Complete Mitochondrial Genome Characterization and Phylogenomics of the Stingless Bee, Heterotrigona itama (Apidae: Meliponini)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Species Identification
2.2. Ethics Statement
2.3. DNA Extraction
2.4. Sequencing, Assembly, Annotation, and Analysis
2.5. Phylogenomic Reconstruction
2.6. Gene Rearrangement Assessment
3. Results
3.1. Organization and Nucleotide Composition
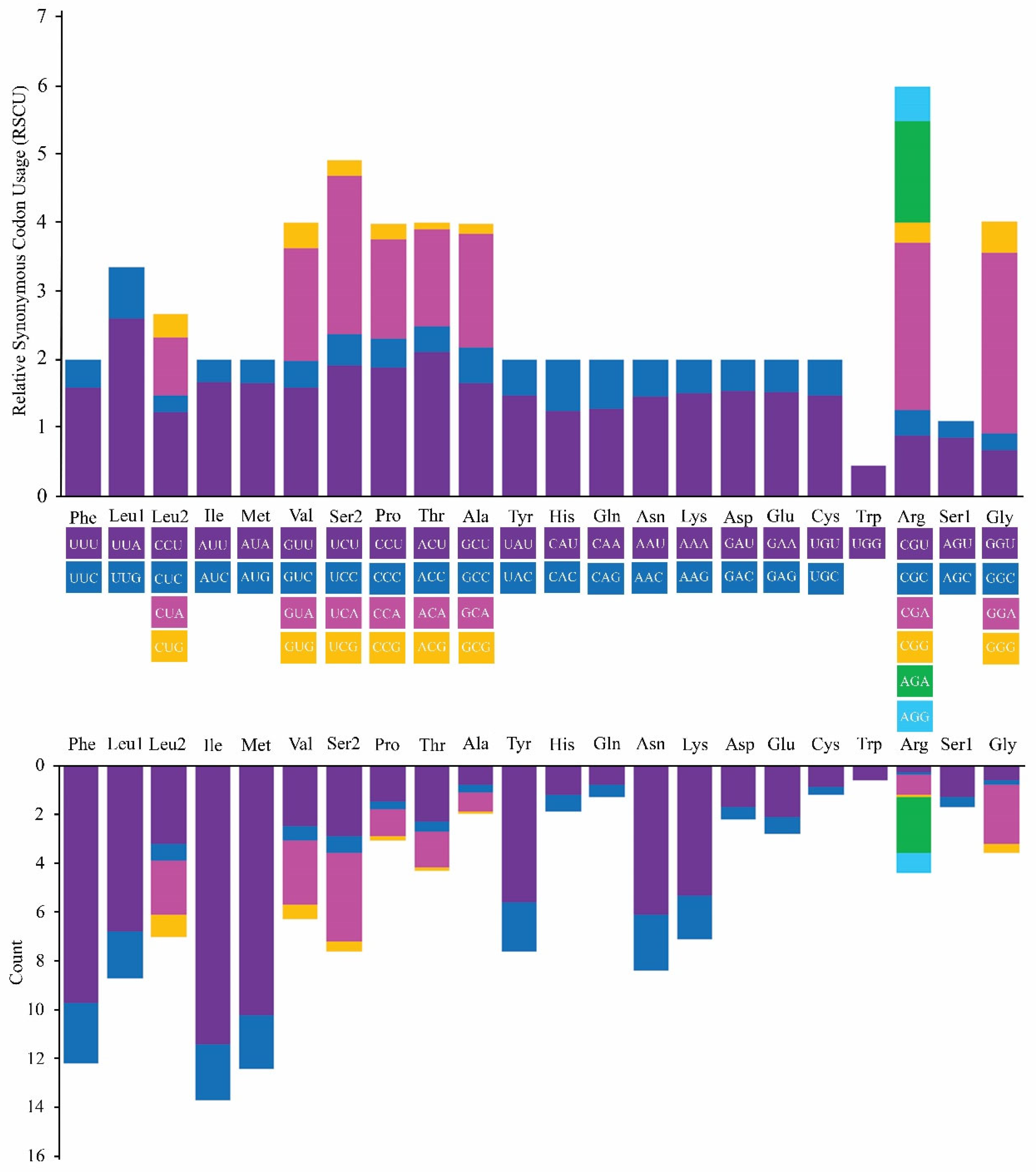
3.2. PCGs and Codon Usage Bias
3.3. Transfer RNAs and Ribosomal RNAs
3.4. Intergenic Spacer and Overlapping Region
3.5. Phylogenomic Relationship
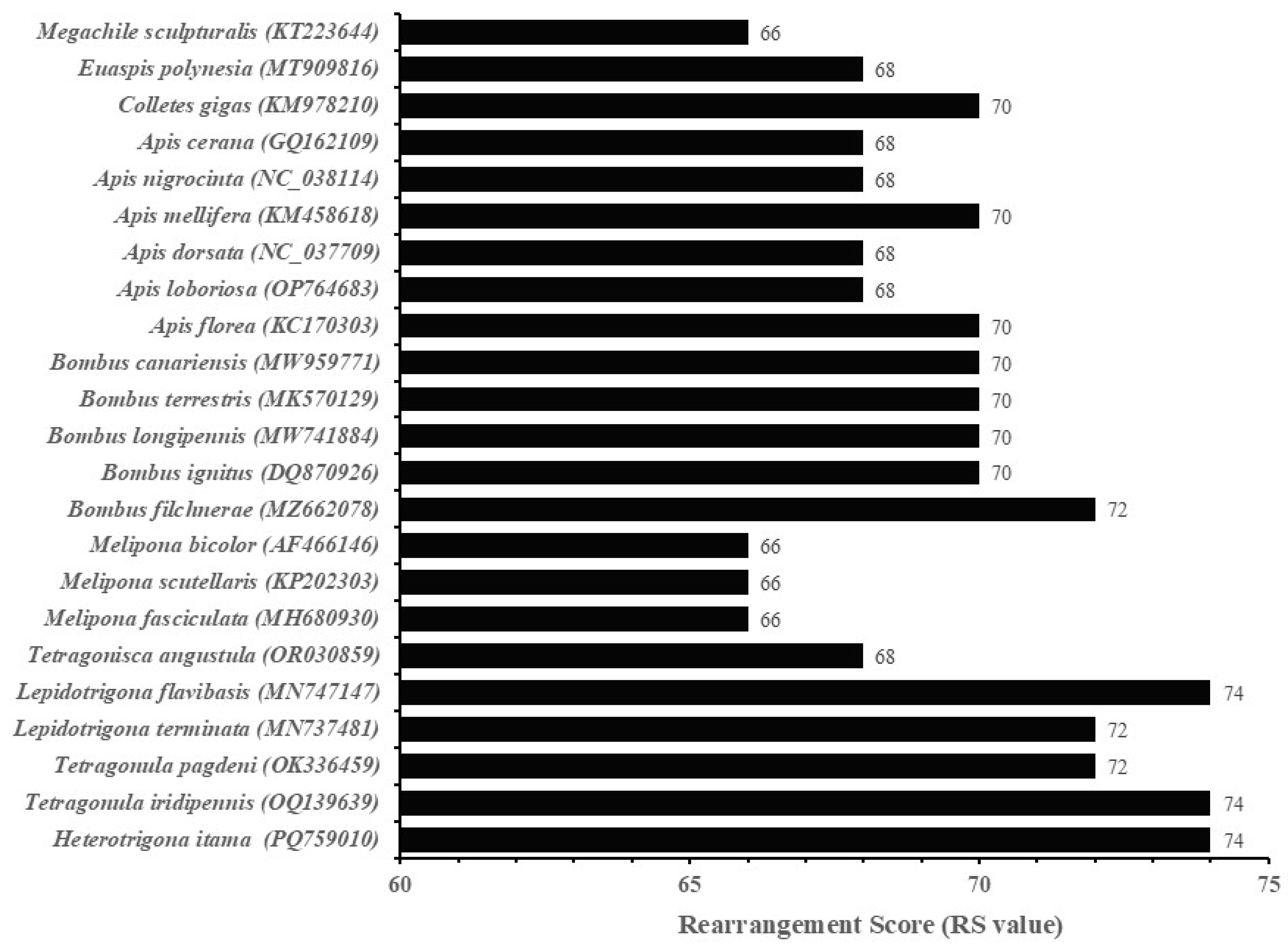
3.6. Gene Rearrangement
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gruter, C. Stingless Bees: Their Behaviour, Ecology and Evolution; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Rattanawannee, A.; Duangphakdee, O. Southeast Asian meliponiculture for sustainable livelihood. In Modern Beekeeping; Ranz, R.E.R., Ed.; IntechOpen: London, UK, 2019; pp. 1–17. [Google Scholar] [CrossRef]
- Bartelli, B.F.; Santos, A.O.R.; Nogueira-Ferreira, F.H. Colony performance of Melipona Quadrifasciata (Hymenoptera, Meliponina) in a greenhouse of Lycopersicon esculentum (Solanaceae). Sociobiology 2014, 61, 60–67. [Google Scholar] [CrossRef]
- da Silva, M.A.; da Silva Ferreira, N.; da Silva Teixeira-Souza, V.H.; Maia-Silva, C.; de Assis de Oliveira, F.; Hrmcir, M. On the thermal limits for the use of stingless bees as pollinators in commercial greenhouses. J. Apic. Res. 2017, 56, 81–90. [Google Scholar] [CrossRef]
- Wongsa, K.; Duangphakdee, O.; Rattanawannee, A. Pollination efficacy of stingless bees, Tetragonula pagdeni Schwarz (Apidae: Meliponini), on greenhouse tomatoes (Solanum lycopersicum Linnaeus). PeerJ 2023, 11, e15367. [Google Scholar] [CrossRef] [PubMed]
- Al-Hatamleh, M.A.; Boer, J.C.; Wilson, K.L.; Plebanski, M.; Mohamud, R.; Mustafa, M.Z. Antioxidant-based medicinal properties of stingless bee products: Recent progress and future directions. Biomolecules 2020, 10, 923. [Google Scholar] [CrossRef]
- Rattanawannee, A.; Chanchao, C. Bee diversity in Thailand and the applications of bee products. In Changing Diversity in Changing Environment; Grillo, O., Venora, G., Eds.; InTech: London, UK, 2011; pp. 133–162. [Google Scholar] [CrossRef][Green Version]
