genes Related to Flight and Genome Stability. Genome Biol. Evol. 2018, 10, 1956–1969. [Google Scholar] [CrossRef] [PubMed]
- Orengo, D.J.; Puerma, E.; Aguadé, M. The Molecular Characterization of Fixed Inversions Breakpoints Unveils the Ancestral Character of the Drosophila Guanche Chromosomal Arrangements. Sci. Rep. 2019, 9, 1706. [Google Scholar] [CrossRef]
- Karageorgiou, C.; Gámez-Visairas, V.; Tarrío, R.; Rodríguez-Trelles, F. Long-Read Based Assembly and Synteny Analysis of a Reference Drosophila subobscura Genome Reveals Signatures of Structural Evolution Driven by Inversions Recombination-Suppression Effects. BMC Genomics 2019, 20, 223. [Google Scholar] [CrossRef]
- Ranz, J.M.; Maurin, D.; Chan, Y.S.; Von Grotthuss, M.; Hillier, L.W.; Roote, J.; Ashburner, M.; Bergman, C.M. Principles of Genome Evolution in the Drosophila melanogaster Species Group. PLoS Biol. 2007, 5, e152. [Google Scholar] [CrossRef] [PubMed]
- Orengo, D.J.; Puerma, E.; Cereijo, U.; Aguadé, M. The Molecular Genealogy of Sequential Overlapping Inversions Implies Both Homologous Chromosomes of a Heterokaryotype in an Inversion Origin. Sci. Rep. 2019, 9, 17009. [Google Scholar] [CrossRef] [PubMed]
- Karageorgiou, C.; Tarrío, R.; Rodríguez-Trelles, F. The Cyclically Seasonal Drosophila Subobscura Inversion O7 Originated From Fragile Genomic Sites and Relocated Immunity and Metabolic Genes. Front. Genet. 2020, 11, 565836. [Google Scholar] [CrossRef]
- Cridland, J.M.; Thornton, K.R. Validation of Rearrangement Break Points Identified by Paired-End Sequencing in Natural Populations of Drosophila Melanogaster. Genome Biol. Evol. 2010, 2, 83–101. [Google Scholar] [CrossRef]
