Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Accession Numbers
2.3. Domain Search of HP1
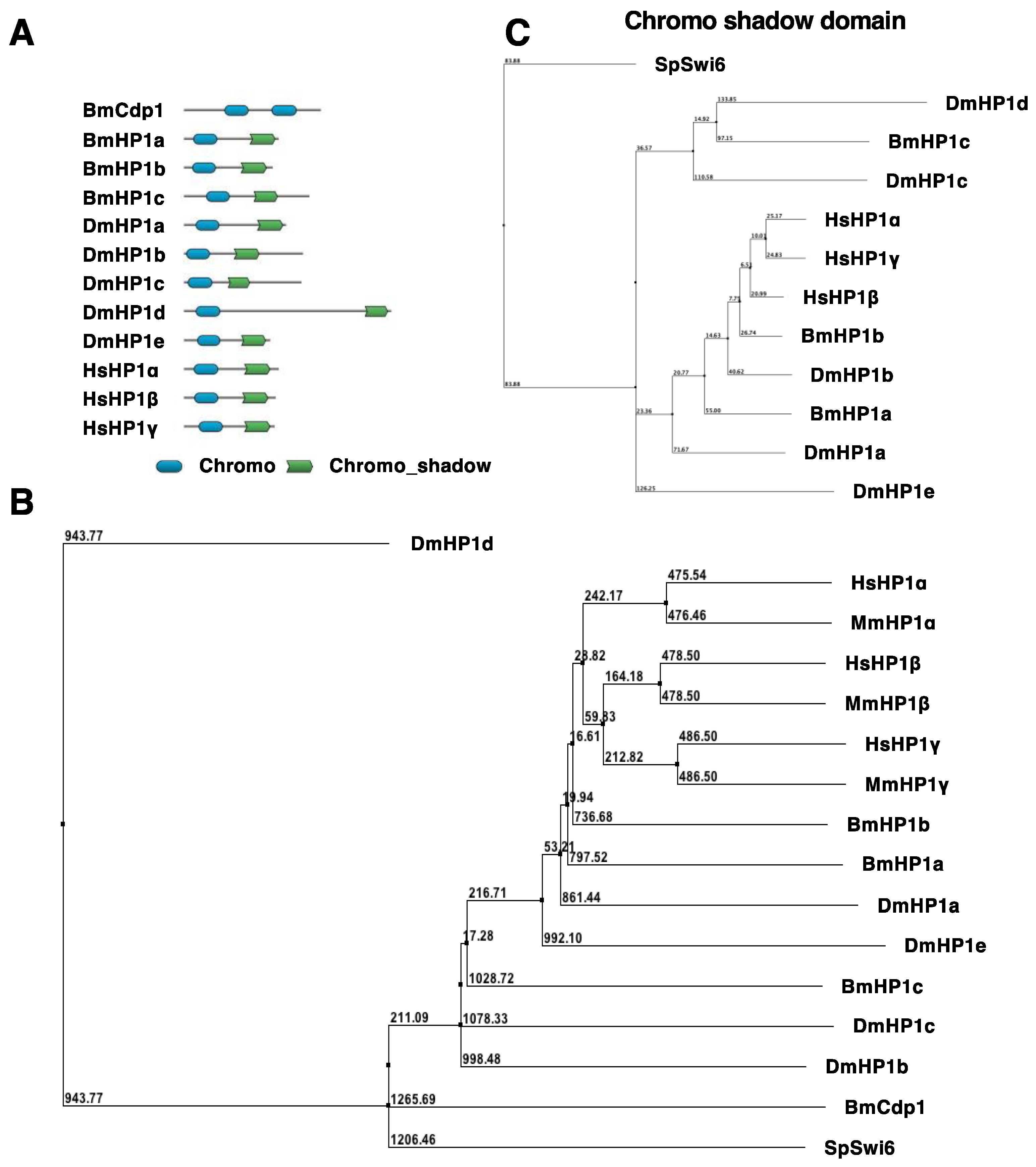
2.4. Phylogenetic Tree
2.5. Transcription Repression Assay
2.6. Co-Immunoprecipitation Assay
2.7. Western Blotting
2.8. Semi-Quantitative RT-PCR
2.9. Fluorescence Microscopic Observation
2.10. RNA Interference
2.11. Flow Cytometry
3. Results
3.1. BmHP1c Was Identified as a Novel HP1 Gene
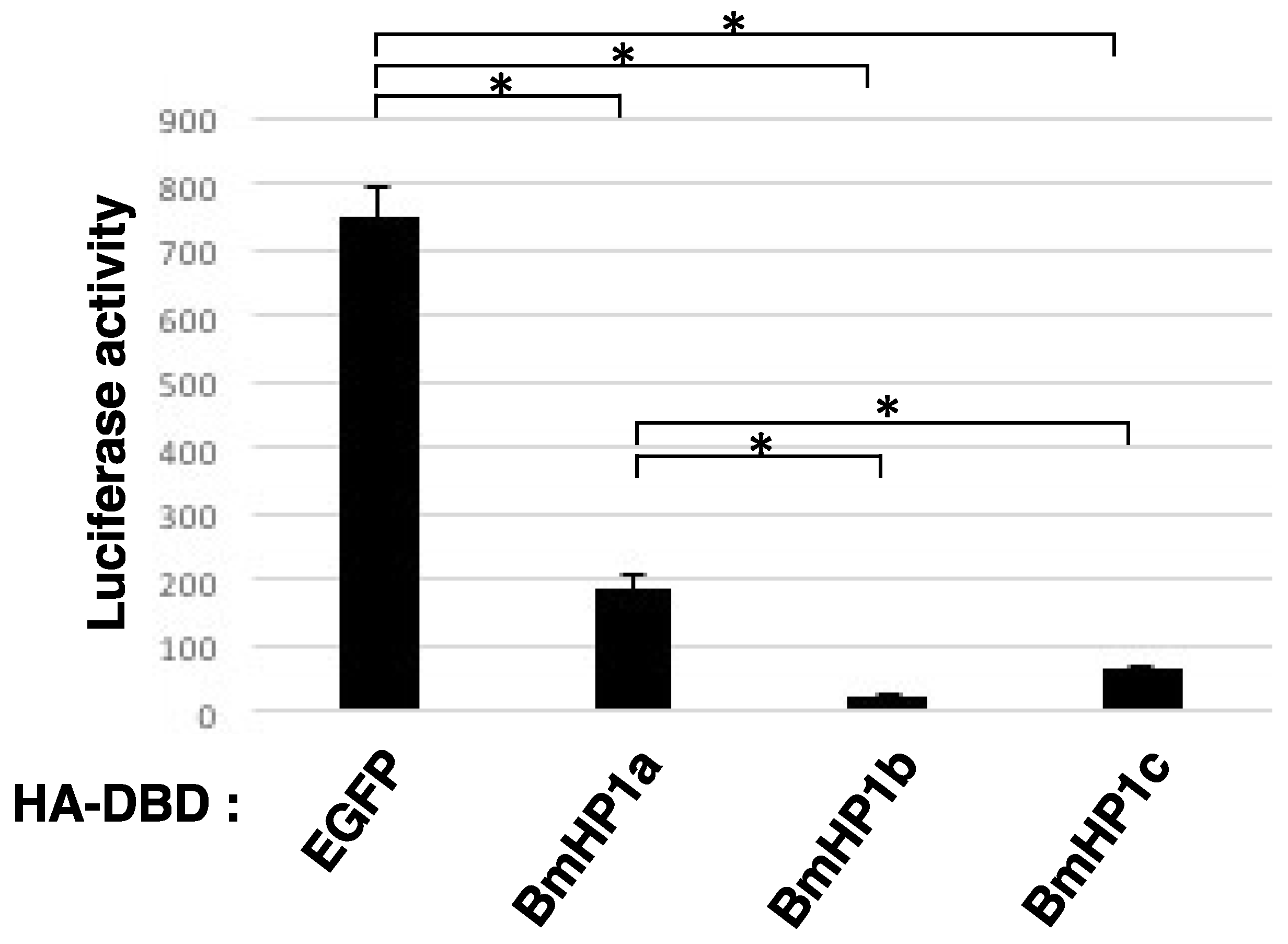
3.2. BmHP1c Has Transcriptional Repression Activity
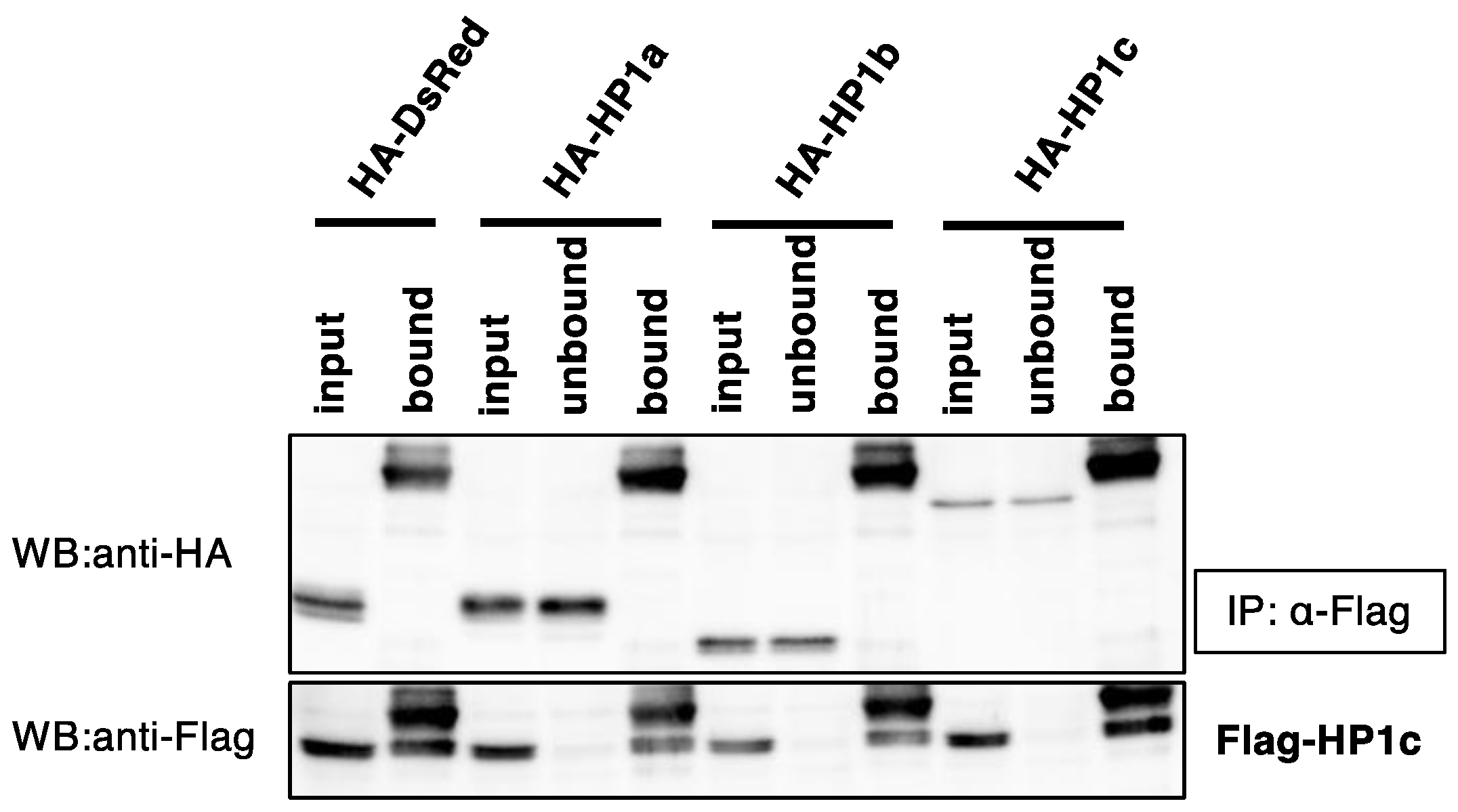
3.3. BmHP1c Does Not Form Homo- and Heterodimers with the Other BmHP1s
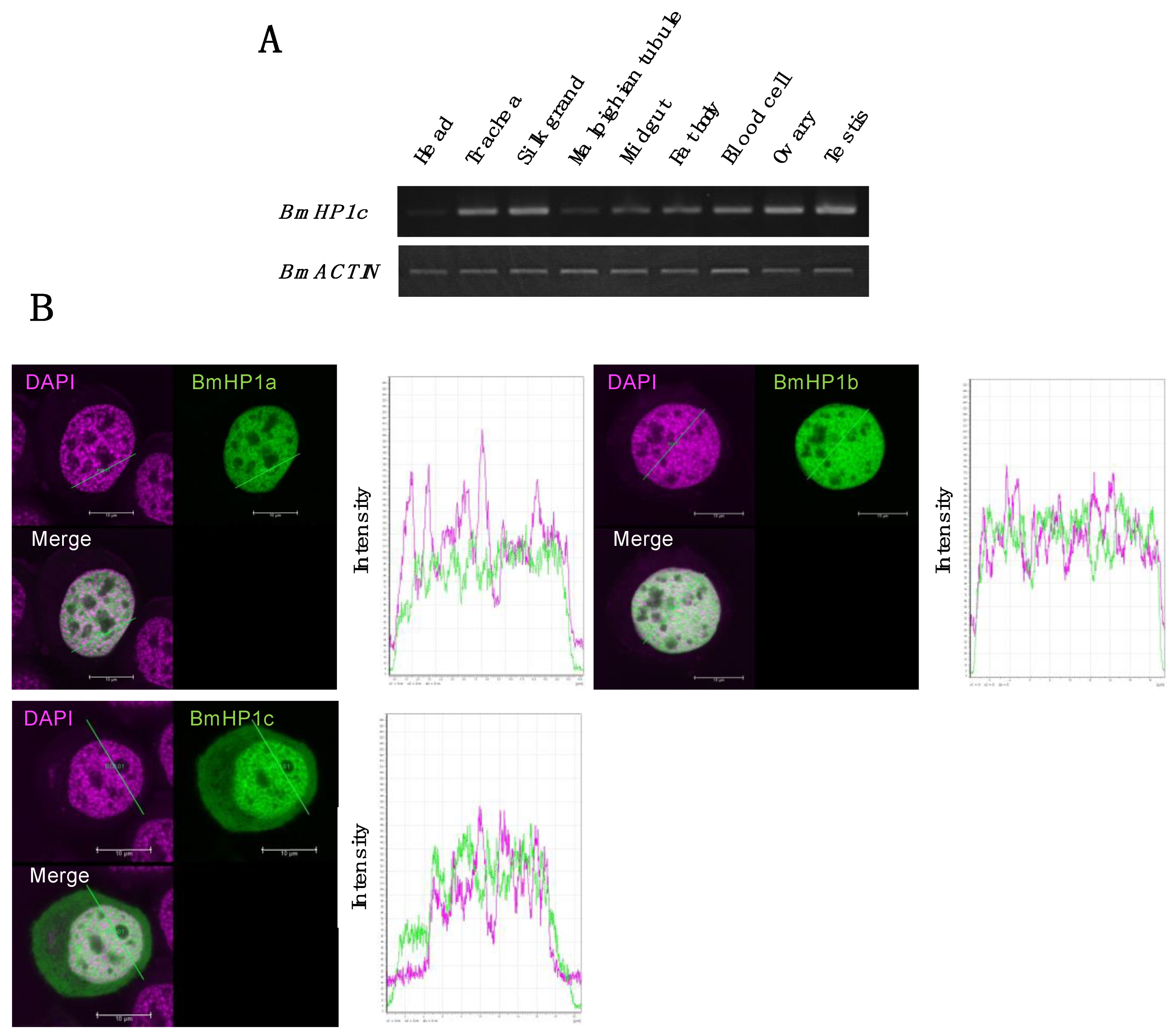
3.4. Tissue-Specific Expression and Subcellular Localization of the BmHP1c in Silkworm Cultured Cell
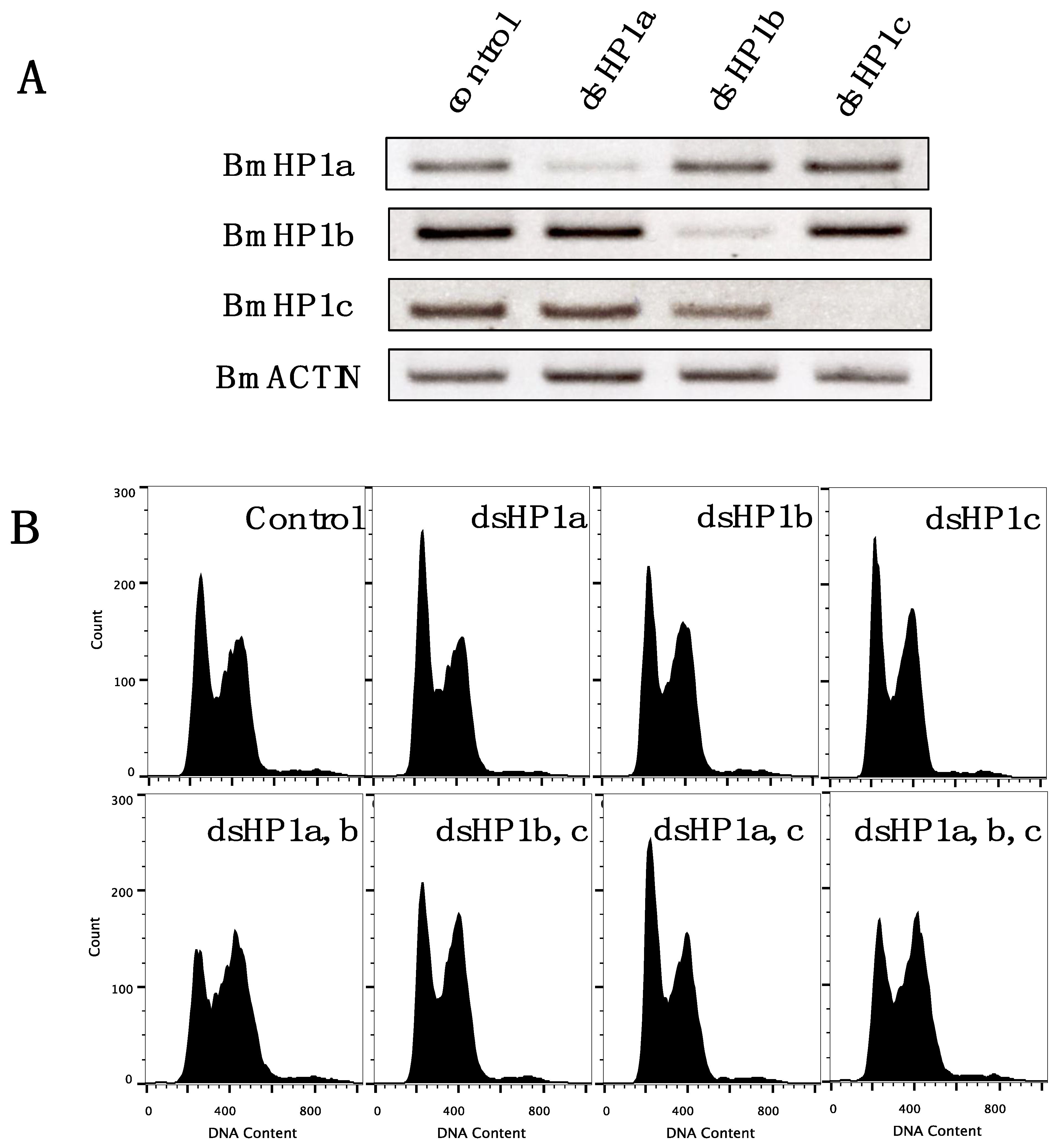
3.5. Knockdown of BmHP1c Does Not Affect Cell Cycle Progression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Filion, G.J.; van Bemmel, J.G.; Braunschweig, U.; Talhout, W.; Kind, J.; Ward, L.D.; Brugman, W.; de Castro, I.J.; Kerkhoven, R.M.; Bussemaker, H.J.; et al. Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells. Cell 2010, 143, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Rhind, N.R.; Gilbert, D.M. DNA Replication Timing. Cold Spring Harb. Perspect. Biol. 2013, 5, a010132. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Jacobs, S.A.; Taverna, S.D.; Zhang, Y.; Briggs, S.D.; Li, J.; Eissenberg, J.C.; Allis, C.D.; Khorasanizadeh, S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001, 20, 5232–5241. [Google Scholar] [CrossRef]
- Brasher, S.V.; Smith, B.; Fogh, R.H.; Nietlispach, D.; Thiru, A.; Nielsen, P.R.; Broadhurst, B.; Ball, L.J.; Murzina, N.V.; Laue, E.D. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000, 19, 1587–1597. [Google Scholar] [CrossRef]
- Cowieson, N.P.; Partridge, J.; Allshire, R.; McLaughlin, P.J. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000, 10, 517–525. [Google Scholar] [CrossRef]
- Smothers, J.F.; Henikoff, S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 2000, 10, 27–30. [Google Scholar] [CrossRef]
- Lomberk, G.; Bensi, D.; Fernandez-Zapico, M.E.; Urrutia, R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 2006, 8, 407–415. [Google Scholar] [CrossRef]
- Smothers, J.F.; Henikoff, S. The Hinge and Chromo Shadow Domain Impart Distinct Targeting of HP1-Like Proteins. Mol. Cell. Biol. 2001, 21, 2555–2569. [Google Scholar] [CrossRef] [PubMed]