- Wongsa, K.; Meemongkolkiat, T.; Duangphakdee, O.; Prasongsuk, S.; Rattanawannee, A. Physicochemical properties, phenolic, flavonoid contents and antioxidant potential of stingless bee (Heterotrigona itama) honey from Thailand. Curr. Res. Nutr. Food Sci. 2023, 11, 246–257. [Google Scholar] [CrossRef]
- Quezada-Euán, J.J.G. Stingless Bees of Mexico: The Biology, Management and Conservation of an Ancient Heritage; Springer: New York, NY, USA, 2018. [Google Scholar]
- Rasmussen, C. Catalog of the Indo-Malayan/Australasians stingless bees (Hymenoptera: Apidae: Meliponini). Zootaxa 2008, 1935, 1–80. [Google Scholar] [CrossRef]
- Wongsa, K.; Duangphakdee, O.; Poolprasert, P.; Rattanawannee, A. External morphometric and microscopic analysis of the reproductive system in in- vitro reared stingless bee queens, Heterotrigona itama, and their mating frequency. PLoS ONE 2024, 19, e0306085. [Google Scholar] [CrossRef]
- Wongsa, K.; Jeratthitikul, E.; Poolprasert, P.; Duangphakdee, O.; Rattanawannee, A. Genetic structure of the commercial stingless bee Heterotrigona itama (Apidae: Meliponini) in Thailand. PLoS ONE 2024, 19, e0312386. [Google Scholar] [CrossRef]
- Hrncir, M.; Maia-Silva, C. On the diversity of foraging-related traits in stingless bees. In Pot-Honey; Vit, P., Pedro, S., Roubik, D., Eds.; Springer: New York, NY, USA, 2013; pp. 201–215. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhao, M.; Wang, S.J.; Xu, H.I.; Yang, Y.M.; Liu, L.N.; Feng, Y. The complete mitochondrial genome of Lepidotrigona flavibasis (Hymenoptera: Meliponini) and high gene rearrangement in Lepidotrigona mitogenomes. J. Insect Sci. 2021, 21, 1–7. [Google Scholar] [CrossRef]
- Rasmussen, C.; Cameron, S.A. Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal. Biol. J. Linn. Soc. 2010, 99, 206–232. [Google Scholar] [CrossRef]
- Samsudin, S.F.; Mamat, M.R.; Hazmi, I.R. “Taxonomic study on selected species of stingless bee (Hymenoptera: Apidae: Meliponini) in Peninsular Malaysia. Serangga 2018, 23, 203–258. [Google Scholar]
- Trianto, M.; Arisuryanti, T.; Purwanto, H.; Ubaidillah, R. Updated species check-list of the Indonesian stingless bees (Hymenoptera, Apidae, Apinae, Meliponini). J. Trop. Biodivers. Biotechnol. 2023, 8, 77–160. [Google Scholar] [CrossRef]