- Corbett-Detig, R.B.; Cardeno, C.; Langley, C.H. Sequence-Based Detection and Breakpoint Assembly of Polymorphic Inversions. Genetics 2012, 192, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, P.; Stanciu, M.; Brudno, M. Computational Methods for Discovering Structural Variation with Next-Generation Sequencing. Nat. Methods 2009, 6, S13. [Google Scholar] [CrossRef]
- Bracewell, R.; Chatla, K.; Nalley, M.J.; Bachtrog, D. Dynamic Turnover of Centromeres Drives Karyotype Evolution in Drosophila. Elife 2019, 8, e49002. [Google Scholar] [CrossRef]
- Menozzi, P.; Krimbas, C.B. The Inversion Polymorphism of D. Subobscura Revisited: Synthetic Maps of Gene Arrangement Frequencies and Their Interpretation. J. Evol. Biol. 1992, 5, 625–641. [Google Scholar] [CrossRef]
- Rego, C.; Balanyà, J.; Fragata, I.; Matos, M.; Rezende, E.L.; Santos, M. Clinal Patterns of Chromosomal Inversion Polymorphisms in Drosophila subobscura Are Partly Associated with Thermal Preferences and Heat Stress Resistance. Evolution 2010, 64, 385–397. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kapun, M.; Fabian, D.K.; Goudet, J.; Flatt, T. Genomic Evidence for Adaptive Inversion Clines in Drosophila Melanogaster. Mol. Biol. Evol. 2016, 33, 1317–1336. [Google Scholar] [CrossRef]
- Douglas, A.E. The Drosophila Model for Microbiome Research. Lab Anim. 2018, 47, 157–164. [Google Scholar] [CrossRef]
