Embryonic Development of Parthenogenetic and Sexual Eggs in Lower Termites
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of Female–Female Colonies and Female–Male Colonies in the Laboratory
2.2. Ovary Photography
2.3. Micropyle Measurement
2.4. Egg Nucleus Staining
2.5. Paraffin Section of Termite Eggs
2.6. Collection and Processing of Single-Cell Transcriptome Sequencing Materials
2.7. Single-Cell Transcriptome Sequencing Process
2.8. Data Analysis
2.9. RNA Isolation, cDNA Library Construction, and Quantitative Real-Time PCR
3. Results and Analysis
3.1. Ovary Measurements
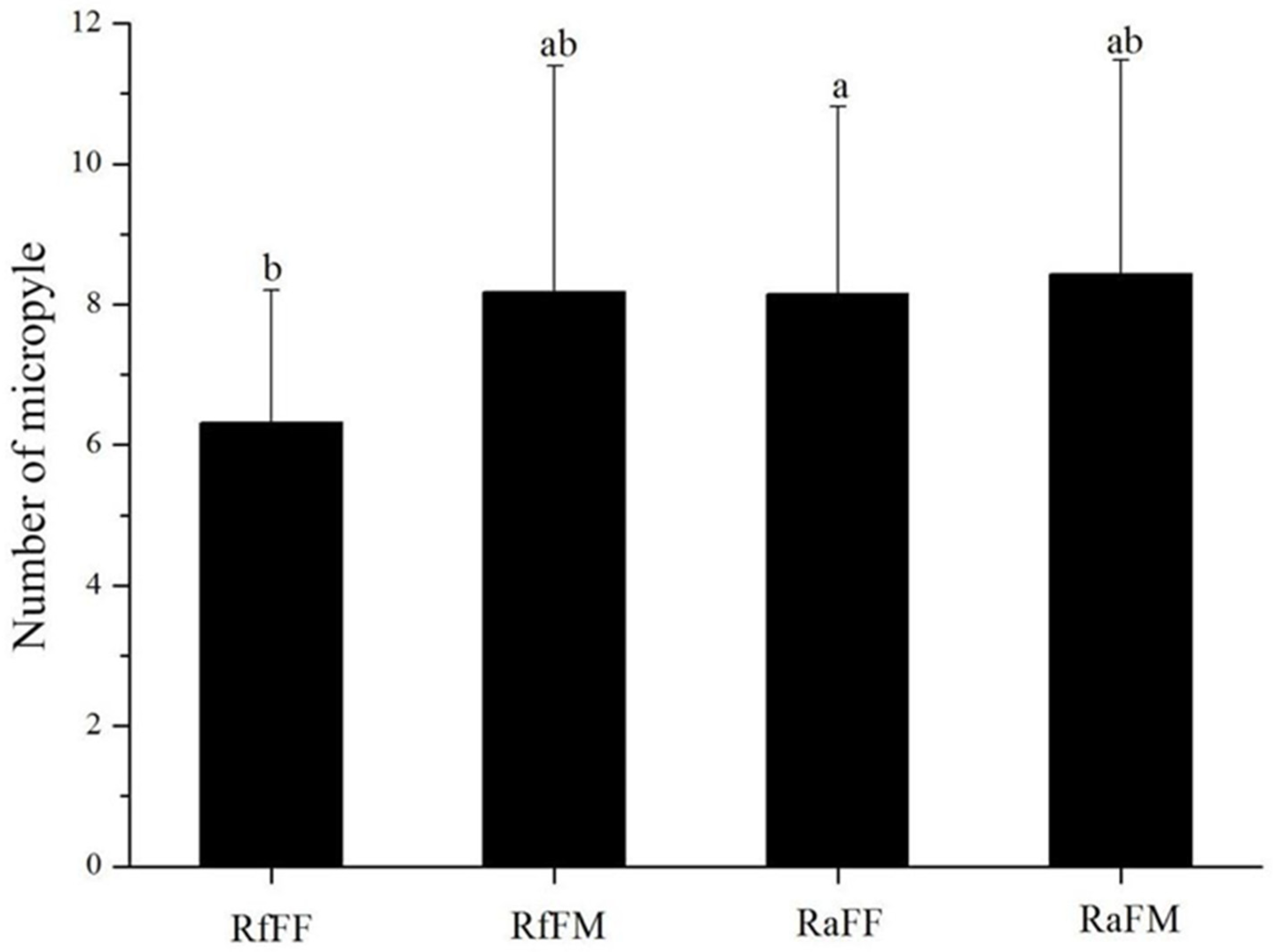
3.2. Micropyles
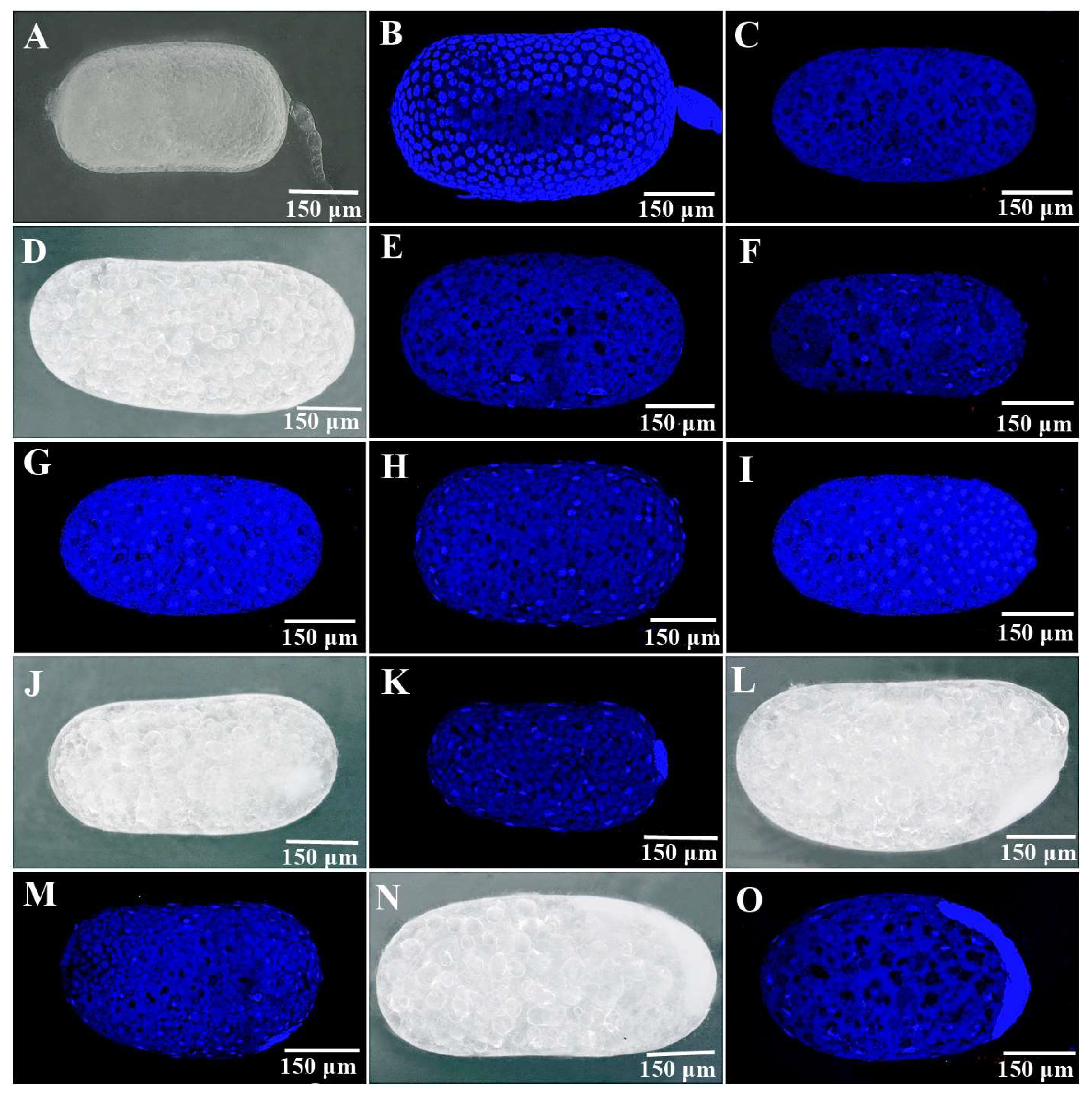
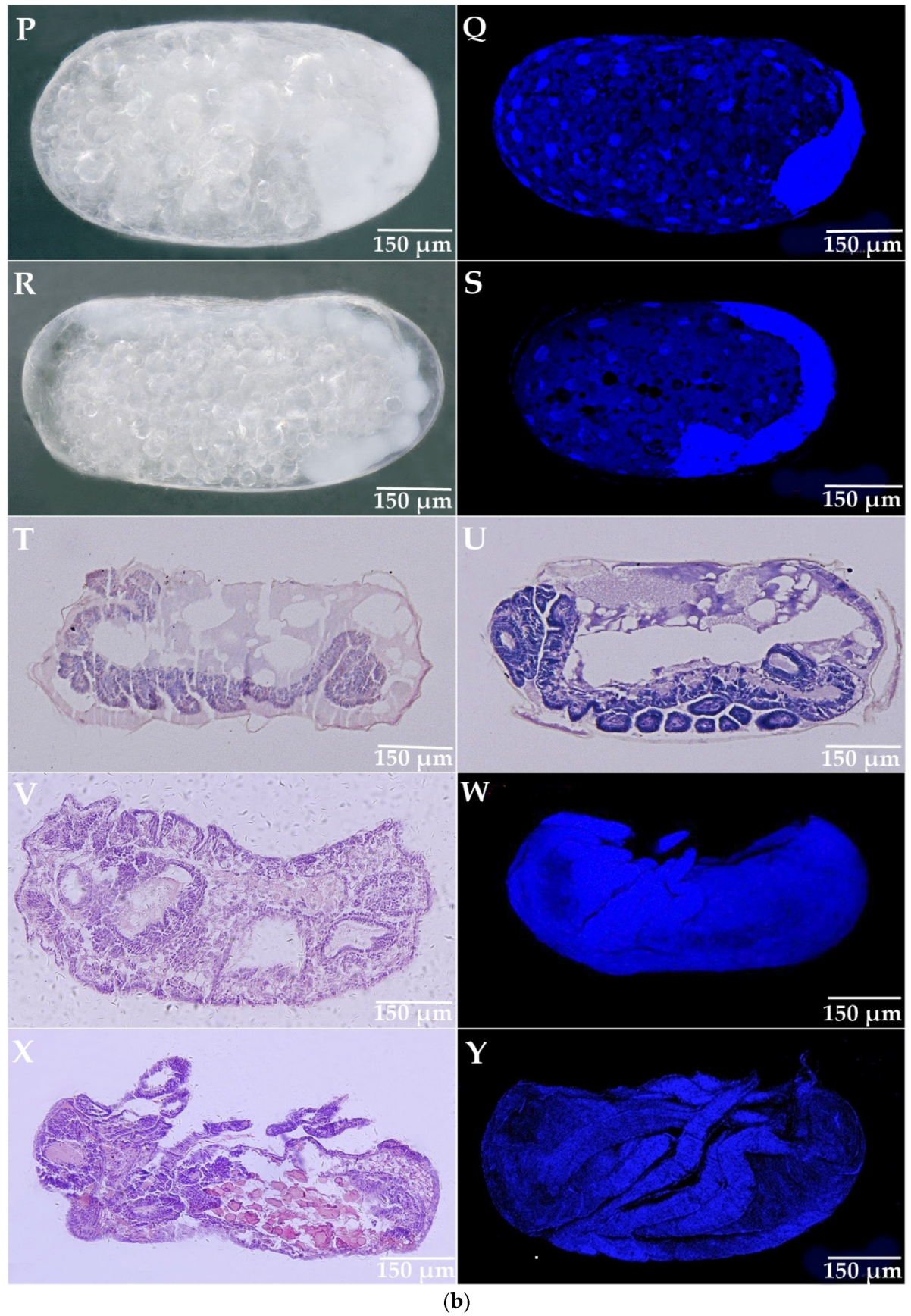
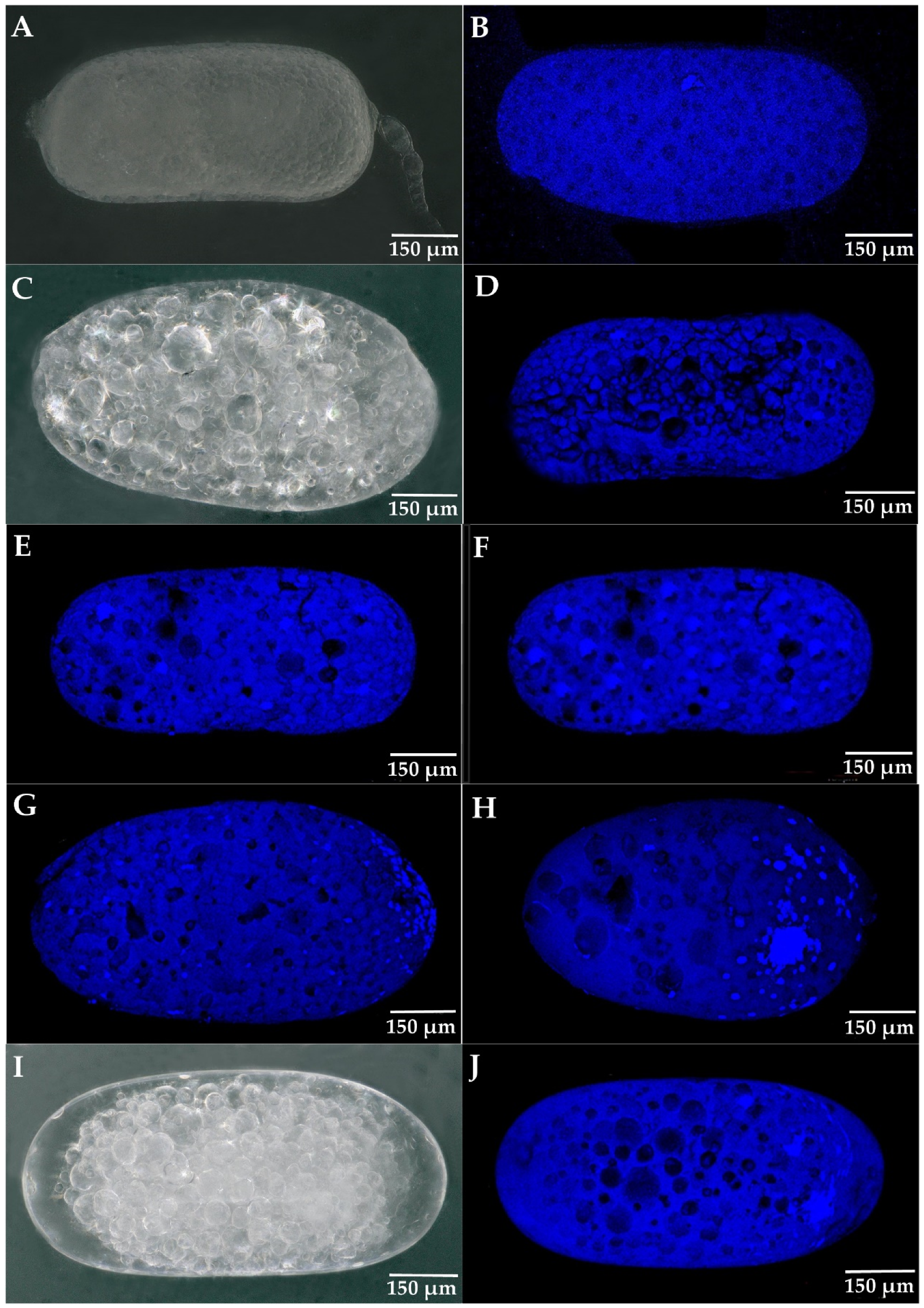
3.3. The Development of Parthenogenetic Embryos in R. aculabialis
3.4. The Development of Parthenogenetic Embryos in R. flaviceps
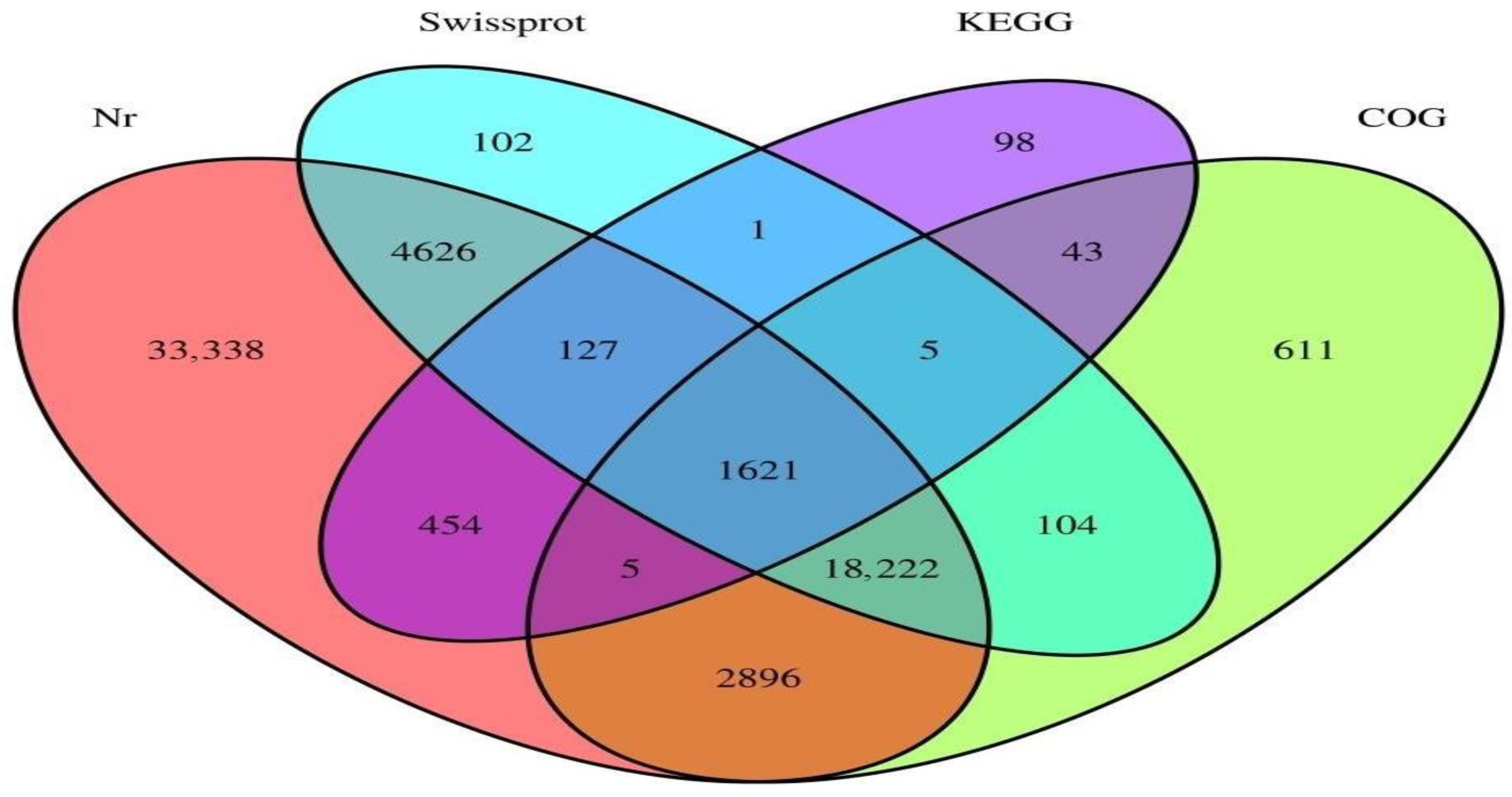
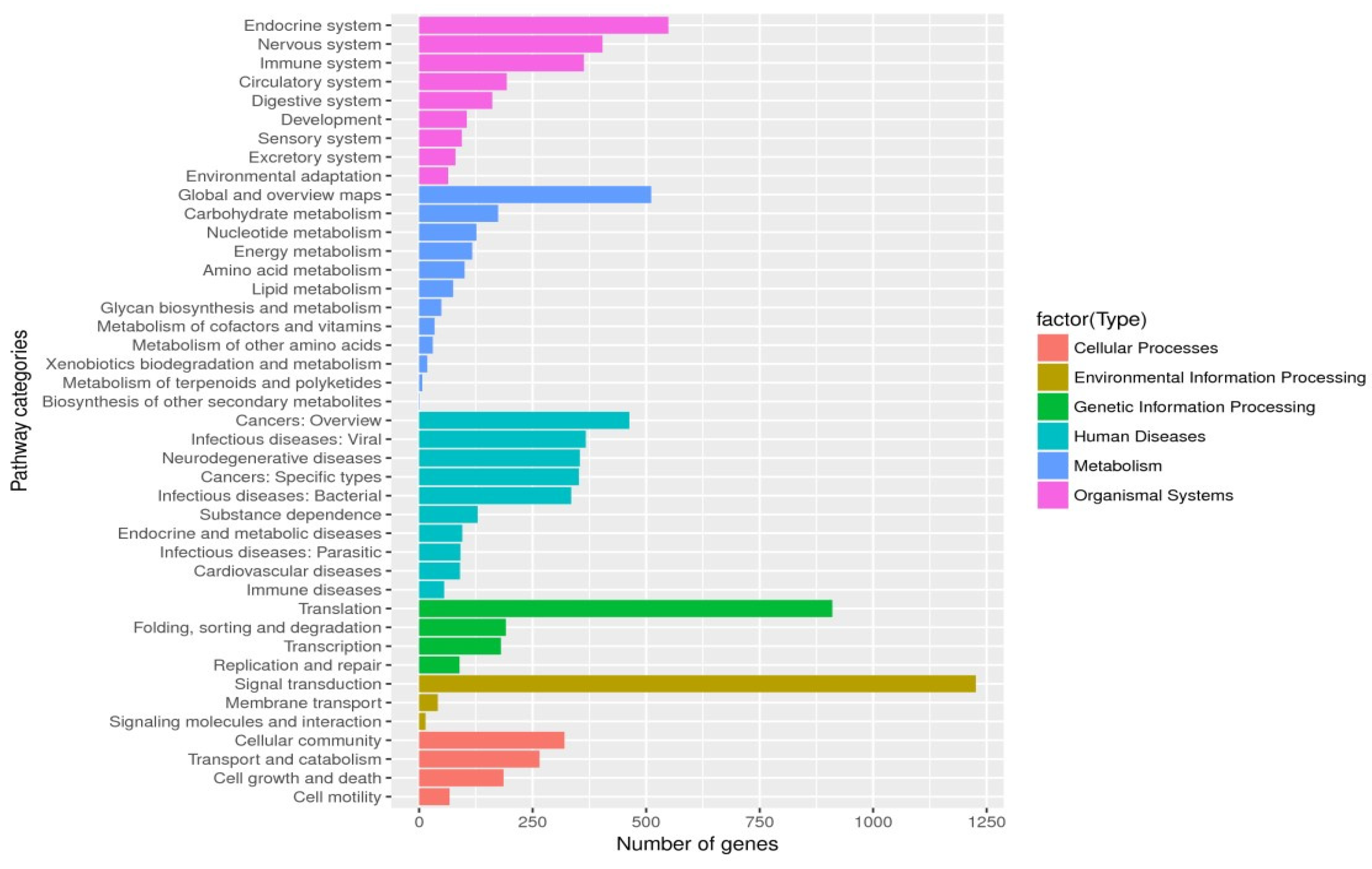
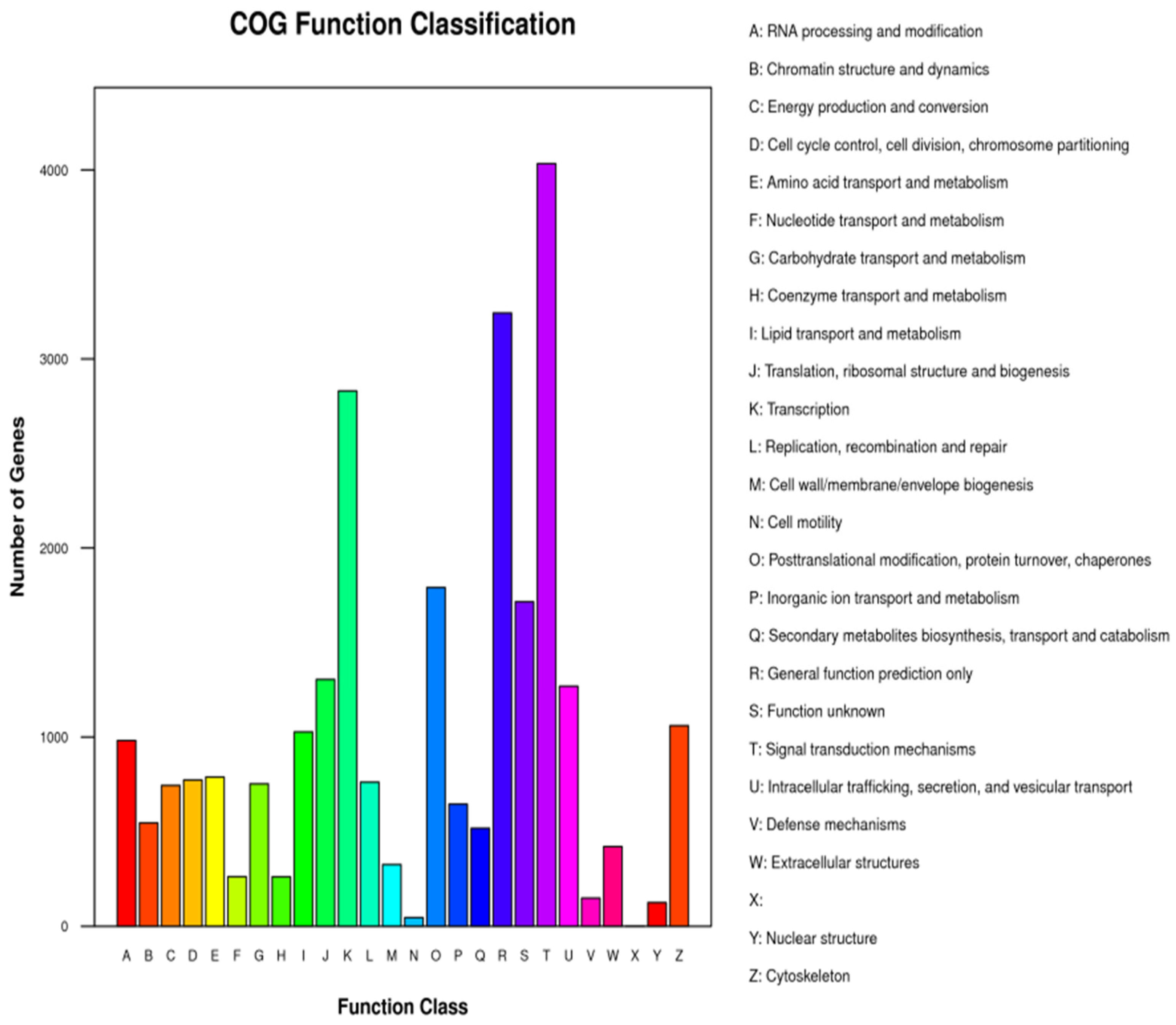
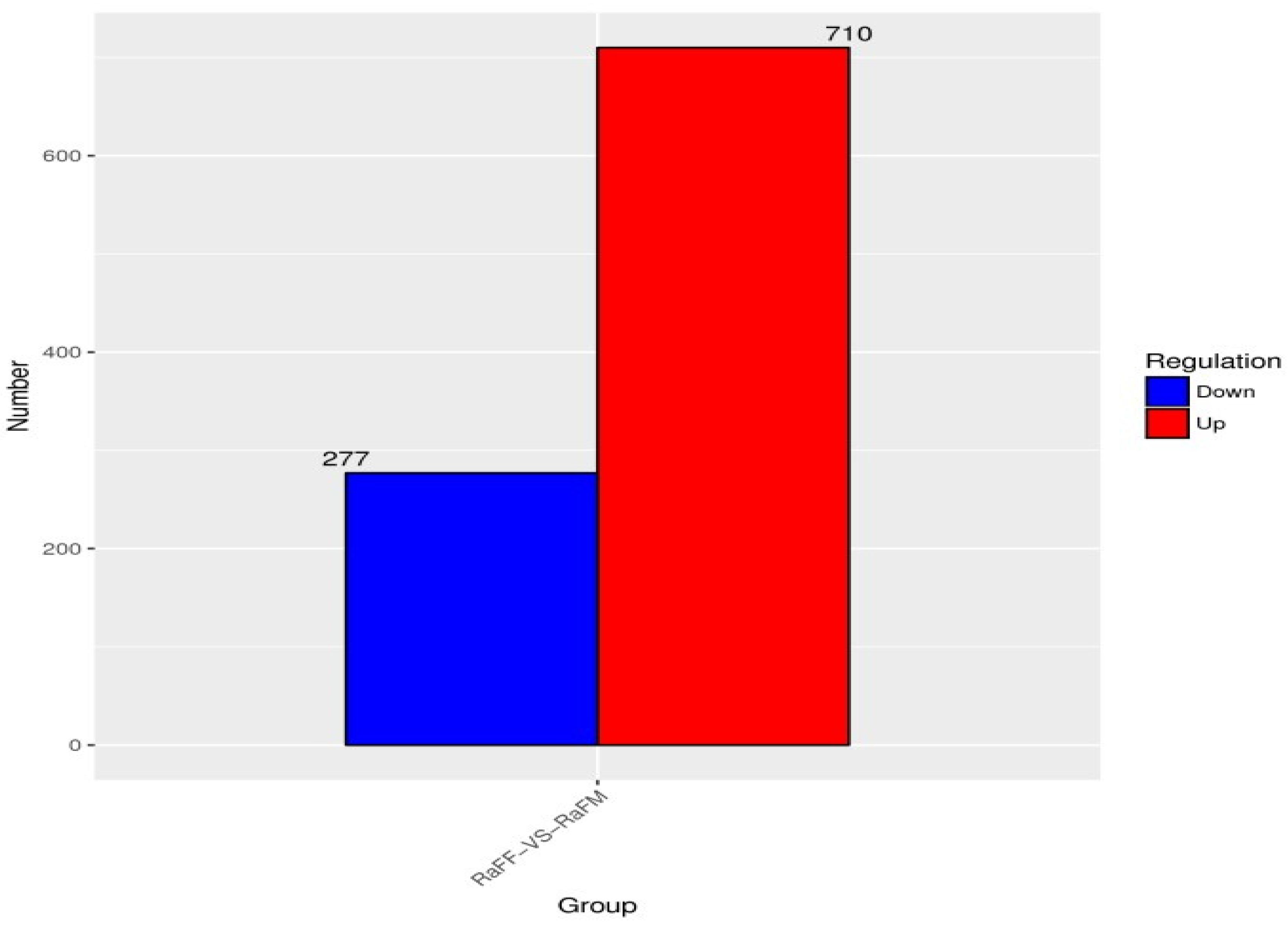
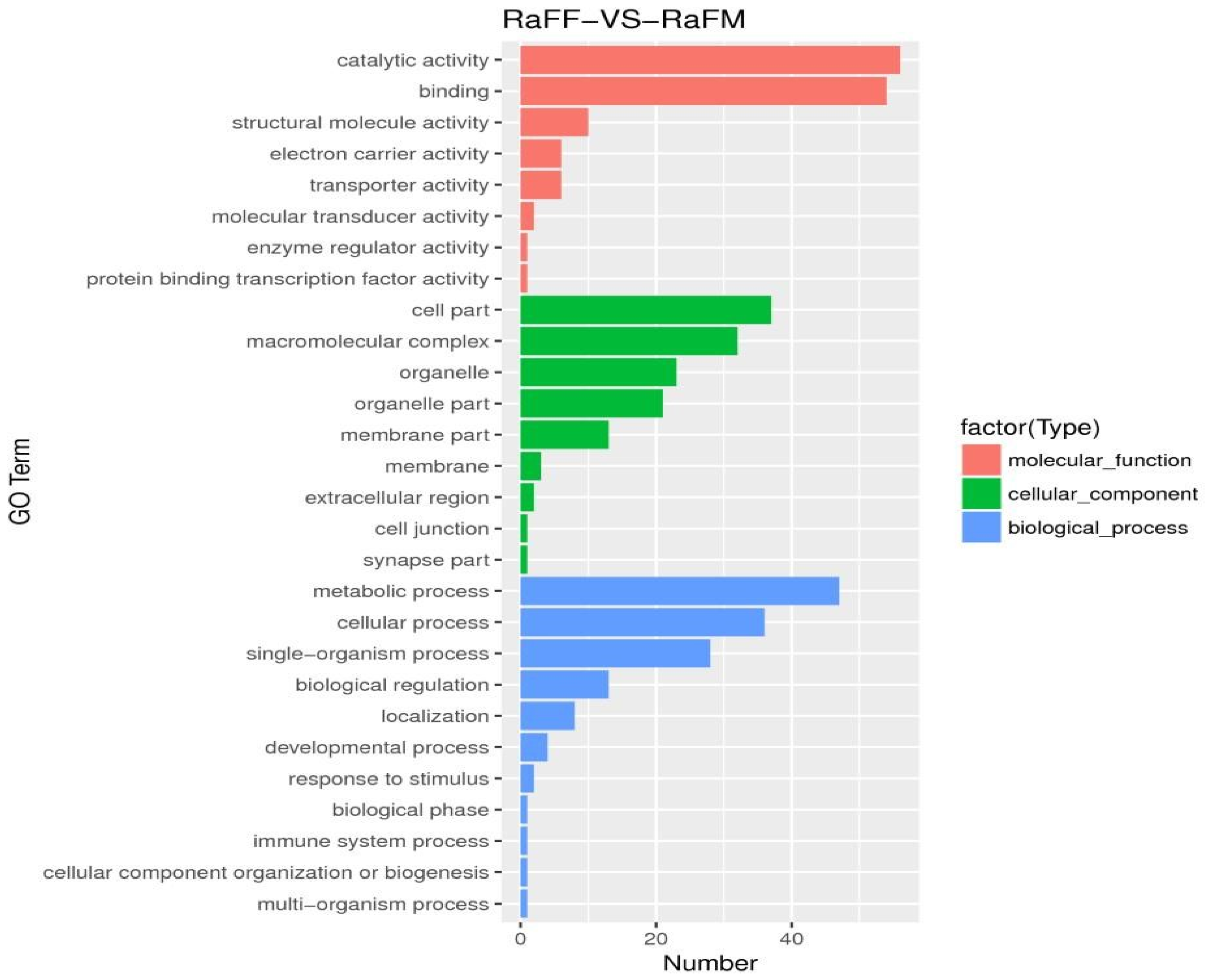
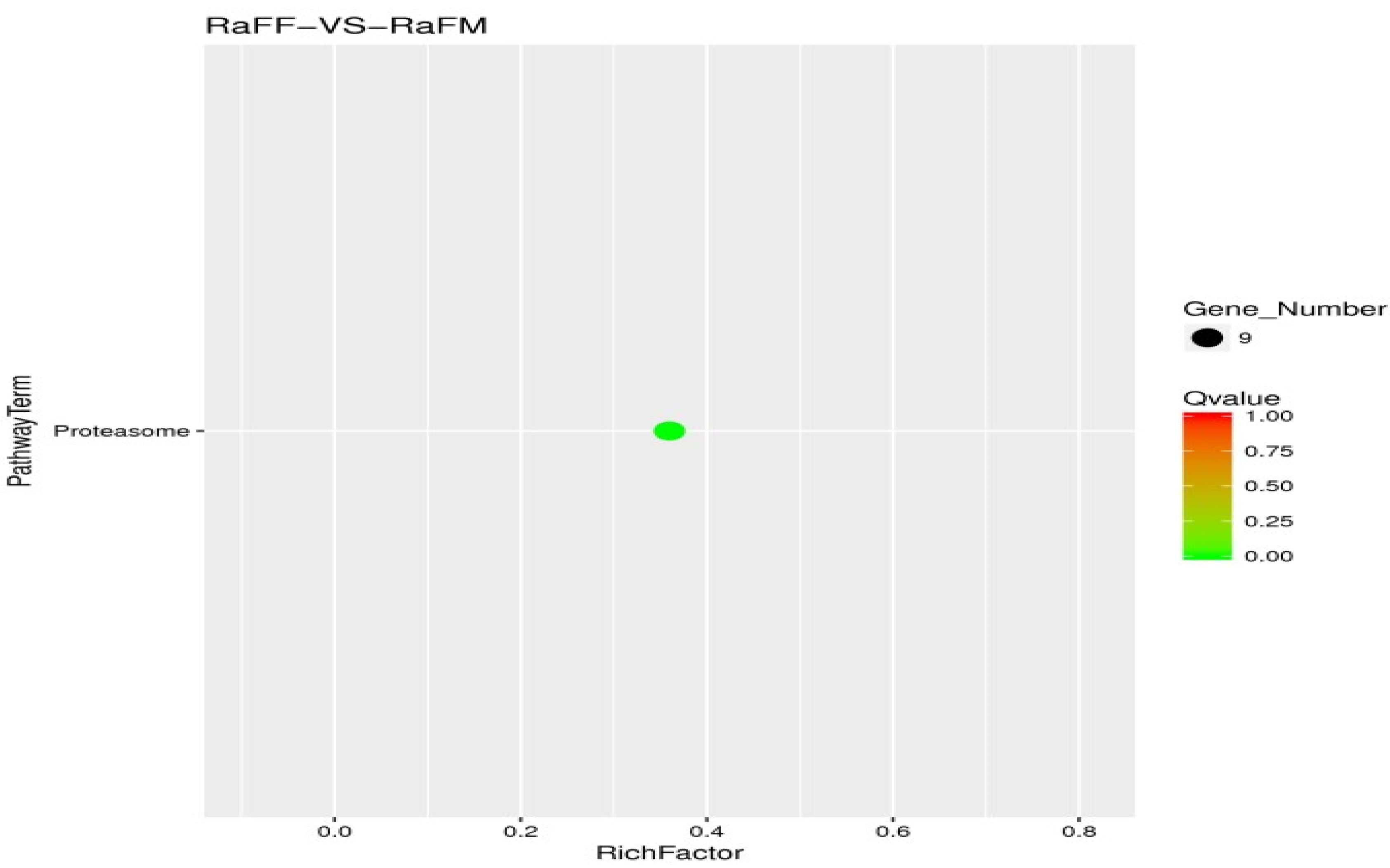
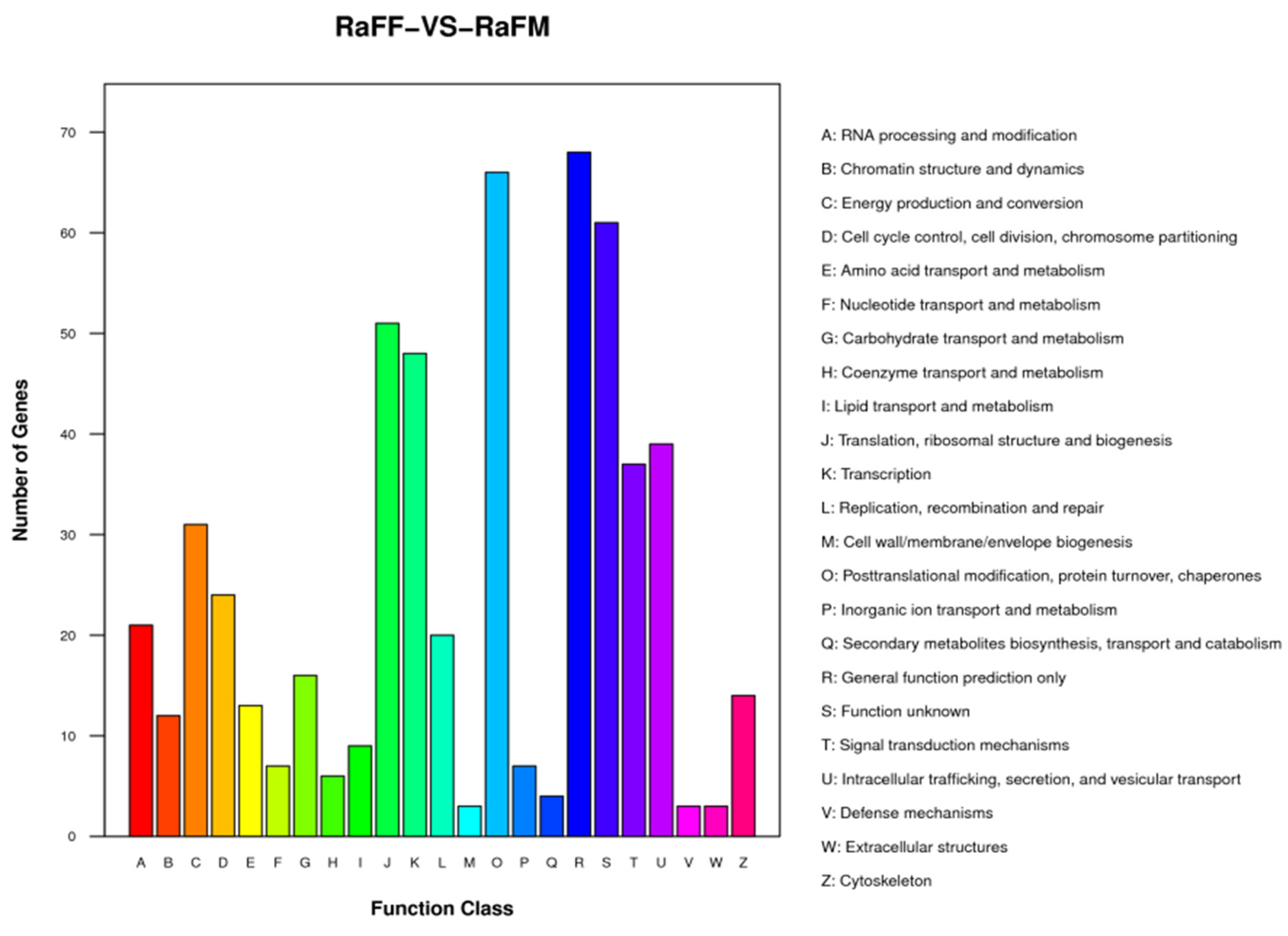
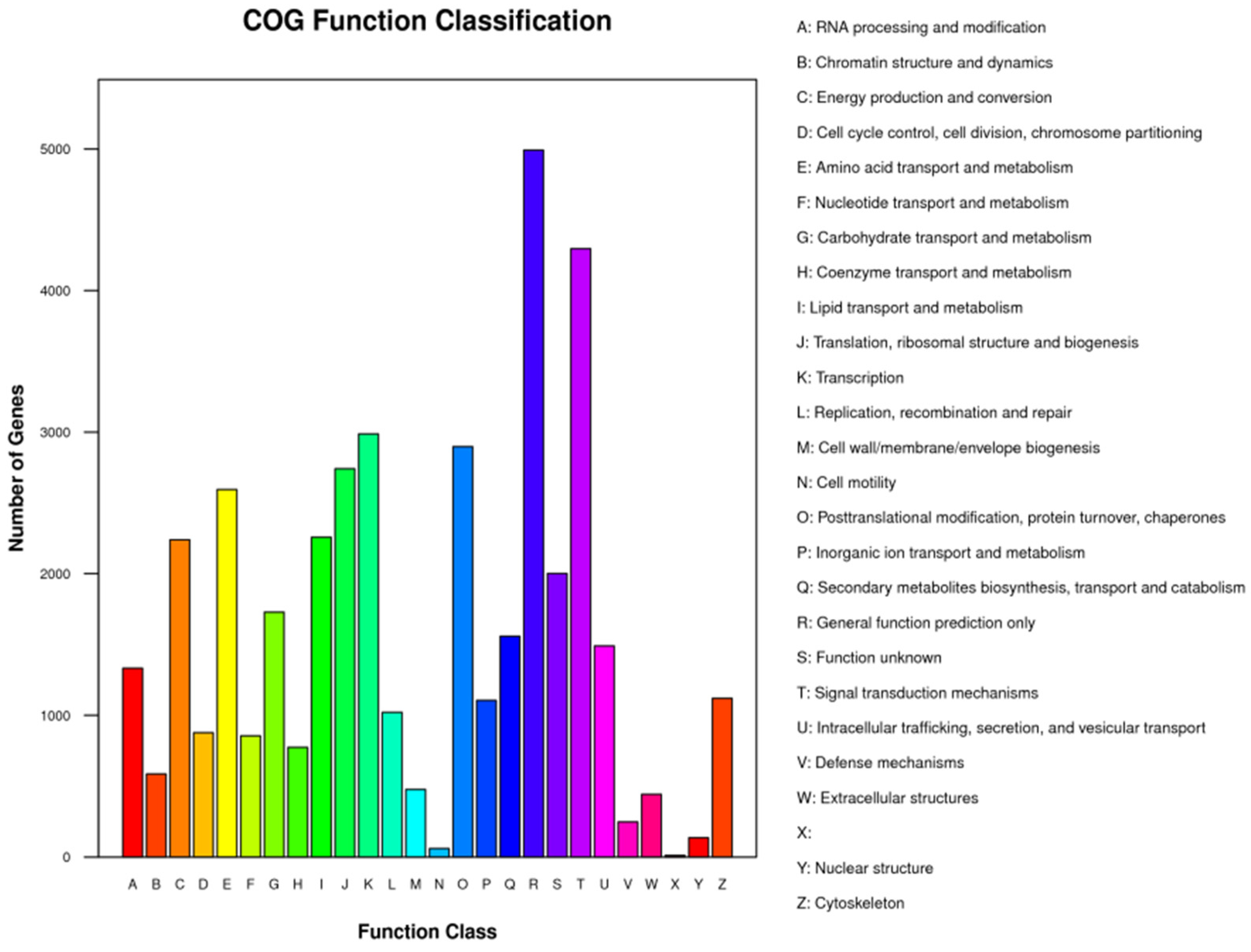
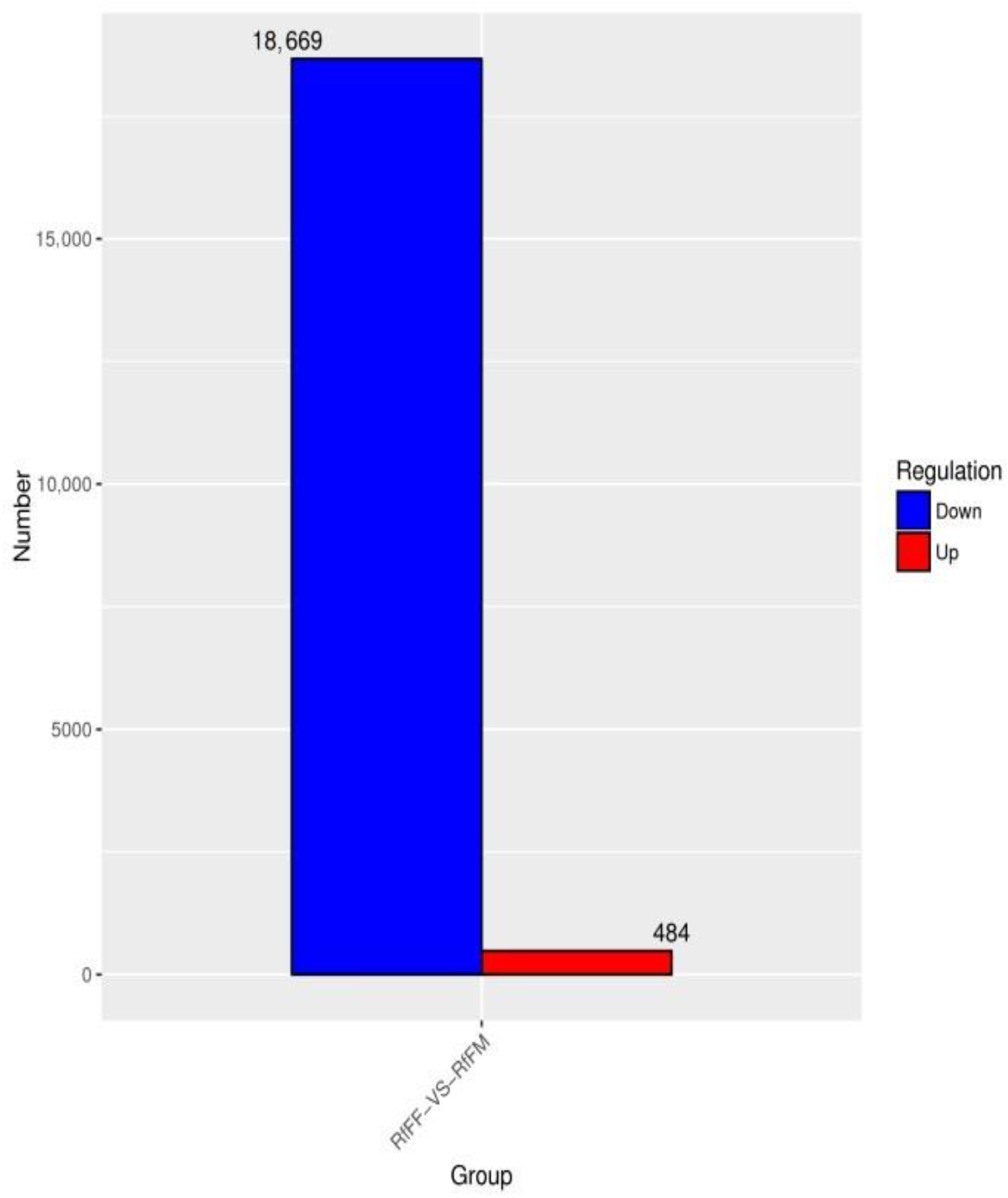
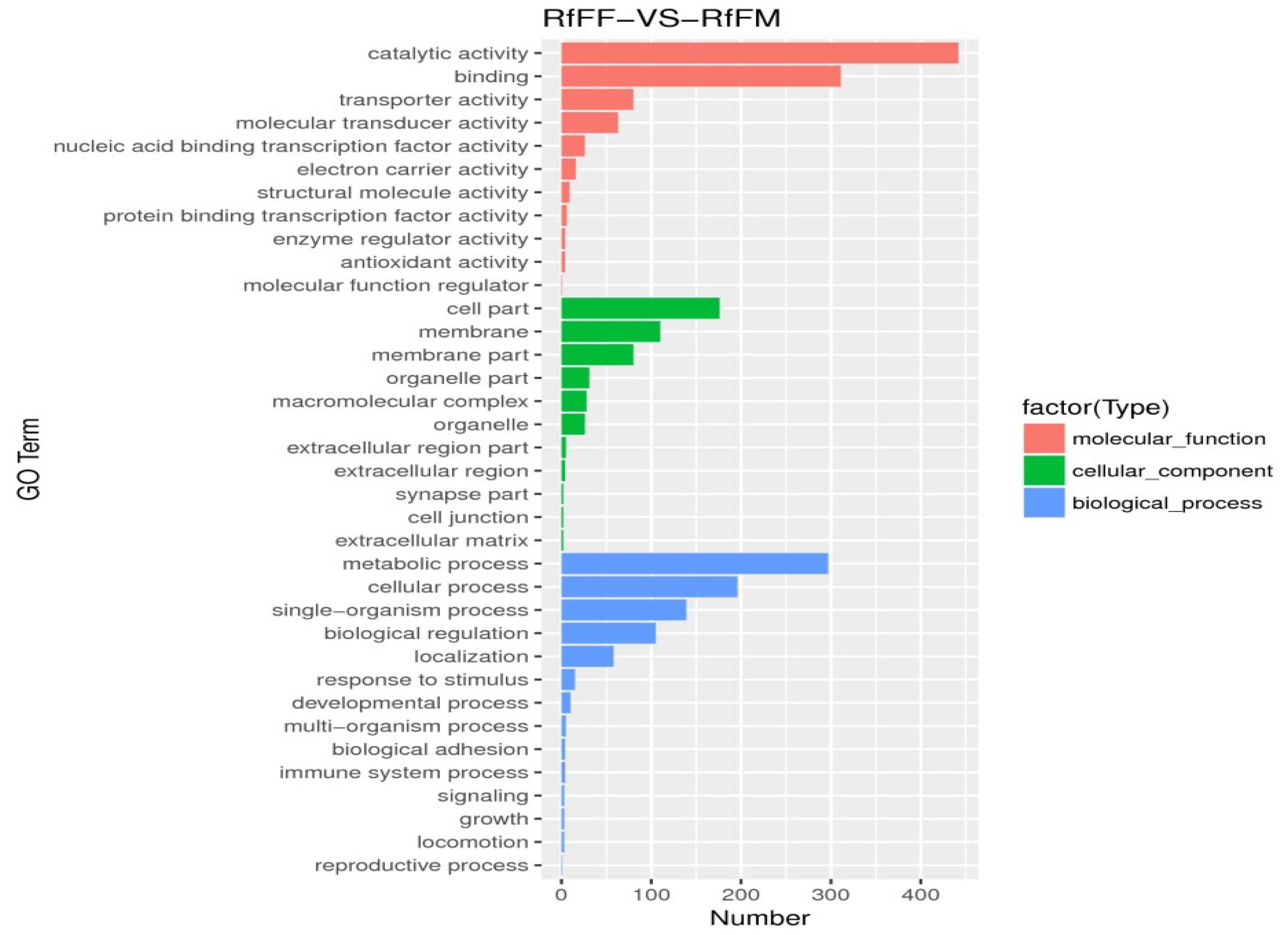
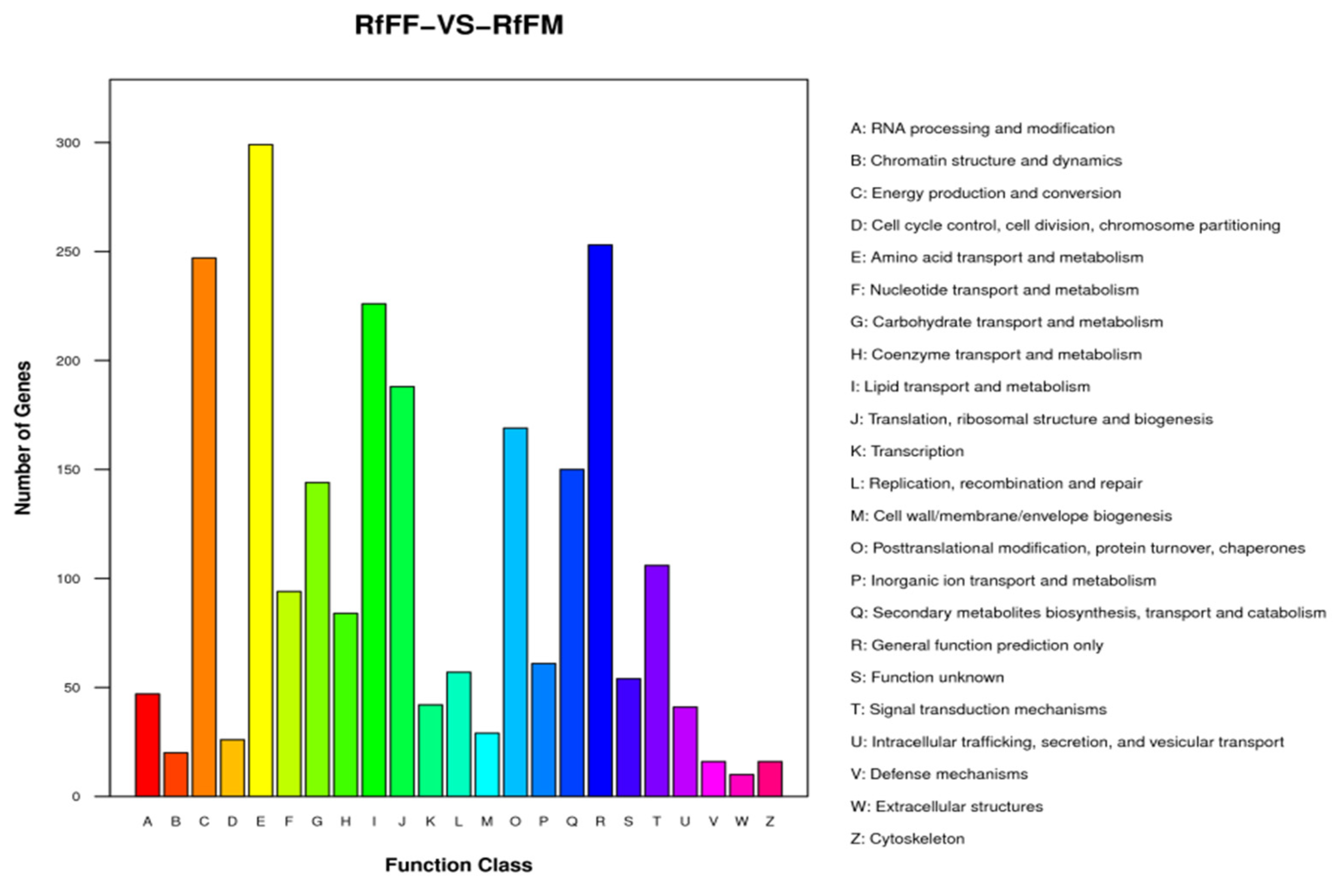
3.5. Single-Cell Transcriptome Sequencing of Parthenogenetic and Sexual Eggs of R. aculabialis
3.6. Single-Cell Transcriptome Sequencing of Parthenogenetic and Sexual Eggs of R. flaviceps
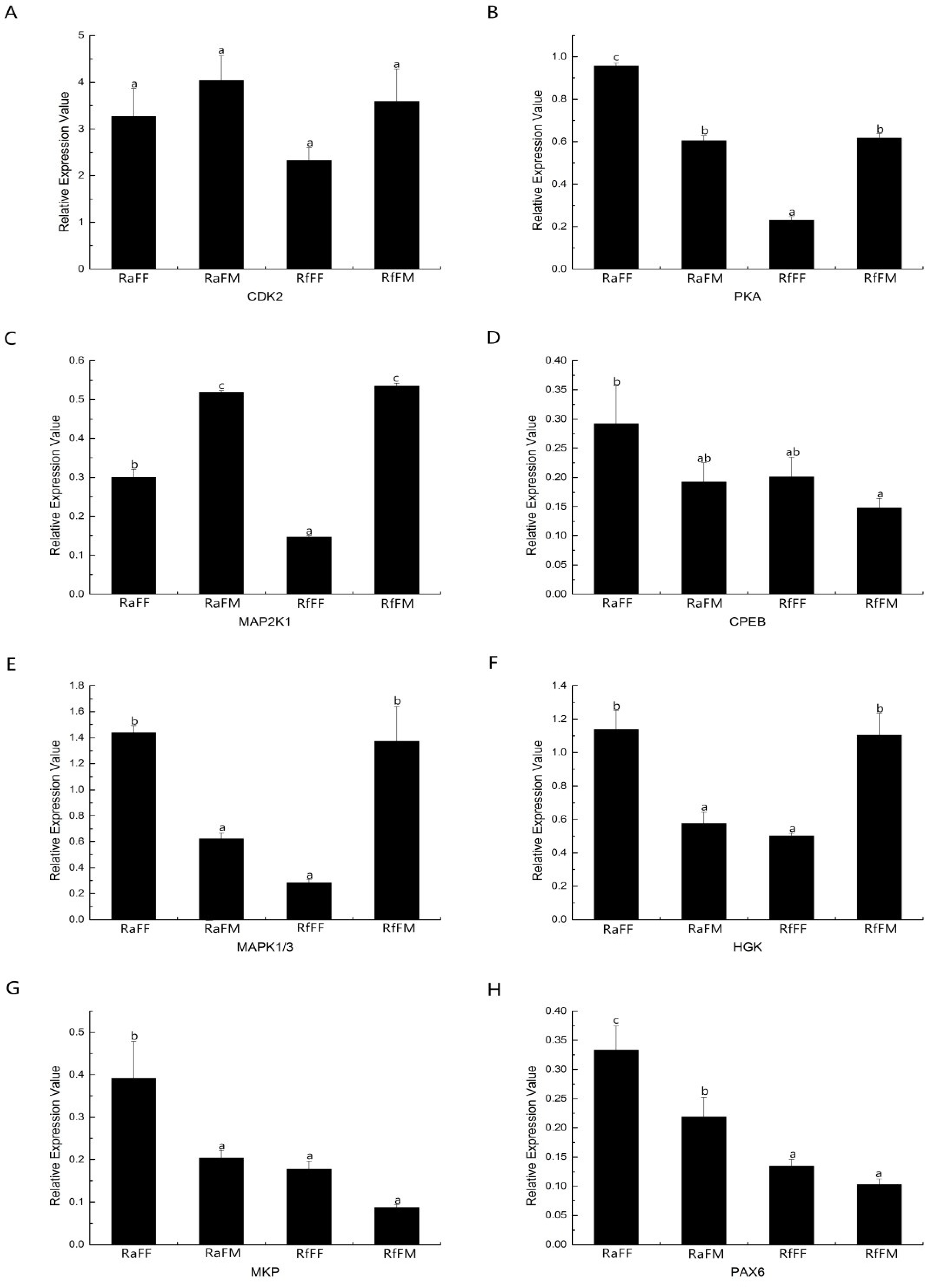
3.7. QRT-PCR Verification of the Gene Expression Analysis of Embryonic Development
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, Z.; Zhang, M.; Meng, Y.-F.; Zhao, J.; Kong, X.; Su, X.-H.; Xing, L.-X. Alates of the termite Reticulitermes flaviceps feed independently during their 5-month residency in the natal colony. Insectes Soc. 2019, 66, 425–433. [Google Scholar] [CrossRef]