- Karuppaiah, V.; Gadge, A.S.; Shirsat, D.V.; Soumia, P.S.; Mainkar, P.; Kumar, S.; Jaiswa, D.K.; Mahajan, V. The complete mitochondrial genome of the Indian dammer bee, Tetragonula iridipennis, and the phylogenomics of meliponini. Front. Ecol. Evol. 2023, 11, 1171242. [Google Scholar] [CrossRef]
- Bleidorn, C.; Henze, K. A new primer pair for barcoding of bees (Hymenoptera: Anthophila) without amplifying the orthologous coxa gene of Wolbachia bacteria. BMC Res. Notes 2021, 14, 427. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zheng, R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and hylogenetic relationship of dicroglossidae. BMC Ecol. Evol. 2018, 18, 26. [Google Scholar] [CrossRef]
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Wu, X.; Li, D.; Yan, P.; Wu, X. The complete mitochondrial genome of Rhacophorus dennysi (Anura: Rhacophoridae) with novel gene arrangements and its phylogenetic implications. Pak. J. Zool. 2021, 53, 2013–2019. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Chen, L.; Chen, P.Y.; Xue, X.F.; Hua, H.Q.; Li, Y.X.; Zhang, F.; Wei, S.J. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae). Sci. Rep. 2018, 8, 7034. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. Mitoz: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partition finder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. Iq-Tree 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- San Mauro, D.; Agorreta, A. Molecular systematics: A synthesis of the common methods and the state of knowledge. Cell. Mol. Biol. Lett. 2010, 15, 311–341. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. Phylosuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Res. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Zhang, J.; Kan, X.; Miao, G.; Hu, S.; Sun, Q.; Tian, W. QMGR: A new approach for quantifying mitochondrial genome rearrangement. Mitochondrion 2020, 52, 20–23. [Google Scholar] [CrossRef]
- Crozier, R.H.; Crozier, Y.C. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics 1993, 133, 97–117. [Google Scholar] [CrossRef]
