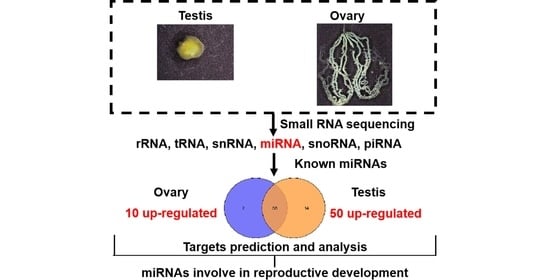
Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Gonad Observation and Collection of Tissue Samples
2.3. RNA Extraction and Library Construction
2.4. Small RNA Sequence Analysis
2.5. Identification and Expression Analysis of miRNAs
2.6. RT-qPCR Validation of 11 Gonad-Biased Candidate miRNAs
2.7. Target Prediction and Enrichment Analysis of 8 Validated Gonad-Biased miRNAs
2.8. Data Analysis
3. Results
3.1. Gonadal Development
3.2. Analysis of Small RNA Sequencing Data
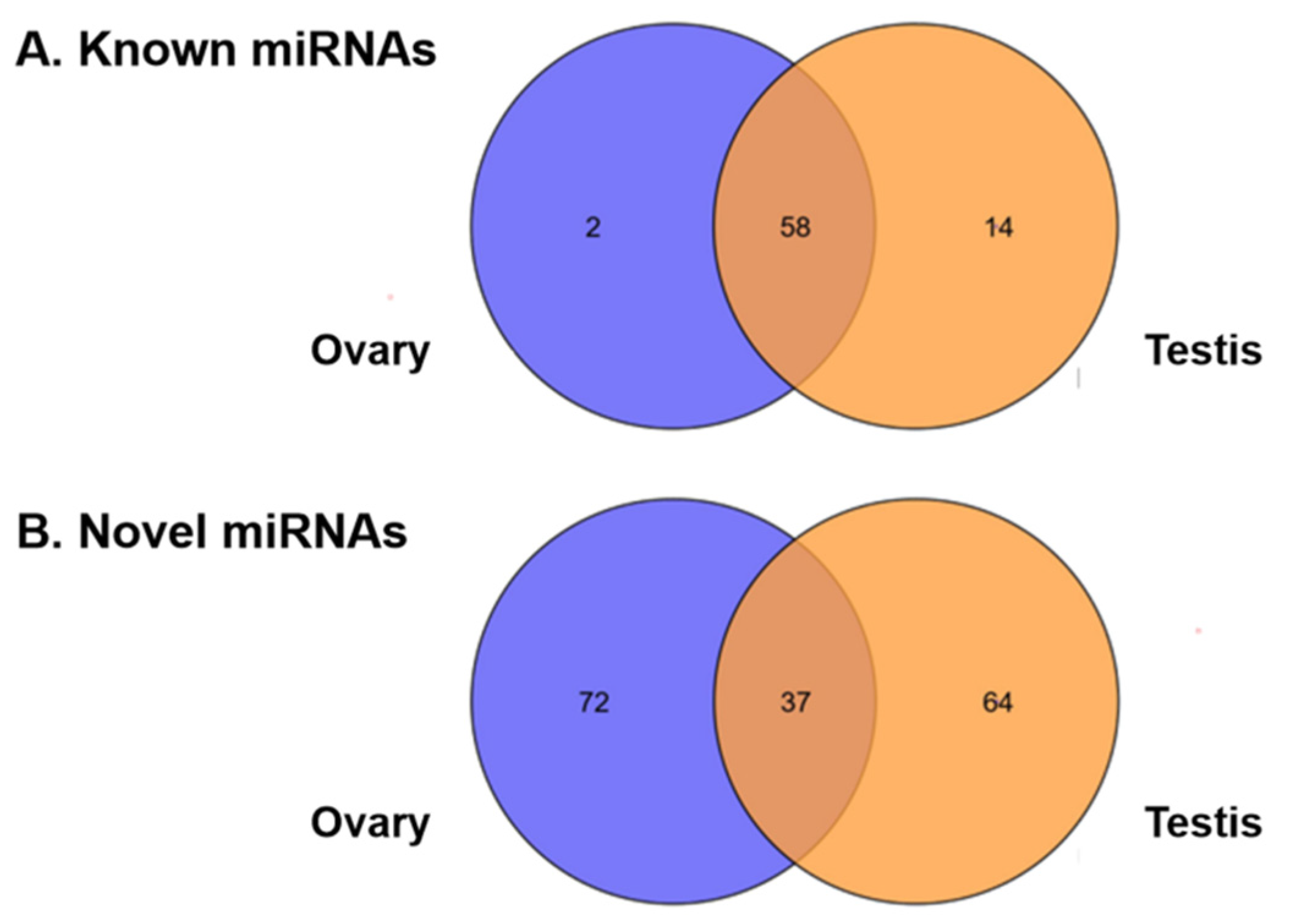
3.3. Identification and Expression of miRNAs in the Testis and Ovary from H. armigera
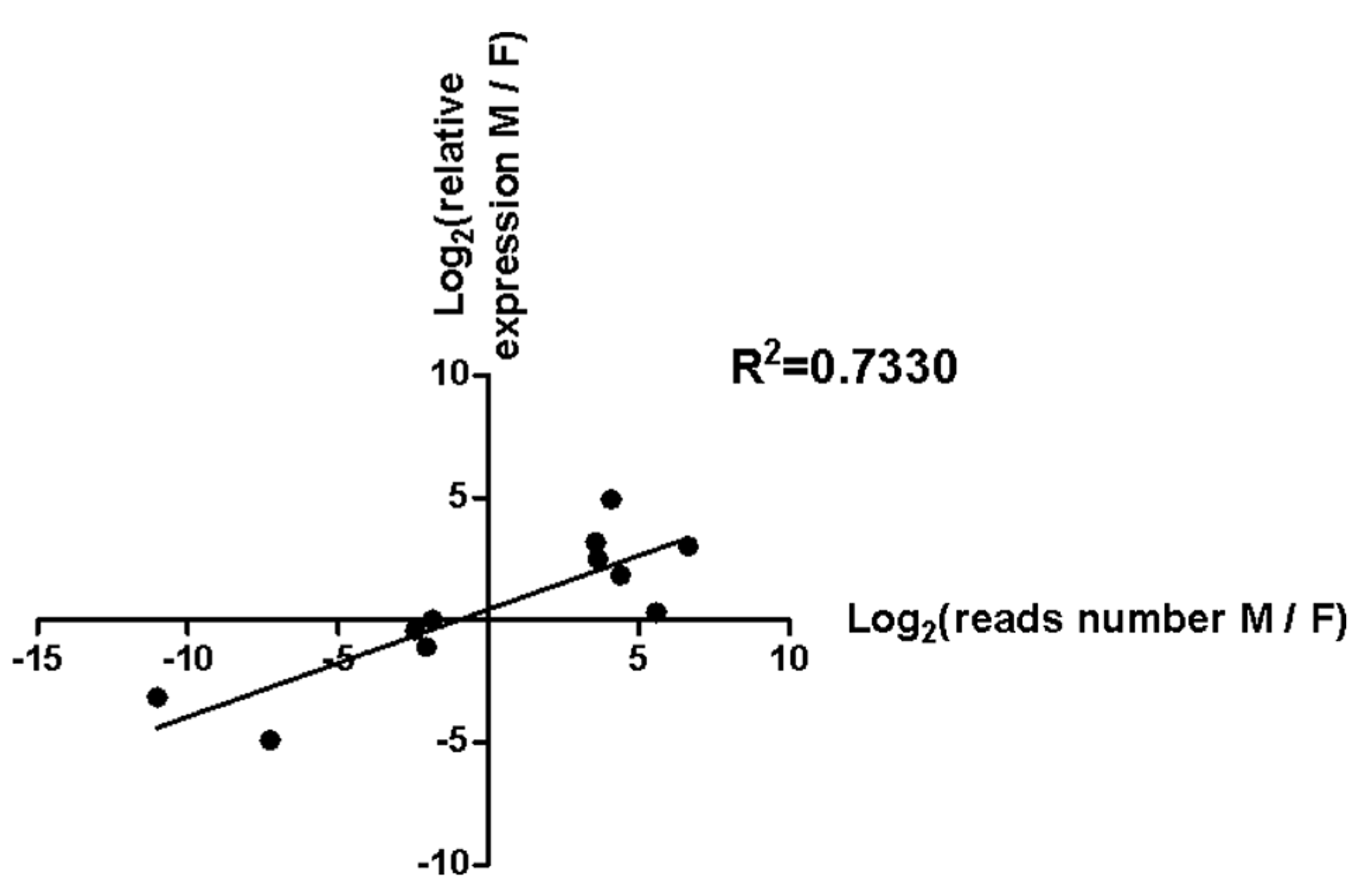
3.4. Validation of 11 Sex-Biased Candidate miRNAs Using RT-qPCR
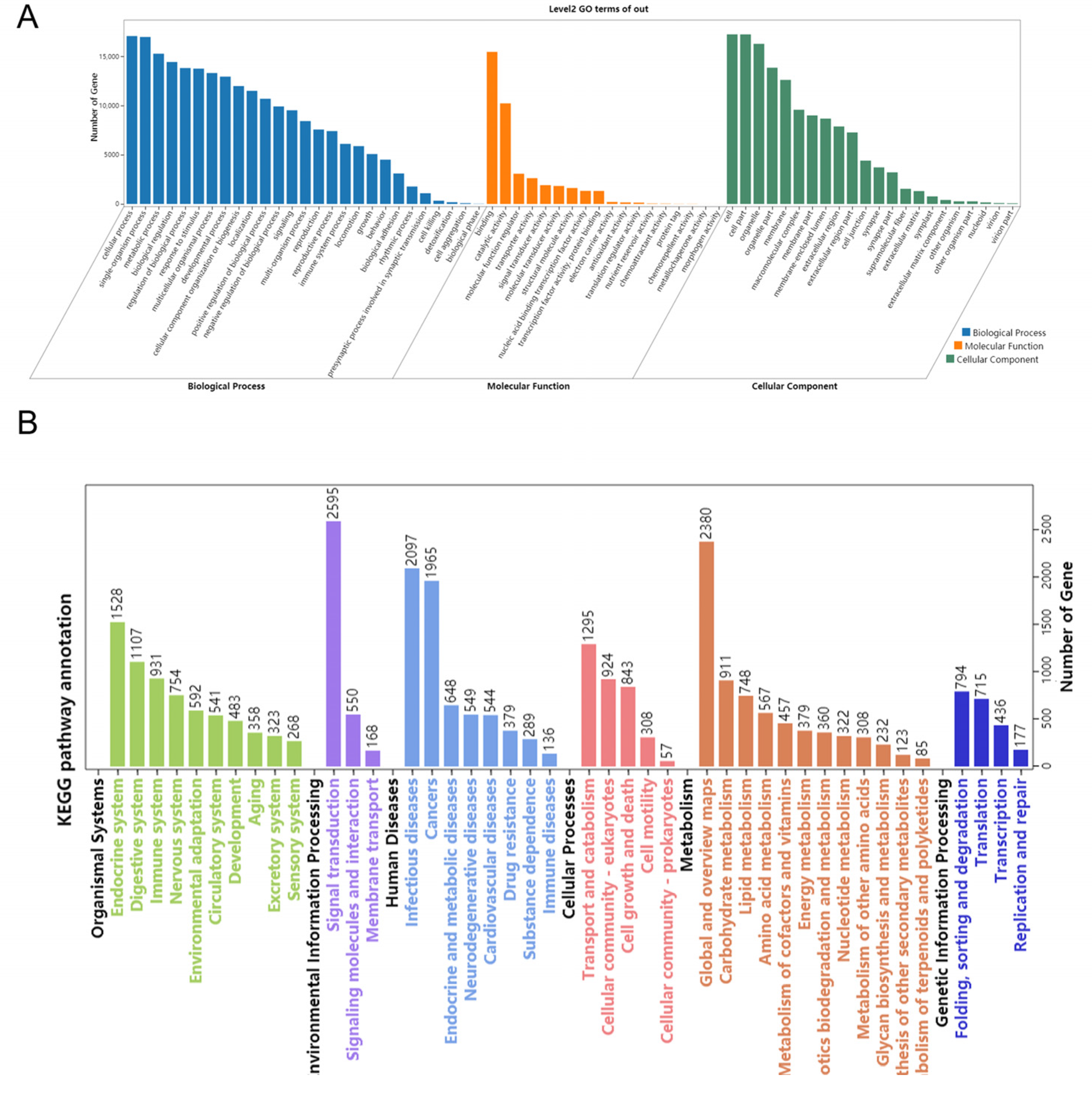
3.5. Target Prediction and Function Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [PubMed]
- Xing, Y.H.; Chen, L.L. Processing and roles of snoRNA-ended long noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 596–606. [Google Scholar] [CrossRef]
- Yang, J.X.; Rastetter, R.H.; Wilhelm, D. Non-coding RNAs: An introduction. Adv. Exp. Med. Biol. 2016, 886, 13–32. [Google Scholar]
- Yuan, Z.H.; Zhao, Y.M. The regulatory functions of piRNA/PIWI in spermatogenesis. Yi Chuan 2017, 39, 683–691. [Google Scholar]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013, 43, 24–38. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Yang, M.; Wei, Y.; Jiang, F.; Wang, Y.; Guo, X.; He, J.; Kang, L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014, 10, e1004206. [Google Scholar] [CrossRef] [PubMed]