- Khan, Z.; Li, Y.-X.; Liu, Q.; Su, X.-H.; Xing, L.-X. The first description of alate and supplementary description of soldier of Reticulitermes aculabialis Tsai et Hwang (Isoptera, Rhinotermitidae). Int. J. Trop. Insect Sci. 2021, 41, 2643–2648. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, M.S.; Suleman; Muhammad, N.; Haroon; Su, X.-H.; Xing, L.-X. Morphology of testis, sperm, and spermatheca in two capable hybridized termite species indicates no interspecific reproductive isolation. Int. J. Trop. Insect Sci. 2022, 42, 2909–2926. [Google Scholar] [CrossRef]
- Matsuura, K.; Nishida, T. Comparison of colony foundation success between sexual pairs and female asexual units in the termite Reticulitermes speratus (Isoptera: Rhinotermitidae). Popul. Ecol. 2001, 43, 119–124. [Google Scholar] [CrossRef]
- Xing, L.-X.; Liu, M.; Kong, X.; Liu, X.; Su, X.-H.; Yin, L.; Tan, J. Parthenogenetic reproductive behavior and initial colony foundation in the termite, Reticulitermes aculabialis. Chin. J. Appl. Entomol. 2013, 50, 1671–1678. [Google Scholar]
- Büning, J. The Insect Ovary: Ultrastructure, Previtellogenic Growth and Evolution; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1994; pp. 1–400. [Google Scholar]
- Heming, B.S. Insect Development and Evolution; Cornell University Press: Ithaca, NY, USA, 2018; p. 464. [Google Scholar]
- Page, A.W.; Orr-Weaver, T.L. Stopping and starting the meiotic cell cycle. Curr. Opin. Genet. Dev. 1997, 7, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, Y.; Yu, J.; Wolfner, M.F. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 2001, 234, 416–424. [Google Scholar] [CrossRef]
- Edgar, B.A.; Sprenger, F.; Duronio, R.J.; Leopold, P.; O’Farrell, P.H. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994, 8, 440–452. [Google Scholar] [CrossRef]