- Wang, C.Y.; Yang, P.L.; Zhao, M.; Xu, H.L.; Liu, L.N.; Feng, Y.; Wang, S.J. Unusual mitochondrial tRNA rearrangements in stingless bee Tetragonula pagdeni and phylogenetic analysis. Entomol. Sci. 2022, 25, e12526. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Maximizing transcription efficiency causes codon usage bias. Genetics 1996, 144, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Silvestre, D.; Dowton, M.; Arias, M.C. The mitochondrial genome of the stingless bee Melipona bicolor (Hymenoptera, Apidae, Meliponini): Sequence, gene organization and a unique tRNA translocation event conserved across the tribe Meliponini. Genet. Mol. Biol. 2008, 31, 451–460. [Google Scholar] [CrossRef]
- Pereira, U.D.P.; Bonetti, A.M.; Goulart, L.R.; Santos, A.R.D.; Oliveira, G.C.D.; Cuadros-Orellana, S.; Ueira-Vieira, C. Complete mitochondrial genome sequence of Melipona scutellaris, a Brazilian stingless bee. Mitochondrial DNA Part A 2016, 27, 3387–3388. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhao, M.; Xu, H.L.; Zhang, F.L.; Zhong, Y.H.; Fengaan, Y.; Wang, S.J. Complete mitochondrial genome of the stingless bee Lepidotrigona terminate (Hymenoptera: Meliponinae) and phylogenetic analysis. Mitochondrial DNA Part B 2020, 5, 752–753. [Google Scholar] [CrossRef]
- Tan, H.W.; Liu, G.H.; Dong, X.; Lin, R.Q.; Song, H.Q.; Huang, S.Y.; Yuan, Z.G.; Zhao, G.H.; Zhu, X.Q. The complete mitochondrial genome of the asiatic cavity-nesting honeybee Apis cerana (Hymenoptera: Apidae). PLoS ONE 2011, 6, e23008. [Google Scholar] [CrossRef]
- Yatawara, L.; Wickramasinghea, S.; Rajapakse, R.P.; Agatsuma, T. The complete mitochondrial genome of Setaria digitata (Nematoda: Filarioidea): Mitochondrial gene content, arrangement and composition compared with other nematodes. Mol. Biochem. Parasitol. 2010, 173, 32–38. [Google Scholar] [CrossRef]
- Castro, L.R.; Ruberu, K.; Dowton, M. Mitochondrial genomes of Vanhornia eucnemidarum (Apocrita: Vanhorniidae) and Primeuchroeus spp. (Aculeata: Chrysididae): Evidence of rearranged mitochondrial genomes within the Apocrita (Insecta: Hymenoptera). Genome 2006, 49, 752–766. [Google Scholar] [CrossRef]
- Cha, S.Y.; Yoon, H.J.; Lee, E.M.; Yoon, M.H.; Hwang, J.S.; Jin, B.R.; Han, Y.S.; Kim, I. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignites (Hymenoptera: Apidae). Gene 2007, 392, 206–220. [Google Scholar] [CrossRef]