- Font-Burgada, J.; Rossell, D.; Auer, H.; Azorín, F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 2008, 22, 3007–3023. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, F.; Ni, J.-Q.; Vaillant, C.; Sun, F.-L. HP1 modulates the transcription of cell-cycle regulators in Drosophila melanogaster. Nucleic Acids Res. 2005, 33, 2852–2858. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Ryu, H.W.; Kim, G.W.; Kwon, S.H. Comparison of three heterochromatin protein 1 homologs of Drosophila. J. Cell Sci. 2019, 132, jcs.222729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, D.; Sun, F. Drosophila melanogaster heterochromatin protein HP1b plays important roles in transcriptional activation and development. Chromosoma 2010, 120, 97–108. [Google Scholar] [CrossRef]
- Lee, D.H.; Li, Y.; Shin, D.-H.; Yi, S.A.; Bang, S.-Y.; Park, E.K.; Han, J.-W.; Kwon, S.H. DNA microarray profiling of genes differentially regulated by three heterochromatin protein 1 (HP1) homologs in Drosophila. Biochem. Biophys. Res. Commun. 2013, 434, 820–828. [Google Scholar] [CrossRef]
- Cryderman, D.E.; Grade, S.K.; Li, Y.; Fanti, L.; Pimpinelli, S.; Wallrath, L.L. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 2005, 232, 767–774. [Google Scholar] [CrossRef]
- Mitsunobu, H.; Izumi, M.; Mon, H.; Tatsuke, T.; Lee, J.M.; Kusakabe, T. Molecular characterization of heterochromatin proteins 1a and 1b from the silkworm, Bombyx mori. Insect Mol. Biol. 2011, 21, 9–20. [Google Scholar] [CrossRef]