- Guilhot, R.; Xuéreb, A.; Lagmairi, A.; Olazcuaga, L.; Fellous, S. Microbiota Acquisition and Transmission in Drosophila Flies. iScience 2023, 26, 107656. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Chen, K.; Wallis, J.W.; Mclellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; Mcgrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An Algorithm for High Resolution Mapping of Genomic Structural Variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef]
- Papaceit, M.; Aguadé, M.; Segarra, C. Chromosomal Evolution of Elements B and C in the Sophophora Subgenus of Drosophila: Evolutionary Rate and Polymorphism. Evolution 2006, 60, 768. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 2020, 70, e102. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene Sequence Analysis Software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar]
- Miller, W.J.; Nagel, A.; Bachmann, J.; Bachmann, L. Evolutionary Dynamics of the SGM Transposon Family in the Drosophila Obscura Species Group. Mol. Biol. Evol. 2000, 17, 1597–1609. [Google Scholar] [CrossRef][Green Version]
- Bachmann, L.; Raab, M.; Sperlich, D. Satellite DNA and Speciation: A Species Specific Satellite DNA of Drosophila Guanche. J. Zool. Syst. Evol. Res. 1989, 27, 84–93. [Google Scholar] [CrossRef]
- Balanyà, J.; Solé, E.; Oller, J.M.; Sperlich, D.; Serra, L. Long-Term Changes in the Chromosomal Inversion Polymorphism of Drosophila subobscura. II. European Populations. J. Zool. Syst. Evol. Res. 2004, 42, 191–201. [Google Scholar] [CrossRef]
- Rezende, E.L.; Balanyà, J.; Rodríguez-Trelles, F.; Rego, C.; Fragata, I.; Matos, M.; Serra, L.; Santos, M. Climate Change and Chromosomal Inversions in Drosophila subobscura. Clim. Res. 2010, 43, 103–114. [Google Scholar] [CrossRef]