- Wei, S.J.; Shi, M.; He, J.H.; Sharkey, M.; Chen, X.X. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 2009, 52, 308–319. [Google Scholar] [CrossRef]
- Nakao, M.; Abmed, D.; Yamasaki, H.; Ito, A. Mitochondrial genomes of the human broad tapeworms Diphyllobothrium latum and Diphyllobothrium nihonkaiense (Cestoda: Diphyllobothriidae). Parasitol. Res. 2007, 101, 233–236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, S.J.; Tang, P.; Zheng, L.H.; Shi, M.; Chen, X.X. The Complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A+T content and a long intergenic spacer between atp8 and atp6. Mol. Biol. Rep. 2010, 37, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Cameron, S.L.; Austin, A.D.; Whiting, M.F. Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera: A lineage with both rapidly and slowly evolving mitochondrial genomes. Mol. Phylogenetics Evol. 2009, 52, 512–529. [Google Scholar] [CrossRef]
- Ramírez, S.R.; Nieh, J.C.; Quental, T.B.; Roubik, D.W.; Imperatriz-Fonseca, V.L.; Pierce, N.E. A molecular phylogeny of the stingless bee genus Melipona (Hymenoptera: Apidae). Mol. Phylogenetics Evol. 2010, 56, 519–525. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Z.; Huang, J.; Liang, C.; An, J.; Sun, C. Complete mitochondrial genome of Bombus breviceps (Hymenoptera: Apidae). Mitochondrial DNA Part B 2017, 2, 604–606. [Google Scholar] [CrossRef]
- Shao, R.; Dowton, M.; Murrell, A.; Barker, S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003, 20, 1612–1619. [Google Scholar] [CrossRef]
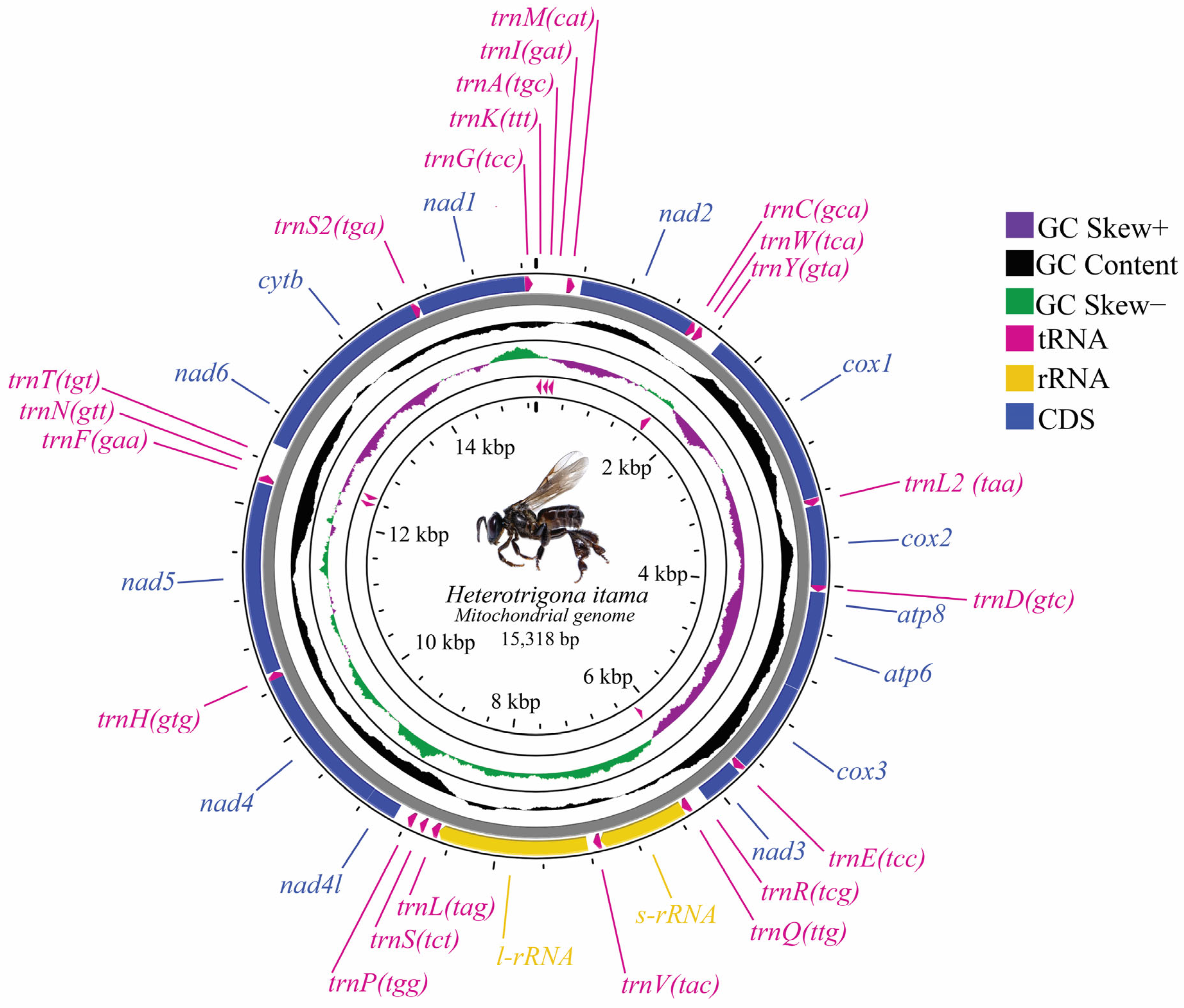




| Locus | Full Name and Function | Position | Length (bp) | Strand | Intergenic Spacer | Codon | Anti- Codon | ||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | Stop | ||||||
| tRNA-Lys (ttt) | Transfer RNA for Lysine | 1 | 69 | 69 | − | 22 | TTT | ||
| tRNA-Ala (tgc) | Transfer RNA for Alanine | 92 | 156 | 65 | − | 8 | TGC | ||
| tRNA-Ile (gat) | Transfer RNA for Isoleucine | 165 | 231 | 67 | − | 35 | GAT | ||
| tRNA-Met (cat) | Transfer RNA for Methionine | 267 | 335 | 69 | + | 56 | CAT | ||
| nad2 | NADH dehydrogenase subunit 2 | 392 | 1383 | 992 | + | 0 | ATC | TAA | |
| tRNA-Cys (gca) | Transfer RNA for Cysteine | 1384 | 1448 | 65 | + | 8 | GCA | ||