- Hu, X.P.; Xu, Y. Morphological embryonic development of the eastern subterranean termite, Reticulitermes flavipes (Isoptera: Rhinotermitidae). Sociobiology 2005, 45, 573–586. [Google Scholar]
- Matsuura, K.; Kobayashi, N. Size, hatching rate, and hatching period of sexually and asexually produced eggs in the facultatively parthenogenetic termite Reticulitermes speratus (Isoptera: Rhinotermitidae). Appl. Entomol. Zool. 2007, 42, 241–246. [Google Scholar] [CrossRef]
- Kishimoto, T. Cell-cycle control during meiotic maturation. Curr. Opin. Cell Biol. 2003, 15, 654–663. [Google Scholar] [CrossRef]
- Sisinni, L.; Maddalena, F.; Condelli, V.; Pannone, G.; Simeon, V.; Li Bergolis, V.; Lopes, E.; Piscazzi, A.; Matassa, D.S.; Mazzoccoli, C. TRAP1 controls cell cycle G2–M transition through the regulation of CDK1 and MAD2 expression/ubiquitination. J. Pathol. 2017, 243, 123–134. [Google Scholar] [CrossRef]
- Ray, L.B.; Sturgill, T.W. Insulin-stimulated microtubule-associated protein kinase is phosphorylated on tyrosine and threonine in vivo. Proc. Natl. Acad. Sci. USA 1988, 85, 3753–3757. [Google Scholar] [CrossRef]
- Gehring, W.J. The genetic control of eye development and its implications for the evolution of the various eye-types. Int. J. Dev. Biol. 2003, 46, 65–73. [Google Scholar] [CrossRef]
- Pichaud, F.; Desplan, C. Pax genes and eye organogenesis. Curr. Opin. Genet. Dev. 2002, 12, 430–434. [Google Scholar] [CrossRef]
- Masami, S.; Chisako, N.; Hiroyuki, O.; Hitoshi, I. Effect of an insulin sensitizer, pioglitazone, on hypertension in fructose-drinking rats. Jpn. J. Pharmacol. 1997, 74, 297–302. [Google Scholar] [CrossRef]
- Gustincich, S.; Sandelin, A.; Plessy, C.; Katayama, S.; Simone, R.; Lazarevic, D.; Hayashizaki, Y.; Carninci, P. The complexity of the mammalian transcriptome. J. Physiol. 2006, 575, 321–332. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef]