- Galludo, M.; Canals, J.; Pineda-Cirera, L.; Esteve, C.; Rosselló, M.; Balanyà, J.; Arenas, C.; Mestres, F. Climatic Adaptation of Chromosomal Inversions in Drosophila Subobscura. Genetica 2018, 146, 433–441. [Google Scholar] [CrossRef]
- Khadem, M.; Arenas, C.; Balanyà, J.; Mestres, F. Long-Term Changes in the Inversion Chromosomal Polymorphism: Drosophila Subobscura Population from Rasht (North of Iran). J. Genet. 2022, 101, 45. [Google Scholar] [CrossRef]
- Zivanovic, G.; Arenas, C.; Mestres, F. Rate of Change for the Thermal Adapted Inversions in Drosophila subobscura. Genetica 2019, 147, 401–409. [Google Scholar] [CrossRef]
- Zivanovic, G.; Arenas, C.; Mestres, F. Individual Inversions or Their Combinations: Which Is the Main Selective Target in a Natural Population of Drosophila Subobscura? J. Evol. Biol. 2016, 29, 657–664. [Google Scholar] [CrossRef]
- Andolfatto, P.; Wall, J.D.; Kreitman, M. Unusual Haplotype Structure at the Proximal Breakpoint of In(2L)t in a Natural Population of Drosophila melanogaster. Genetics 1999, 153, 1297–1311. [Google Scholar] [CrossRef]
- Matzkin, L.M.; Merritt, T.J.S.; Zhu, C.-T.; Eanes, W.F. The Structure and Population Genetics of the Breakpoints Associated with the Cosmopolitan Chromosomal Inversion In(3R)Payne in Drosophila melanogaster. Genetics 2005, 170, 1143–1152. [Google Scholar] [CrossRef]
- Domínguez, M.I. Localización y Estudio de La Inversión Cromosómica J1 En Drosophila subobscura. Implicaciones Evolutivas y Adaptativas. Master’s Thesis, Universitat Oberta de Catalunya, Barcelona, Spain, 2025. [Google Scholar]