| tRNA-Trp (tca) | Transfer RNA for Tryptophan | 1457 | 1524 | 68 | + | −1 | TCA | ||
| tRNA-Tyr (gta) | Transfer RNA for Tyrosine | 1524 | 1611 | 88 | − | 69 | GTA | ||
| cox1 | Cytochrome c oxidase subunit I | 1681 | 3240 | 1560 | + | 6 | ATT | TAG | |
| tRNA-Leu (taa) | Transfer RNA for Leucine | 3247 | 3312 | 66 | + | 0 | TAA | ||
| cox2 | Cytochrome c oxidase subunit II | 3313 | 3999 | 687 | + | −1 | ATT | TAG | |
| tRNA-Asp (gtc) | Transfer RNA for Aspartic acid | 3999 | 4056 | 58 | + | 2 | GTC | ||
| atp8 | ATP synthase F0 subunit 8 | 4059 | 4226 | 168 | + | 17 | ATT | TAA | |
| atp6 | ATP synthase F0 subunit 6 | 4244 | 4909 | 666 | + | 4 | TTG | TAG | |
| cox3 | Cytochrome c oxidase subunit III | 4914 | 5693 | 780 | + | 8 | ATG | TAG | |
| tRNA-Glu(ttc) | Transfer RNA for Glutamic acid | 5702 | 5769 | 68 | + | 0 | TTC | ||
| nad3 | NADH dehydrogenase subunit 3 | 5770 | 6123 | 354 | + | 0 | ATA | TAA | |
| tRNA-Arg(tcg) | Transfer RNA for Arginine | 6124 | 6185 | 62 | − | 87 | TCG | ||
| tRNA-Gln(ttg) | Transfer RNA for Glutamine | 6273 | 6340 | 68 | + | 1 | TTG | ||
| s-rRNA | 12S ribosomal RNA | 6342 | 7102 | 761 | + | −2 | |||
| tRNA-Val(tac) | Transfer RNA for Valine | 7101 | 7165 | 65 | + | 53 | TAC | ||
| l-rRNA | 16S ribosomal RNA | 7219 | 8513 | 1295 | + | −2 | |||
| tRNA-Leu(tag) | Transfer RNA for Leucine | 8512 | 8579 | 68 | + | 55 | TAG | ||
| tRNA-Ser(tct) | Transfer RNA for Serine | 8635 | 8691 | 57 | + | 52 | TCT | ||
| tRNA-Pro(tgg) | Transfer RNA for Proline | 8744 | 8807 | 64 | + | 100 | TGG | ||
| nad4l | NADH dehydrogenase subunit 4L | 8908 | 9180 | 273 | + | 2 | ATT | TAA | |
| nad4 | NADH dehydrogenase subunit 4 | 9183 | 10485 | 1303 | + | 0 | ATG | T (* AA) | |
| tRNA-His(gtg) | Transfer RNA for Histidine | 10486 | 10551 | 66 | + | 0 | GTG | ||
| nad5 | NADH dehydrogenase subunit 5 | 10552 | 12204 | 1653 | + | 16 | ATT | TAA | |
| tRNA-Phe(gaa) | Transfer RNA for Phenylalanine | 12221 | 12286 | 66 | + | 20 | GAA | ||
| tRNA-Asn(gtt) | Transfer RNA for Asparagine | 12307 | 12375 | 69 | − | 30 | GTT | ||
| tRNA-Thr(tgt) | Transfer RNA for Threonine | 12406 | 12471 | 66 | − | 87 | TGT | ||
| nad6 | NADH dehydrogenase subunit 6 | 12559 | 13077 | 519 | + | −1 | ATA | TAA | |
| cytb | Cytochrome b | 13077 | 14225 | 1149 | + | −1 | ATG | TAA | |