- Shoji, K.; Hara, K.; Kawamoto, M.; Kiuchi, T.; Kawaoka, S.; Sugano, S.; Shimada, T.; Suzuki, Y.; Katsuma, S. Silkworm HP1a transcriptionally enhances highly expressed euchromatic genes via association with their transcription start sites. Nucleic Acids Res. 2014, 42, 11462–11471. [Google Scholar] [CrossRef][Green Version]
- Shoji, K.; Kiuchi, T.; Hara, K.; Kawamoto, M.; Kawaoka, S.; Arimura, S.-I.; Tsutsumi, N.; Sugano, S.; Suzuki, Y.; Shimada, T.; et al. Characterization of a novel chromodomain-containing gene from the silkworm, Bombyx mori. Gene 2013, 527, 649–654. [Google Scholar] [CrossRef]
- Hino, M.; Kawanami, T.; Xu, J.; Morokuma, D.; Hirata, K.; Yamashita, M.; Karasaki, N.; Tatsuke, T.; Mon, H.; Iiyama, K.; et al. High-level expression and purification of biologically active human IL-2 using silkworm-baculovirus expression vector system. J. Asia-Pacific Entomol. 2016, 19, 313–317. [Google Scholar] [CrossRef]
- Mon, H.; Kobayashi, I.; Ohkubo, S.; Tomita, S.; Lee, J.; Sezutsu, H.; Tamura, T.; Kusakabe, T. Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol. 2012, 9, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Vermaak, D.; Henikoff, S.; Malik, H.S. Positive Selection Drives the Evolution of rhino, a Member of the Heterochromatin Protein 1 Family in Drosophila. PLoS Genet. 2005, 1, e9. [Google Scholar] [CrossRef] [PubMed]