- Jääger, K.; Islam, S.; Zajac, P.; Linnarsson, S.; Neuman, T. RNA-seq analysis reveals different dynamics of differentiation of human dermis-and adipose-derived stromal stem cells. PLoS ONE 2012, 7, e38833. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Yu, W.; Zhao, S.; Gong, Y. Identification of genes related to white and black plumage formation by RNA-Seq from white and black feather bulbs in ducks. PLoS ONE 2012, 7, e36592. [Google Scholar] [CrossRef]
- Sangwan, R.S.; Tripathi, S.; Singh, J.; Narnoliya, L.K.; Sangwan, N.S. De novo sequencing and assembly of Centella asiatica leaf transcriptome for mapping of structural, functional and regulatory genes with special reference to secondary metabolism. Gene 2013, 525, 58–76. [Google Scholar] [CrossRef] [PubMed]
- Mutz, K.-O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.-G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Sultan, M.; Schulz, M.H.; Richard, H.; Magen, A.; Klingenhoff, A.; Scherf, M.; Seifert, M.; Borodina, T.; Soldatov, A.; Parkhomchuk, D. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008, 321, 956–960. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Haroon; Li, Y.-X.; Ye, C.-X.; Su, J.; Nabi, G.; Su, X.-H.; Xing, L.-X. De Novo Transcriptome Assembly and Analysis of Longevity Genes Using Subterranean Termite (Reticulitermes chinensis) Castes. Int. J. Mol. Sci. 2022, 23, 13660. [Google Scholar] [CrossRef]
- Tan, Y.; Dang, Y.; Zhang, H.; Guo, X.; Yan, X.; Su, X.; Xing, L. A comparative study of embryonic development between gamogenesis and parthenogenesis in two Reticulitermes termites (Isoptera: Rhinotermitidae). Acta Entomol. Sin. 2016, 59, 438–445. [Google Scholar]
- Tojo, K.; Machida, R. Early embryonic development of the mayfly Ephemera japonica McLachlan (Insecta: Ephemeroptera, Ephemeridae). J. Morphol. 1998, 238, 327–335. [Google Scholar] [CrossRef]