- Fan, X.; Abbott, T.E.; Larson, D.; Chen, K. BreakDancer: Identification of Genomic Structural Variation from Paired-End Read Mapping. Curr. Protoc. Bioinformatics 2014, 45, 15.6.1–15.6.11. [Google Scholar] [CrossRef]
- Aulard, S.; Vaudin, P.; Ladevèze, V.; Chaminade, N.; Périquet, G.; Lemeunier, F. Maintenance of a Large Pericentric Inversion Generated by the Hobo Transposable Element in a Transgenic Line of Drosophila melanogaster. Heredity 2004, 92, 151–155. [Google Scholar] [CrossRef]
- Engels, W.R.; Preston, C.R. Formation of Chromosome Rearrangements by P Factors in Drosophila. Genetics 1984, 107, 657–678. [Google Scholar] [CrossRef]
- Lyttle, T.W.; Haymer, D.S. The Role of the Transposable Element Hobo in the Origin of Endemic Inversions in Wild Populations of Drosophila melanogaster. Genetica 1992, 86, 113–126. [Google Scholar] [CrossRef]
- Kusakabe, S.; Harada, K.; Mukai, T. The Rare Inversion with a P Element at the Breakpoint Maintained in a Natural Population of Drosophila Melanogaster. Genetica 1990, 82, 111–115. [Google Scholar] [CrossRef]
- Casals, F.; Cáceres, M.; Ruiz, A. The Foldback-like Transposon Galileo Is Involved in the Generation of Two Different Natural Chromosomal Inversions of Drosophila buzzatii. Mol. Biol. Evol. 2003, 20, 674–685. [Google Scholar] [CrossRef]
- Cáceres, M.; Ranz, J.M.; Barbadilla, A.; Long, M.; Ruiz, A. Generation of a Widespread Drosophila Inversion by a Transposable Element. Science 1999, 285, 415–418. [Google Scholar] [CrossRef]
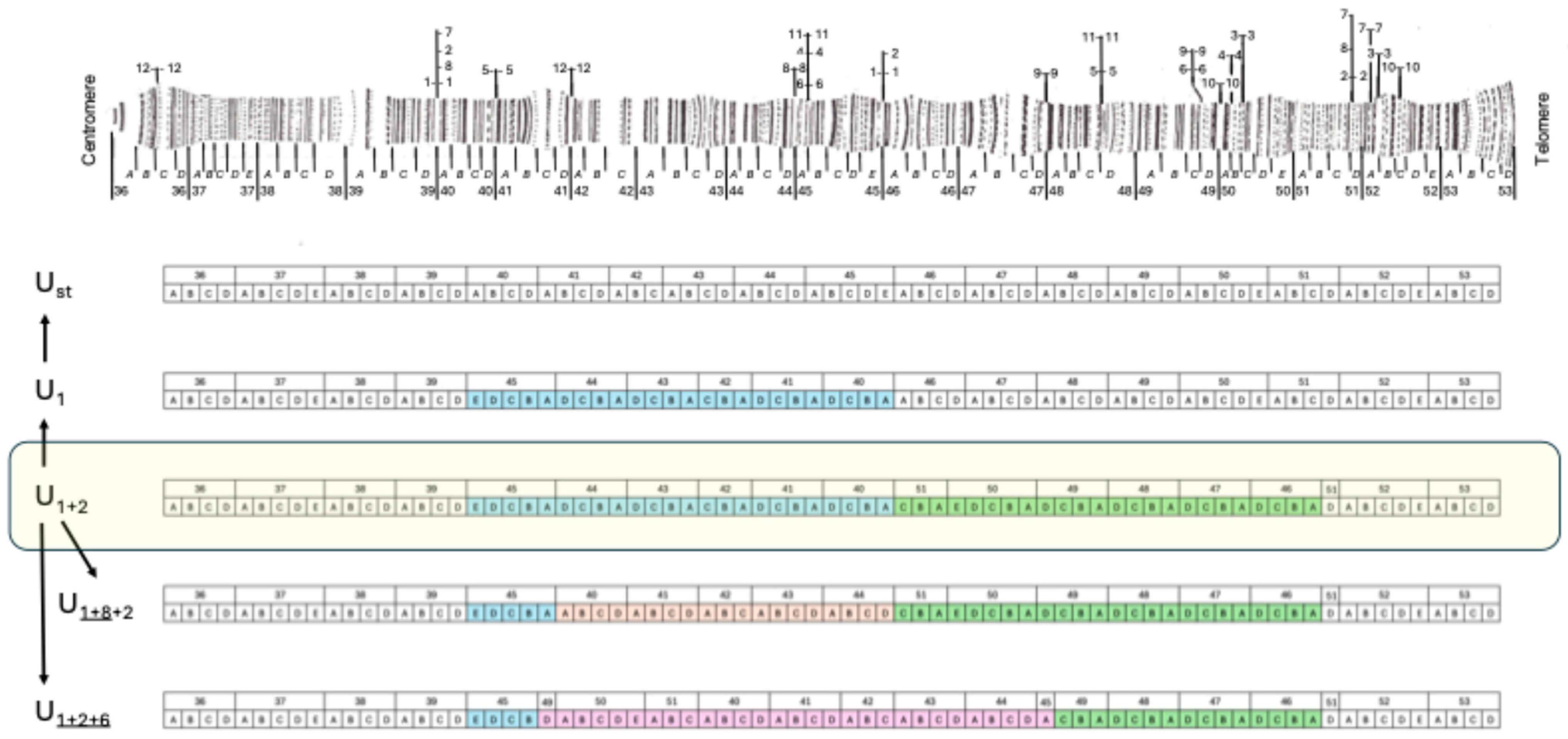
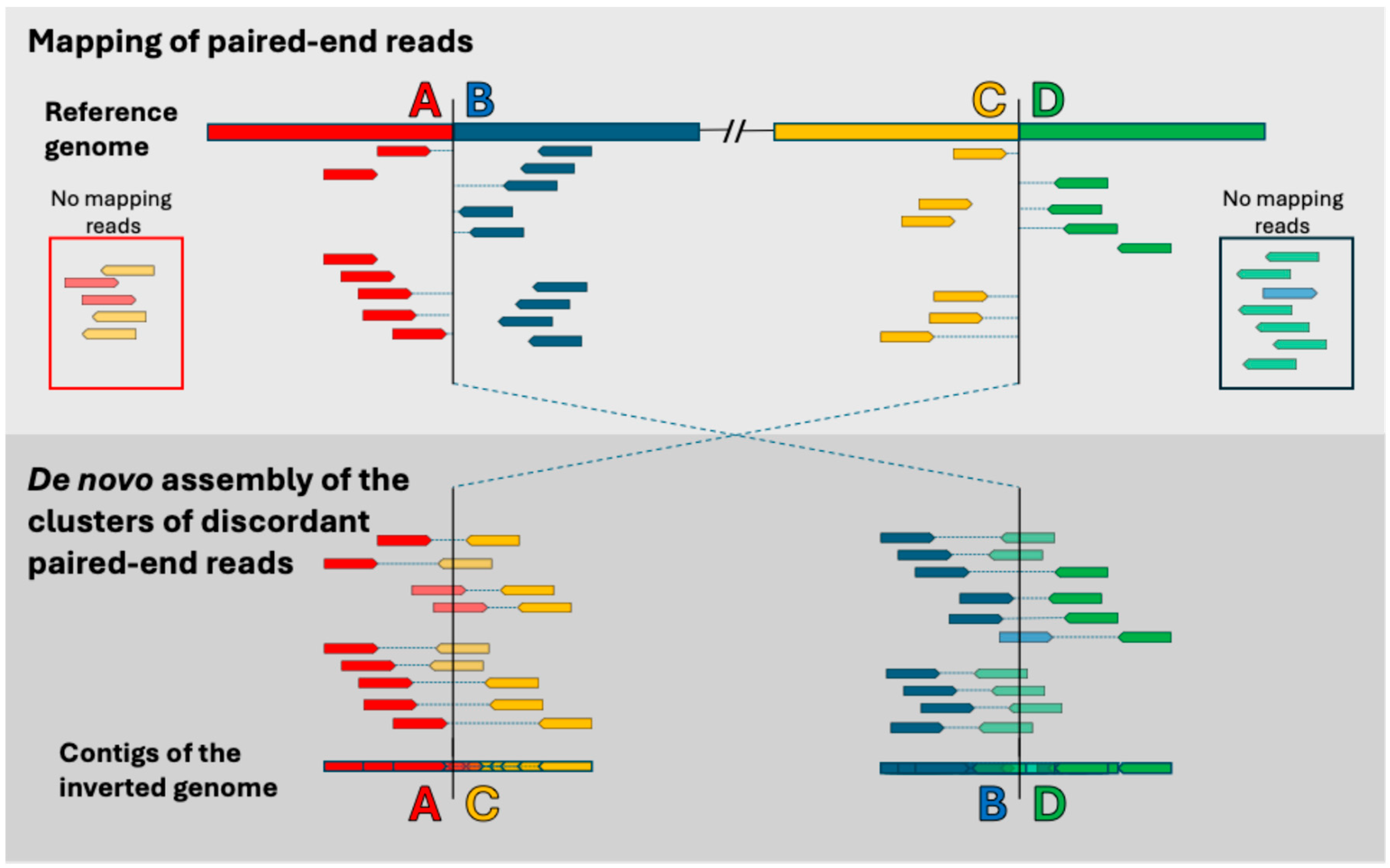
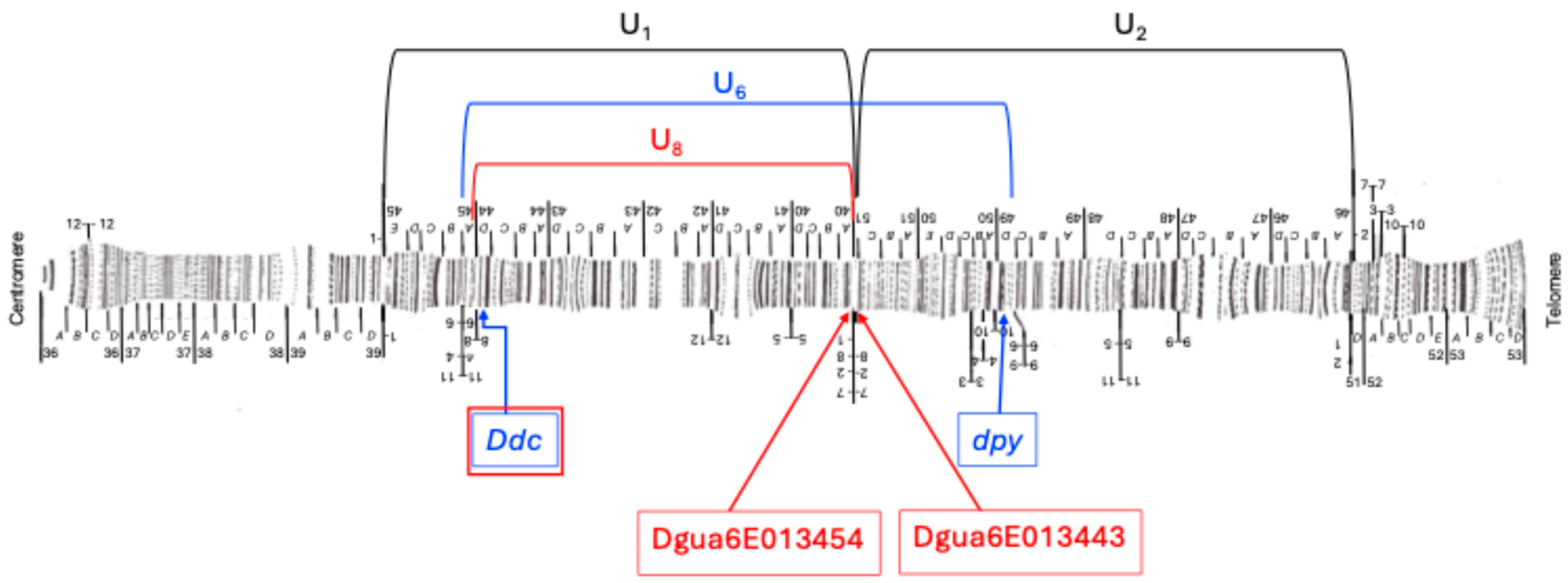