- Nakahigashi, K.; Jasencakova, Z.; Schubert, I.; Goto, K. The Arabidopsis HETEROCHROMATIN PROTEIN1 Homolog (TERMINAL FLOWER2) Silences Genes Within the Euchromatic Region but not Genes Positioned in Heterochromatin. Plant Cell Physiol. 2005, 46, 1747–1756. [Google Scholar] [CrossRef]
- Charó, N.L.; Galigniana, N.M.; Piwien-Pilipuk, G. Heterochromatin protein (HP)1γ is not only in the nucleus but also in the cytoplasm interacting with actin in both cell compartments. Biochim. Biophys. Acta 2018, 1865, 432–443. [Google Scholar] [CrossRef]
- Ruddock-D’Cruz, N.T.; Prashadkumar, S.; Wilson, K.J.; Heffernan, C.; Cooney, M.A.; French, A.J.; Jans, D.A.; Verma, P.J.; Holland, M.K. Dynamic changes in localization of chromobox (CBX) family members during the maternal to embryonic transition. Mol. Reprod. Dev. 2007, 75, 477–488. [Google Scholar] [CrossRef]
- Yi, S.A.; Lee, D.H.; Kim, G.W.; Ryu, H.-W.; Park, J.W.; Lee, J.; Han, J.; Park, J.H.; Oh, H.; Lee, J.; et al. HPV-mediated nuclear export of HP1γ drives cervical tumorigenesis by downregulation of p53. Cell Death Differ. 2020, 27, 2537–2551. [Google Scholar] [CrossRef]