- Wang, W.; Wang, S.; Zhang, H.; Yuan, C.; Yan, R.; Song, X.; Xu, L.; Li, X. Galectin Hco-gal-m from Haemonchus contortus modulates goat monocytes and T cell function in different patterns. Parasites Vectors 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, P.; Zhou, X.; Lei, C. Characterization of head transcriptome and analysis of gene expression involved in caste differentiation and aggression in Odontotermes formosanus (Shiraki). PLoS ONE 2012, 7, e50383. [Google Scholar] [CrossRef] [PubMed]
- Van Hiel, M.B.; Van Wielendaele, P.; Temmerman, L.; Van Soest, S.; Vuerinckx, K.; Huybrechts, R.; Broeck, J.V.; Simonet, G. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Liu, H.; Yang, X.; Chen, J.; Zhang, H.; Xing, L.; Zhang, X. Characterization of the transcriptomes and cuticular protein gene expression of alate adult, brachypterous neotenic and adultoid reproductives of Reticulitermes labralis. Sci. Rep. 2016, 6, 34183. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, T.; Matsuura, K. Distribution and phylogenetic analysis of termite egg-mimicking fungi “termite balls” in Reticulitermes termites. Ann. Entomol. Soc. Am. 2007, 100, 532–538. [Google Scholar] [CrossRef]
- Klag, J.; Krzysztofowicz, A.; Jura, C.; Kisiel, E. Predetermination of cytokinesis and transition from holoblastic to partial superficial cleavage in early embryonic development of Tetrodontophora bielanensis (Collembola). Folia Biol.-Krakow 2000, 48, 137–142. [Google Scholar]
- Yashiro, T.; Matsuura, K. Termite queens close the sperm gates of eggs to switch from sexual to asexual reproduction. Proc. Natl. Acad. Sci. USA 2014, 111, 17212–17217. [Google Scholar] [CrossRef]