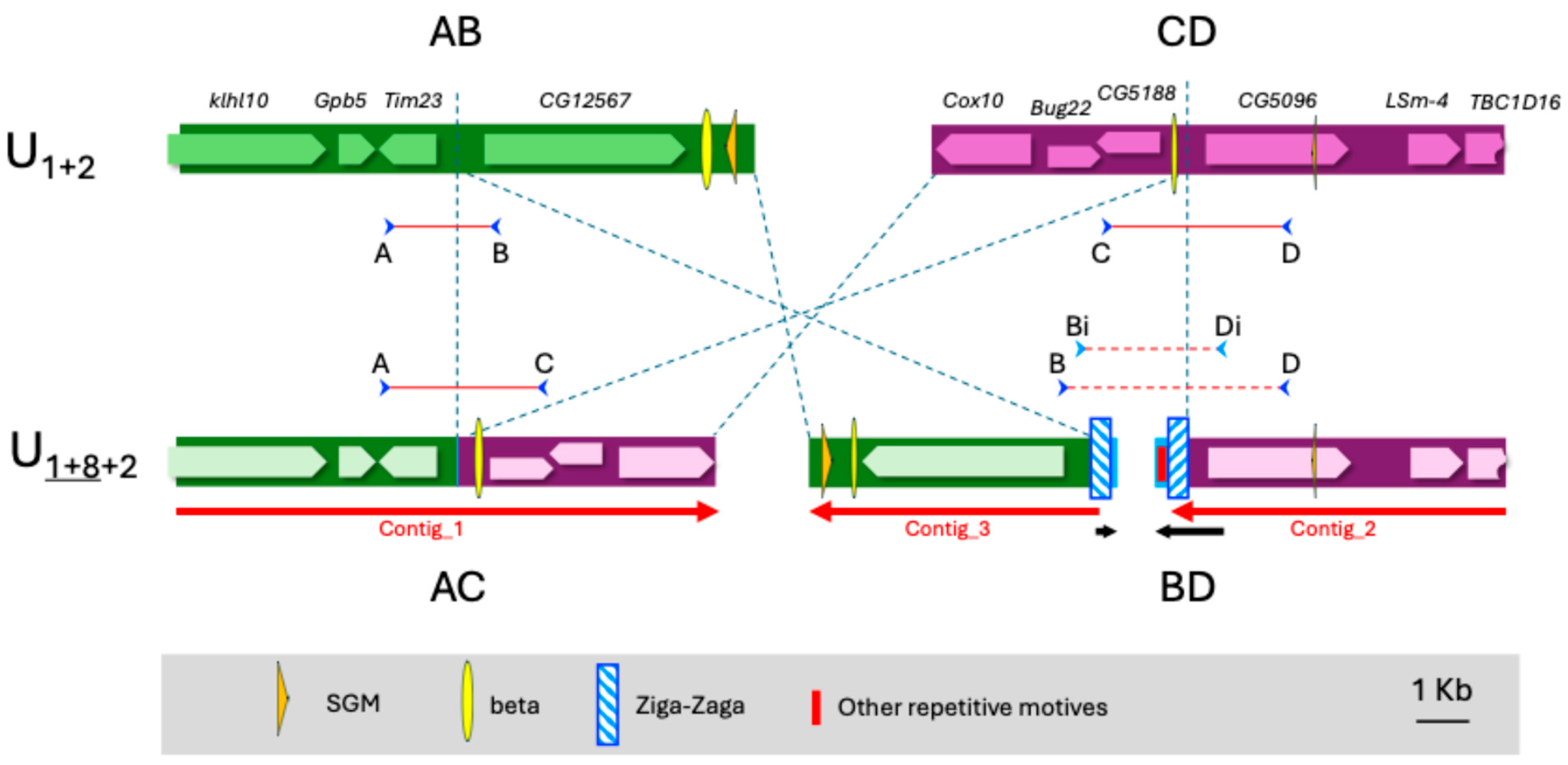
| Inversion | Marker | Band | Reference | BLAST |
|---|---|---|---|---|
| U6 | Proximal BP. | 45B/45A | [8] | ¿? |
| Ddc | 44D | [39] | 7,888,695–7,890,041 | |
| dpy | 49D | [20] | 16,420,251–16,503,087 | |
| Distal BP. | 49D/49C | [8] | ¿? | |
| U8 | Proximal BP. | 45A/44D | [8] | ¿? |
| Ddc | 44D | [8] | 7,888,695–7,890,041 | |
| Proximal BP. U1 (Dgua6E013454) | 40A | [8,22] | 14,589,270–14,593,607 | |
| Distal BP. U2(Dgua6E013443) | 51C | [8,22] | 14,634,213–14,639,290 | |
| Distal breakpoint | 51C/40A | [8] | ¿? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merayo, M.; Delgado, K.M.; Salguero, D.; Orengo, D.J. Chromosomal Inversions in Chromosome U of Drosophila subobscura: A Story from Population Studies to Molecular Level. Insects 2025, 16, 586. https://doi.org/10.3390/insects16060586
Merayo M, Delgado KM, Salguero D, Orengo DJ. Chromosomal Inversions in Chromosome U of Drosophila subobscura: A Story from Population Studies to Molecular Level. Insects. 2025; 16(6):586. https://doi.org/10.3390/insects16060586
Chicago/Turabian StyleMerayo, Mercè, Kenia M. Delgado, David Salguero, and Dorcas J. Orengo. 2025. "Chromosomal Inversions in Chromosome U of Drosophila subobscura: A Story from Population Studies to Molecular Level" Insects 16, no. 6: 586. https://doi.org/10.3390/insects16060586
APA StyleMerayo, M., Delgado, K. M., Salguero, D., & Orengo, D. J. (2025). Chromosomal Inversions in Chromosome U of Drosophila subobscura: A Story from Population Studies to Molecular Level. Insects, 16(6), 586. https://doi.org/10.3390/insects16060586