- Giot, L.; Bader, J.S.; Brouwer, C.; Chaudhuri, A.; Kuang, B.; Li, Y.; Hao, Y.L.; Ooi, C.E.; Godwin, B.; Vitols, E.; et al. A Protein Interaction Map of Drosophila melanogaster. Science 2003, 302, 1727–1736. [Google Scholar] [CrossRef]
- Canzio, D.; Larson, A.; Narlikar, G.J. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014, 24, 377–386. [Google Scholar] [CrossRef]
- Nishibuchi, G.; Nakayama, J.-I. Biochemical and structural properties of heterochromatin protein 1: Understanding its role in chromatin assembly. J. Biochem. 2014, 156, 11–20. [Google Scholar] [CrossRef]
- Ye, Q.; Callebaut, I.; Pezhman, A.; Courvalin, J.-C.; Worman, H.J. Domain-specific Interactions of Human HP1-type Chromodomain Proteins and Inner Nuclear Membrane Protein LBR. J. Biol. Chem. 1997, 272, 14983–14989. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.H.; Workman, J.L. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression. BioEssays 2011, 33, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kirschmann, D.A.; Wallrath, L.L. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. 2002, 99, 16462–16469. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence |
|---|---|
| BmHP1a-RT5 | GGTAAAAAAGAGAAGAAAACGGAGACCAG |
| BmHP1a-RT3 | CGCCTATAATTTTCTCAGCTTTGAGTCC |