- Yurikova, O.Y.; Aisina, D.E.; Niyazova, R.E.; Atambayeva, S.A.; Labeit, S.; Ivashchenko, A.T. The interaction of miRNA-5p and miRNA-3p with the mRNAs of orthologous genes. Mol. Biol. 2019, 53, 692–704. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef]
- Jia, X.; Zhou, M.; Zou, Z.; Lin, P.; Wang, Y.; Zhang, Z. Identification and comparative analysis of the ovary and testis microRNAome of mud crab Scylla paramamosain. Mol. Reprod. Dev. 2018, 85, 519–531. [Google Scholar] [CrossRef]
- Xiao, J.; Zhong, H.; Zhou, Y.; Yu, F.; Gao, Y.; Luo, Y.; Tang, Z.; Guo, Z.; Guo, E.; Gan, X.; et al. Identification and characterization of microRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus) by using solexa sequencing technology. PLoS ONE 2014, 9, e86821. [Google Scholar] [CrossRef]
- Marco, A. Sex-biased expression of microRNAs in Drosophila melanogaster. Open Biol. 2014, 4, 140024. [Google Scholar] [CrossRef][Green Version]
- Fagegaltier, D.; Konig, A.; Gordon, A.; Lai, E.C.; Gingeras, T.R.; Hannon, G.J.; Shcherbata, H.R. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics 2014, 198, 647–668. [Google Scholar] [CrossRef]
- Kugler, J.M.; Verma, P.; Chen, Y.W.; Weng, R.; Cohen, S.M. miR-989 is required for border cell migration in the Drosophila ovary. PLoS ONE 2013, 8, e67075. [Google Scholar] [CrossRef]
- Peng, W.; Tariq, K.; Xie, J.; Zhang, H. Identification and characterization of sex-biased microRNAs in Bactrocera dorsalis (Hendel). PLoS ONE 2016, 11, e0159591. [Google Scholar]