| tRNA-Ser(tga) | Transfer RNA for Serine | 14225 | 14291 | 67 | + | 0 | TGA | ||
| nad1 | NADH dehydrogenase subunit 1 | 14292 | 15218 | 927 | − | 3 | ATT | TAA | |
| tRNA-Gly (tcc) | Transfer RNA for Glycine | 15222 | 15289 | 68 | − | 29 | TCC | ||
| Gene | Length | T% | C% | A% | G% | AT% | GC% | GC Skew | AT Skew |
|---|---|---|---|---|---|---|---|---|---|
| nad2 | 996 | 40.7 | 15.9 | 31.2 | 12.2 | 71.9 | 28.1 | −0.131 | −0.132 |
| cox1 | 1575 | 39.34 | 18.16 | 27.92 | 14.58 | 67.26 | 32.74 | −0.109 | −0.169 |
| cox2 | 676 | 37.66 | 17.7 | 30.94 | 13.7 | 68.6 | 31.4 | −0.127 | −0.098 |
| atp8 | 168 | 39.78 | 15.86 | 36.72 | 7.64 | 76.5 | 23.5 | −0.349 | −0.040 |
| atp6 | 666 | 39.76 | 21.12 | 25.76 | 13.36 | 65.52 | 34.48 | −0.225 | −0.21368 |
| cox3 | 780 | 36.68 | 19.76 | 30.38 | 13.18 | 67.06 | 32.94 | −0.199 | −0.093 |
| nad3 | 354 | 39.4 | 21.2 | 25.82 | 13.58 | 65.22 | 34.78 | −0.219 | −0.208 |
| nad4l | 300 | 48.4 | 5.7 | 36.04 | 9.86 | 84.44 | 15.56 | 0.267 | −0.146 |
| nad4 | 1303 | 46.16 | 8.16 | 36.38 | 9.3 | 82.54 | 17.46 | 0.065 | −0.118 |
| nad5 | 1653 | 44.56 | 7.64 | 39.86 | 7.94 | 84.42 | 15.58 | 0.019 | −0.056 |
| nad6 | 501 | 46.5 | 8.6 | 38.74 | 6.16 | 85.24 | 14.76 | −0.165 | −0.091 |
| cytb | 1149 | 45 | 12.82 | 32.62 | 9.56 | 77.62 | 22.38 | −0.146 | −0.159 |
| nad1 | 924 | 45.68 | 8.44 | 35.86 | 10.02 | 81.54 | 18.46 | 0.086 | −0.120 |
| rrnS | 761 | 35.26 | 8.98 | 40.56 | 15.2 | 75.82 | 24.18 | 0.257 | 0.070 |
| rrnL | 1316 | 38.2 | 9.58 | 39.32 | 12.9 | 77.52 | 22.48 | 0.148 | 0.014 |
| Overall Mitogenome | 15318 | 41.42 | 12.22 | 35.06 | 11.30 | 75.413 | 24.587 | −0.039 | −0.083 |
| Aa | Codon | N | RSCU | Aa | Codon | N | RSCU | Aa | Codon | N | RSCU | Aa | Codon | N | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU(F) | 9.7 | 1.59 | Ser | UCU(S) | 2.9 | 1.92 | Tyr | UAU(Y) | 5.6 | 1.47 | Cys | UGU(C) | 0.9 | 1.48 |
| UUC(F) | 2.5 | 0.41 | UCC(S) | 0.7 | 0.45 | UAC(Y) | 2.0 | 0.53 | UGC(C) | 0.3 | 0.52 | ||||
| Leu | UUA(L) | 6.8 | 2.60 | UCA(S) | 3.6 | 2.32 | End | UAA (*) | 2.1 | 1.55 | End | UGA (*) | 2.1 | 1.56 | |
| UUG(L) | 1.9 | 0.74 | UCG(S) | 0.4 | 0.23 | UAG (*) | 0.6 | 0.45 | Trp | UGG(W) | 0.6 | 0.44 | |||