| BmHP1b-RT5 | GCCGACAAGAAAAAAGAAAATGAACCAG |
| BmHP1b-RT3 | TTCATGAGGAACATGAGTTCACCACTAC |
| BmHP1c-RT5 | CAACACTTGGGAACCAGAAGACAACC |
| BmHP1c-RT3 | CTCGCGATTCAACTGCTTCACTGTTAG |
| BmActin-RT5 | GCATCATACCTTCTACAATGAGC |
| BmActin-RT3 | GAGATCCACATCTGTTGGAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hino, M.; Tatsuke, T.; Morio, A.; Mon, H.; Lee, J.M.; Masuda, A.; Kakino, K.; Tonooka, Y.; Kusakabe, T. Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori. Insects 2022, 13, 631. https://doi.org/10.3390/insects13070631
Hino M, Tatsuke T, Morio A, Mon H, Lee JM, Masuda A, Kakino K, Tonooka Y, Kusakabe T. Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori. Insects. 2022; 13(7):631. https://doi.org/10.3390/insects13070631
Chicago/Turabian StyleHino, Masato, Tsuneyuki Tatsuke, Akihiro Morio, Hiroaki Mon, Jae Man Lee, Akitsu Masuda, Kohei Kakino, Yoshino Tonooka, and Takahiro Kusakabe. 2022. "Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori" Insects 13, no. 7: 631. https://doi.org/10.3390/insects13070631
APA StyleHino, M., Tatsuke, T., Morio, A., Mon, H., Lee, J. M., Masuda, A., Kakino, K., Tonooka, Y., & Kusakabe, T. (2022). Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori. Insects, 13(7), 631. https://doi.org/10.3390/insects13070631