- Ge, W.; Deng, Q.; Guo, T.; Hong, X.; Kugler, J.M.; Yang, X.; Cohen, S.M. Regulation of pattern formation and gene amplification during Drosophila oogenesis by the miR-318 microRNA. Genetics 2015, 200, 255–265. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, S.; Xia, L.; Wang, J.; Wen, S.; Jin, P.; Chen, D. The bantam microRNA is associated with Drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009, 5, e1000444. [Google Scholar] [CrossRef]
- Eun, S.H.; Stoiber, P.M.; Wright, H.J.; McMurdie, K.E.; Choi, C.H.; Gan, Q.; Lim, C.; Chen, X. MicroRNAs downregulate Bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development 2013, 140, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Monahan, A.J.; Starz-Gaiano, M. Socs36E attenuates STAT signaling to optimize motile cell specification in the Drosophila ovary. Dev. Biol. 2013, 379, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Sokol, N.S.; Dubrovsky, E.B.; Berger, E.M.; Ambros, V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 2003, 259, 9–18. [Google Scholar] [CrossRef]
- Harari, A.R.; Sharon, R.; Weintraub, P.G. Chapter 6: Manipulation of insect reproductive systems as a tool in pest control. In Advances in Insect Control and Resistance Management; Springer: Cham, Switzerland, 2016; pp. 93–119. [Google Scholar] [CrossRef]
- Surridge, C. Pest control: Reproductive strategy. Nat. Plants 2017, 3, 17110. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, G.; Zotti, M.; Retnakaran, A. Targeting female reproduction in insects with biorational insecticides for pest management: A critical review with suggestions for future research. Curr. Opin. Insect Sci. 2019, 31, 65–69. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Liesner, L.R.; Ellsworth, P.C.; Unnithan, G.C.; Fabrick, J.A.; Naranjo, S.E.; Li, X.; Dennehy, T.J.; Antilla, L.; Staten, R.T.; et al. Transgenic cotton and sterile insect releases synergize eradication of pink bollworm a century after it invaded the United States. Proc. Natl. Acad. Sci. USA 2021, 118, e2019115118. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Rajamani, V.; Reddy, V.S.; Mukherjee, S.K.; Bhatnagar, R.K. Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: An alternative to Bt-toxin technology. Transgen. Res. 2015, 24, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Yogindran, S.; Rajam, M.V. Host-derived artificial miRNA-mediated silencing of ecdysone receptor gene provides enhanced resistance to Helicoverpa armigera in tomato. Genomics 2021, 113 Pt 2, 736–747. [Google Scholar] [CrossRef]
- Luo, J.; Liang, S.; Li, J.; Xu, Z.; Li, L.; Zhu, B.; Li, Z.; Lei, C.; Lindsey, K.; Chen, L.; et al. A transgenic strategy for controlling plant bugs (Adelphocoris suturalis) through expression of double-stranded RNA homologous to fatty acyl-coenzyme a reductase in cotton. New Phytol. 2017, 215, 1173–1185. [Google Scholar] [CrossRef]
- Fitt, G.P. The ecology of heliothis species in relation to agroecosystems. Annu. Rev. Entomol. 1989, 34, 17–53. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, K.; Jiang, Y.; Guo, Y.; Desneux, N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef]