| CUU(L) | 3.2 | 1.22 | Pro | CCU(P) | 1.5 | 1.88 | His | CAU(H) | 1.2 | 1.25 | Arg | CGU(R) | 0.3 | 0.89 | |
| CUC(L) | 0.7 | 0.26 | CCC(P) | 0.3 | 0.43 | CAC(H) | 0.7 | 0.75 | CGC(R) | 0.1 | 0.37 | ||||
| CUA(L) | 2.2 | 0.84 | CCA(P) | 1.1 | 1.45 | Gln | CAA(Q) | 0.8 | 1.28 | CGA(R) | 0.8 | 2.44 | |||
| CUG(L) | 0.9 | 0.34 | CCG(P) | 0.2 | 0.23 | CAG(Q) | 0.5 | 0.72 | CGG(R) | 0.1 | 0.30 | ||||
| Ile | AUU(I) | 11.4 | 1.66 | Thr | ACU(T) | 2.3 | 2.11 | Asn | AAU(N) | 6.1 | 1.46 | Ser | AGU(S) | 1.3 | 0.85 |
| AUC(I) | 2.3 | 0.34 | ACC(T) | 0.4 | 0.38 | AAC(N) | 2.3 | 0.54 | AGC(S) | 0.4 | 0.25 | ||||
| Met | AUA(M) | 10.2 | 1.65 | ACA(T) | 1.5 | 1.41 | Lys | AAA(K) | 5.3 | 1.50 | Arg | AGA(R) | 2.3 | 1.48 | |
| AUG(M) | 2.2 | 0.35 | ACG(T) | 0.1 | 0.1 | AAG(K) | 1.8 | 0.50 | AGG(R) | 0.8 | 0.50 | ||||
| Val | GUU(V) | 2.5 | 1.59 | Ala | GCU(A) | 0.8 | 1.65 | Asp | GAU(D) | 1.7 | 1.54 | Gly | GGU(G) | 0.6 | 0.68 |
| GUC(V) | 0.6 | 0.39 | GCC(A) | 0.3 | 0.53 | GAC(D) | 0.5 | 0.46 | GGC(G) | 0.2 | 0.23 | ||||
| GUA(V) | 2.6 | 1.64 | GCA(A) | 0.8 | 1.65 | Glu | GAA(E) | 2.1 | 1.52 | GGA(G) | 2.4 | 2.65 | |||
| GUG(V) | 0.6 | 0.38 | GCG(A) | 0.1 | 0.16 | GAG(E) | 0.7 | 0.48 | GGG(G) | 0.4 | 0.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duangphakdee, O.; Poolprasert, P.; Rattanawannee, A. Complete Mitochondrial Genome Characterization and Phylogenomics of the Stingless Bee, Heterotrigona itama (Apidae: Meliponini). Insects 2025, 16, 535. https://doi.org/10.3390/insects16050535
Duangphakdee O, Poolprasert P, Rattanawannee A. Complete Mitochondrial Genome Characterization and Phylogenomics of the Stingless Bee, Heterotrigona itama (Apidae: Meliponini). Insects. 2025; 16(5):535. https://doi.org/10.3390/insects16050535
Chicago/Turabian StyleDuangphakdee, Orawan, Pisit Poolprasert, and Atsalek Rattanawannee. 2025. "Complete Mitochondrial Genome Characterization and Phylogenomics of the Stingless Bee, Heterotrigona itama (Apidae: Meliponini)" Insects 16, no. 5: 535. https://doi.org/10.3390/insects16050535
APA StyleDuangphakdee, O., Poolprasert, P., & Rattanawannee, A. (2025). Complete Mitochondrial Genome Characterization and Phylogenomics of the Stingless Bee, Heterotrigona itama (Apidae: Meliponini). Insects, 16(5), 535. https://doi.org/10.3390/insects16050535