- Li, G.; Liu, L.; Sun, P.; Wu, Y.; Lei, C.; Chen, X.; Huang, Q. Physiological profiles associated with ceasing growth of unfertilized eggs produced by unmated queens in the subterranean termite Reticulitermes chinensis. Biol. Open 2016, 5, 756–763. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, M.; Bawazeer, S.; Bawazeer, N.; Irfan, M.; Rauf, A.; Su, X.-H.; Xing, L.-X. A comprehensive review on the documented characteristics of four Reticulitermes termites (Rhinotermitidae, Blattodea) of China. Braz. J. Biol. 2022, 84, e256354. [Google Scholar] [CrossRef]
- Ishitani, K.; Maekawa, K. Ovarian development of female-female pairs in the termite, Reticulitermes speratus. J. Insect Sci. 2010, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.-i.; Shindome, C.; Kobayashi, Y.; Takeda, M.; Yamashita, O.; Shiomi, K.; Fujiwara, Y. Temperature-dependent activation of ERK/MAPK in yolk cells and its role in embryonic diapause termination in the silkworm Bombyx mori. J. Insect Physiol. 2005, 51, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Fujimoto, M.; Goka, K. Sexual and asexual colony foundation and the mechanism of facultative parthenogenesis in the termite Reticulitermes speratus (Isoptera, Rhinotermitidae). Insectes Soc. 2004, 51, 325–332. [Google Scholar] [CrossRef]
- Cao, W.; Bao, C.; Padalko, E.; Lowenstein, C.J. Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling. J. Exp. Med. 2008, 205, 1491–1503. [Google Scholar] [CrossRef] [PubMed]




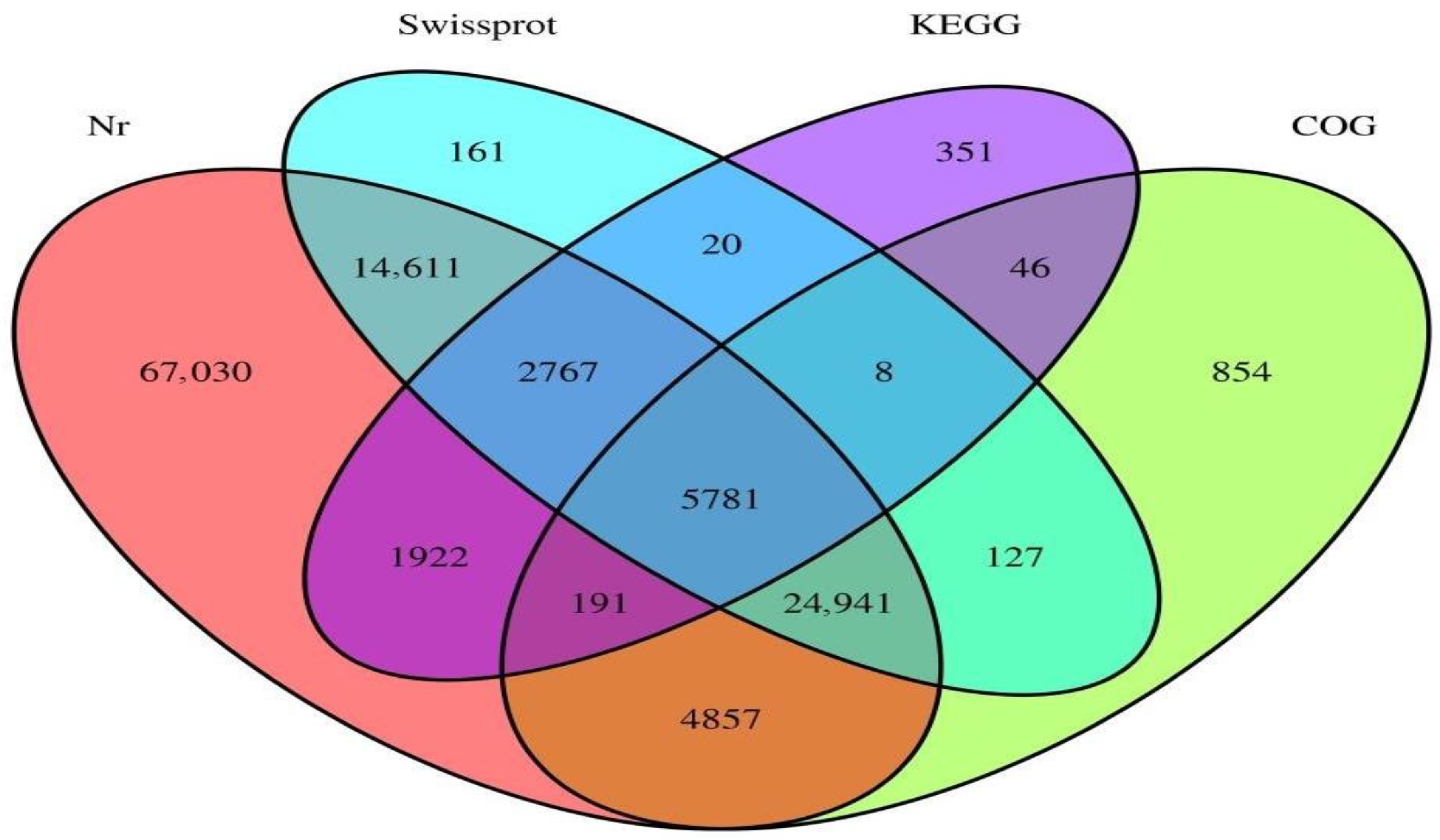
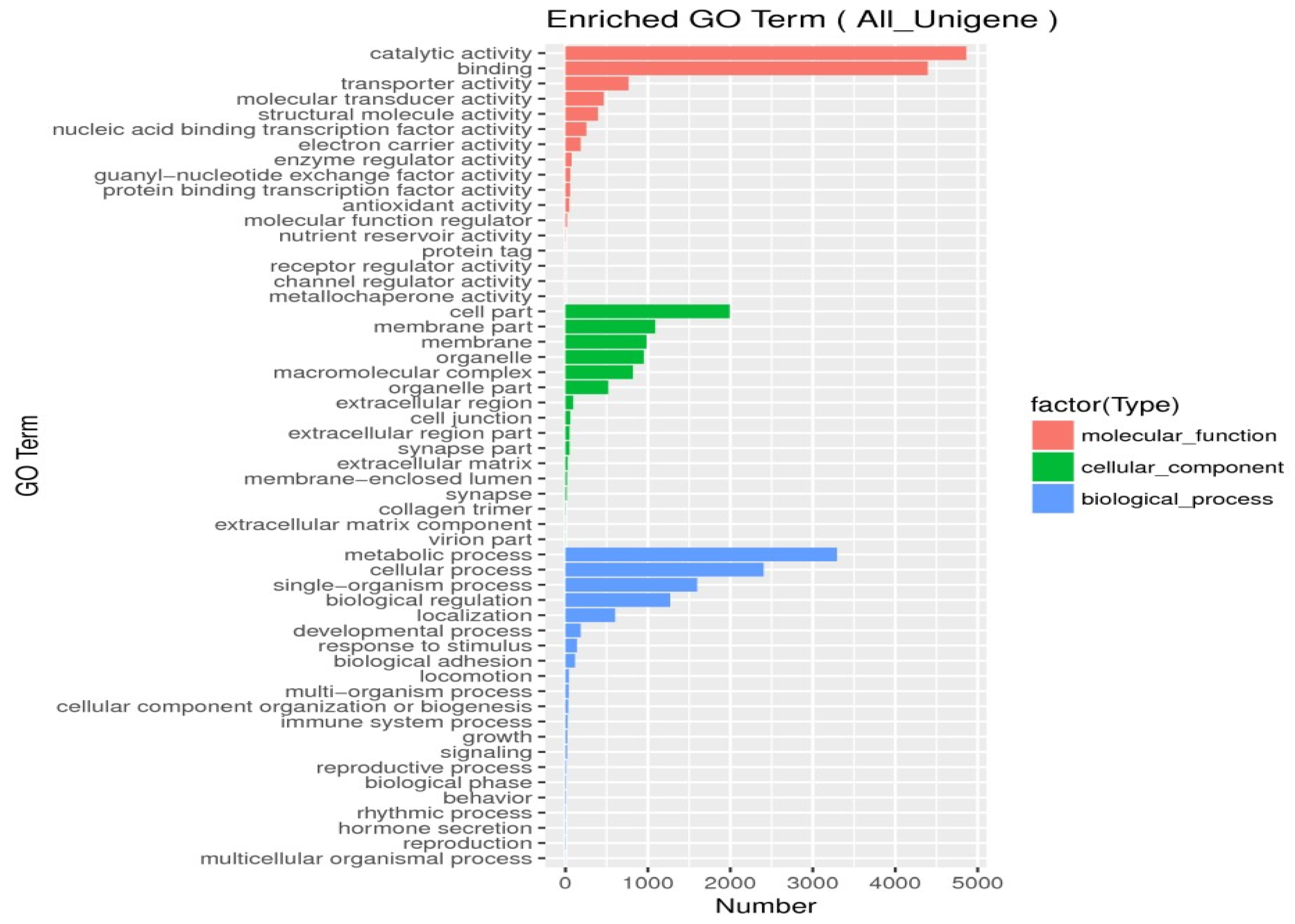
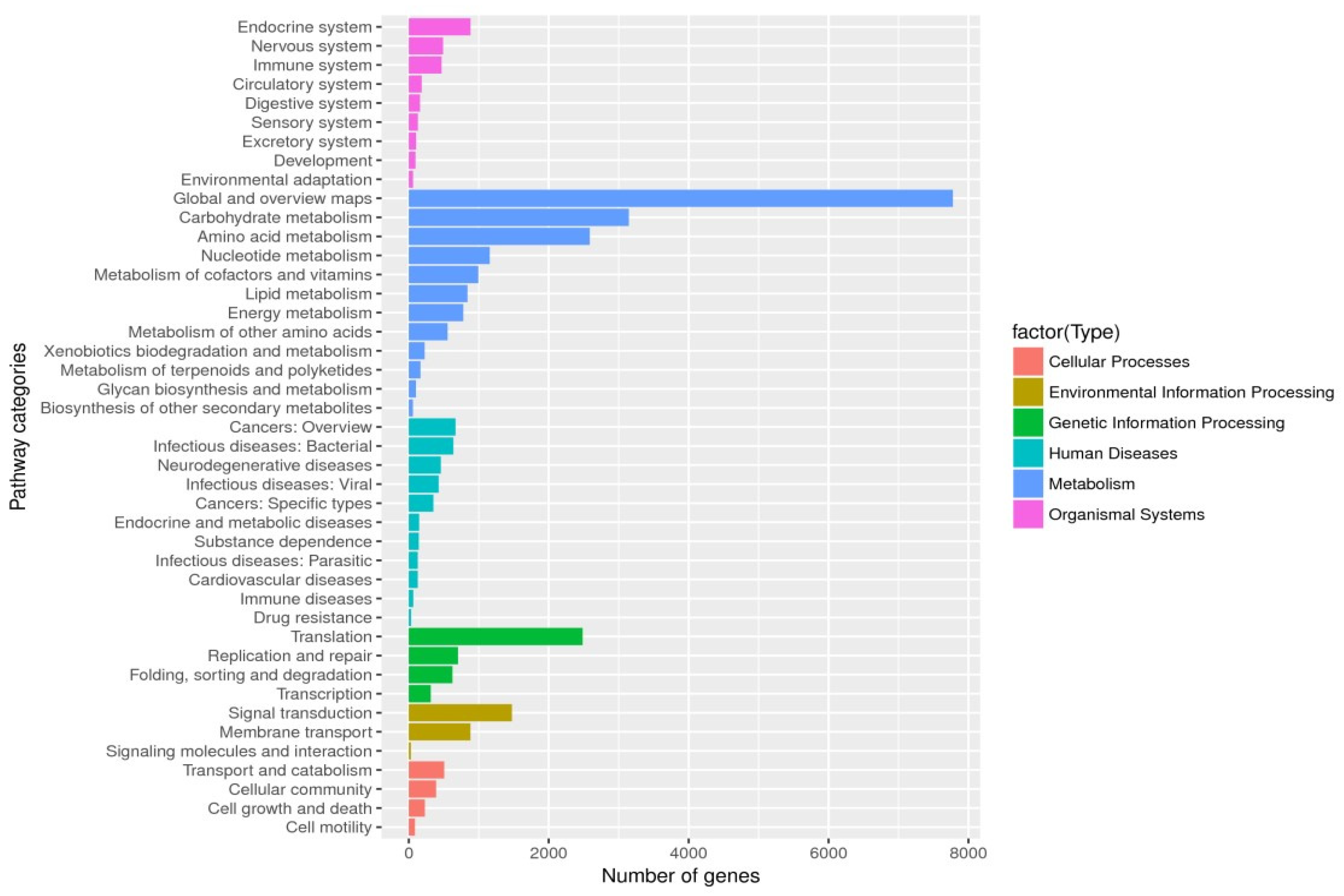
| Short for Gene | Primer Sequences | Annotation |
|---|---|---|
| beta-actin | F:CCCAACACAGCGTCTTACAA R:CAGATGTCCTCAGCTTCACG | |
| CDK2 | F:ACTCTGTGGTACCGAGCACCTG R:CATGGTTGGCTCCAGCTTCTTCAG | cyclin-dependent kinase 2 |
| PKA | F:CTTGGCGCACTTCTCAGTAGACG R:TTGCCTCAACAGACTGGATTGCTG | protein kinase A |
| MAP2K1 | F:CCACTAGAACGATGTCGGACCTTC | mitogen-activated protein kinase |
| R:CAAGCGCATGGAATTCTTCCTGTG | kinase 1 | |
| CPEB | F:GCCATCTACCAGGTGTGAAGCAG | cytoplasmic polyadenylation |
| R:GGAACGCCAACGGTACGAGTC | element-binding protein | |
| MAPK1/3 | F:GGCGTATGGAATGGTGGTATCTGC R:TGTTCGCTGGCAGTATGTCTGATG | mitogen-activated protein kinase 1/3 |
| HGK | F:TGCAGAGTCAAGGTCTACAGCATG | mitogen-activated protein kinase |
| R:TCAAGTCATTCGGTGACCTGATGC | kinase kinase kinase 4 | |
| MKP | F:CGATGCGGCTGACCTCAACAC | dual-specificity MAP kinase |
| R:GCTGCTTGTACGTGATACCACAGG | phosphatase | |
| Pax6 | F:GTGAAGGCGGAGATGAGATTCTGG R:TCTGTACCTGCACCAAGACCAATG | paired box protein 6 |
| Egg Growth, Size, and No | R. aculabialis | R. flaviceps |
|---|---|---|
| Oocyte length at first | 605.8 μm | 657 μm |
| Egg phases | Five | Two |
| Oocyte length at last phase | 685 μm | 687 μm |
| Nuclei in mature oocytes | Visible | Few nuclei visible |
| Before ovulation | Single nucleus | Single nucleus |
| After oviposition | 2–5 nuclei | Not seen |
| 5 h after oviposition | 16–22 nuclei | 6–10 nuclei |
| 10 h after oviposition | 35–42 nuclei | No visible nuclei |
| 24 h after oviposition | 51–60 nuclei | No clear nuclei |
| 48 h later | 67–84 nuclei | 28–37 nuclei |
| 3–5 d after oviposition | Visible blastoderm | |
| At 5–7 d postoviposition | 71–85 nuclei | |
| At 10 d | Blastoderm formation and germinal band growth | 86% embryos stopped developing blastoderm formation and 10% stopped developing at the embryonic formation |
| Embryo rotation | Two time | None |
| Third stage | Germinal formation | None, large cells concentrated at central point and peripheral sites were free |
| Fourth and fifth stages | Development of tissues and organs | Lacked any differentiation of appendages and organs development in eggs |
| Type | Sequences | Bases | Min | Max | Average | N50 | (A + T)% | (C + G)% |
|---|---|---|---|---|---|---|---|---|
| All_Contig | 212,52,163 | 909,041,501 | 25 | 15,261 | 42.77 | 39 | 61.26 | 38.74 |
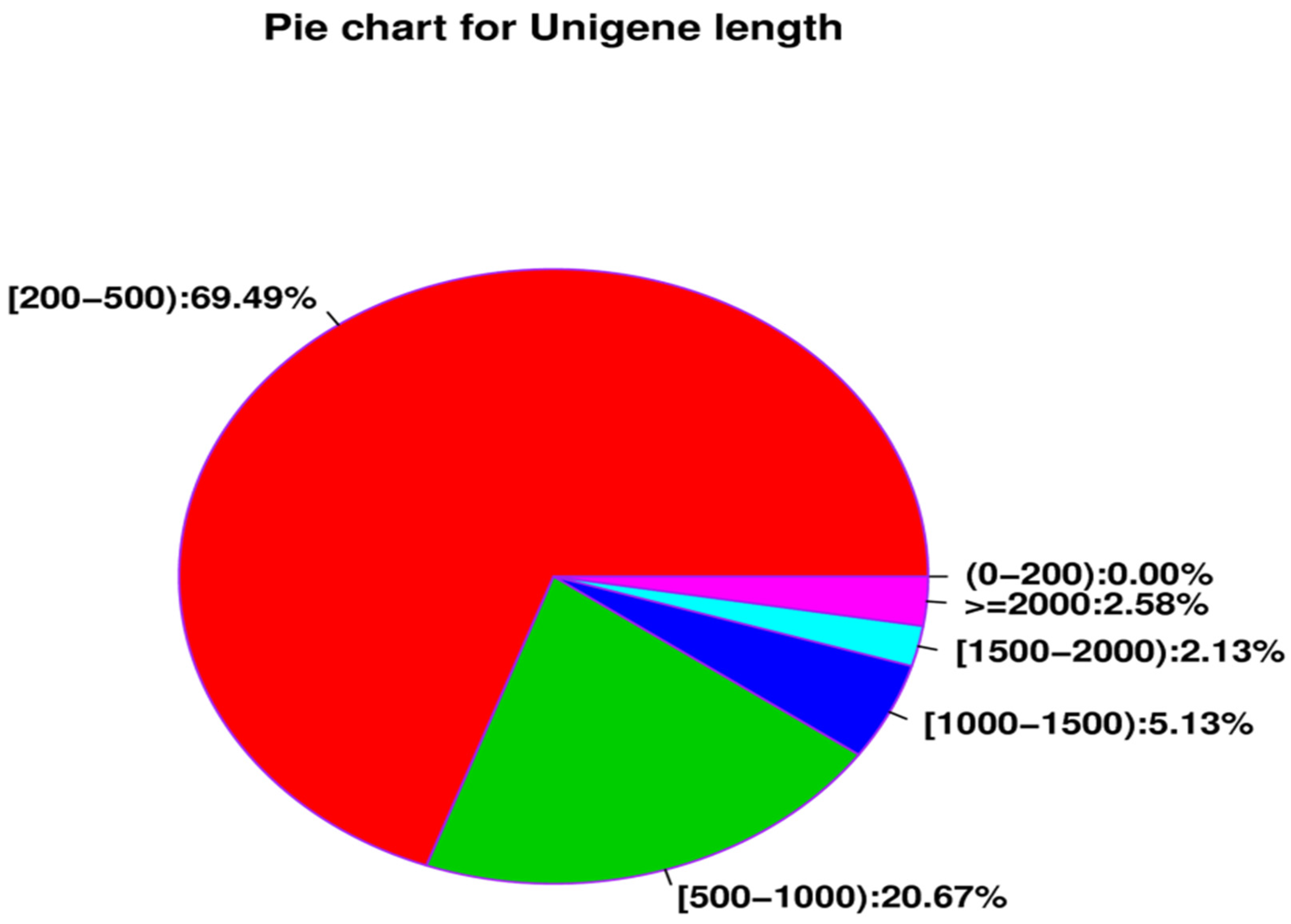
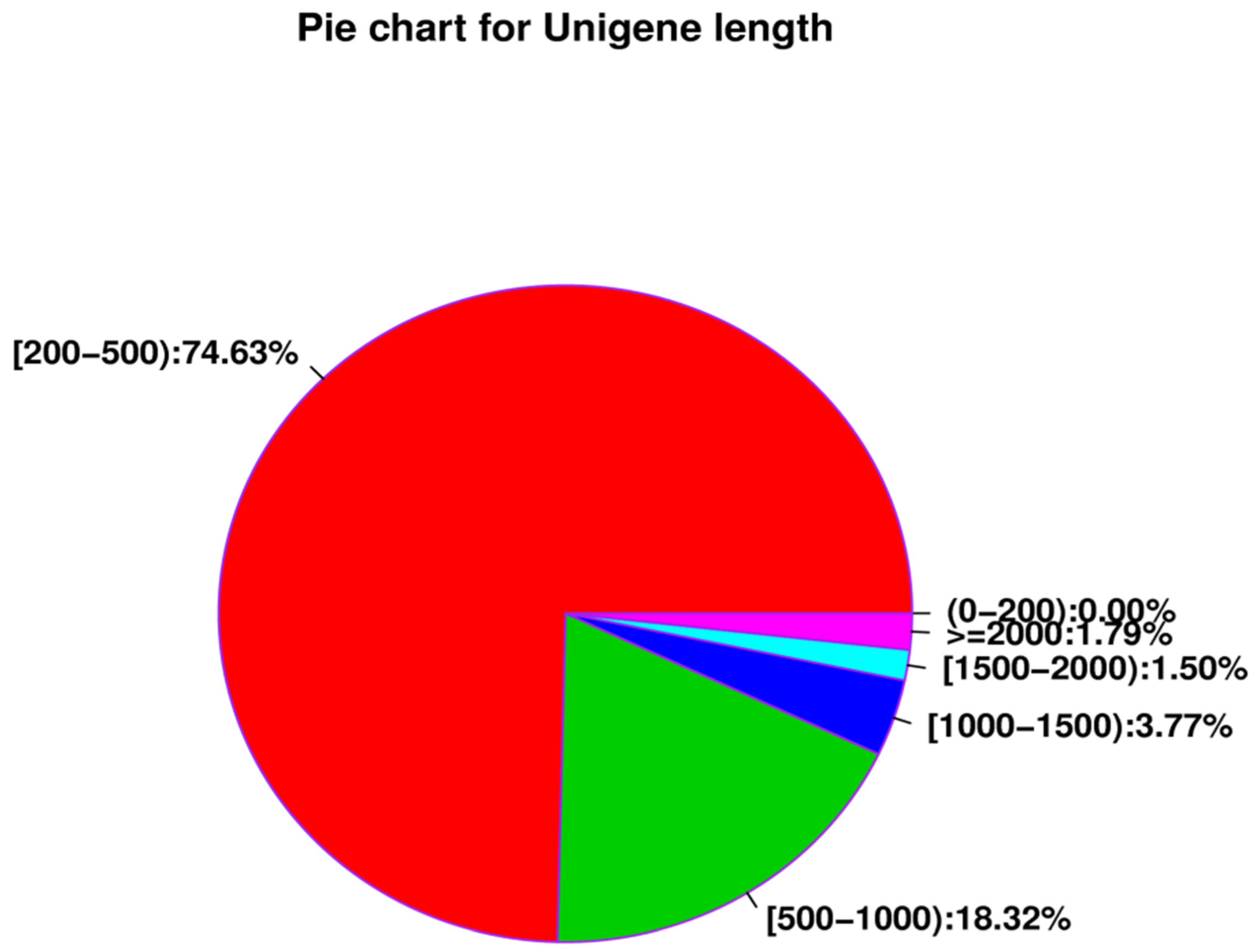
| All_Unigene | 324,454 | 170,772,245 | 201 | 13,549 | 526.34 | 633 | 60.71 | 39.29 |
| Sequence | Quantity |
|---|---|
| Approved SSR | 11,892 |
| Sequence of SSR distribution | 11,883 |
| Single nucleotide | 9649 |
| Dinucleotide | 1236 |
| Trinucleotide | 735 |
| Tetranucleotide | 258 |
| Pentanucleotide | 13 |
| Six nucleotides | 1 |
| Type | Sequences | Bases | Min | Max | Average | N50 | (A + T)% | (C + G)% |
|---|---|---|---|---|---|---|---|---|
| All_Contig | 23,146,174 | 1,012,445,494 | 25 | 143,170 | 43.74 | 39 | 58.72 | 41.28 |
| All_Unigene | 422,213 | 202,603,877 | 201 | 42,295 | 479.86 | 532 | 58.13 | 41.87 |
| Sequence | Quantity |
|---|---|
| Approved SSR | 7196 |
| Sequence of SSR distribution | 7186 |
| Single nucleotide | 6033 |
| Dinucleotide | 588 |
| Trinucleotide | 420 |
| Tetranucleotide | 143 |
| Pentanucleotide | 10 |
| Six nucleotides | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Khan, Z.; Liu, X.-M.; Deng, S.-L.; Fang, Y.-G.; Zhang, M.; Su, X.-H.; Xing, L.-X.; Yan, X.-R. Embryonic Development of Parthenogenetic and Sexual Eggs in Lower Termites. Insects 2023, 14, 640. https://doi.org/10.3390/insects14070640
Peng X, Khan Z, Liu X-M, Deng S-L, Fang Y-G, Zhang M, Su X-H, Xing L-X, Yan X-R. Embryonic Development of Parthenogenetic and Sexual Eggs in Lower Termites. Insects. 2023; 14(7):640. https://doi.org/10.3390/insects14070640
Chicago/Turabian StylePeng, Xin, Zahid Khan, Xiao-Min Liu, Shi-Lin Deng, Yong-Gang Fang, Min Zhang, Xiao-Hong Su, Lian-Xi Xing, and Xing-Rong Yan. 2023. "Embryonic Development of Parthenogenetic and Sexual Eggs in Lower Termites" Insects 14, no. 7: 640. https://doi.org/10.3390/insects14070640
APA StylePeng, X., Khan, Z., Liu, X.-M., Deng, S.-L., Fang, Y.-G., Zhang, M., Su, X.-H., Xing, L.-X., & Yan, X.-R. (2023). Embryonic Development of Parthenogenetic and Sexual Eggs in Lower Termites. Insects, 14(7), 640. https://doi.org/10.3390/insects14070640