- Wu, K.M.; Lu, Y.H.; Feng, H.Q.; Jiang, Y.Y.; Zhao, J.Z. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 2008, 321, 1676–1678. [Google Scholar] [CrossRef] [PubMed]
- Haile, F.; Nowatzki, T.; Storer, N. Overview of pest status, potential risk, and management considerations of Helicoverpa armigera (Lepidoptera: Noctuidae) for U.S. soybean production. J. Integr. Pest Manag. 2021, 12, 3. [Google Scholar] [CrossRef]
- Ge, X.; Zhang, Y.; Jiang, J.; Zhong, Y.; Yang, X.; Li, Z.; Huang, Y.; Tan, A. Identification of microRNAs in Helicoverpa armigera and Spodoptera litura based on deep sequencing and homology analysis. Int. J. Biol. Sci. 2013, 9, 1–15. [Google Scholar] [CrossRef][Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Ahmad, M.; Rasool, B.; Ahmad, M.; Russell, D.A. Resistance and synergism of novel insecticides in field populations of cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) in Pakistan. J. Econ. Entomol. 2019, 112, 859–871. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant. J. 2006, 46, 243–259. [Google Scholar] [CrossRef]
- Dilbar, H.; Muhammad, S.; Ghulam, G.; Muneer, A. Insecticide resistance in field populations of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). J. Entomol. Sci. 2015, 50, 119–128. [Google Scholar]
- Yang, Y.; Li, Y.; Wu, Y. Current status of insecticide resistance in Helicoverpa armigera after 15 years of Bt cotton planting in China. J. Econ. Entomol. 2013, 106, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Wu, K.; Wu, Y. Early detection of field-evolved resistance to Bt cotton in China: Cotton bollworm and pink bollworm. J. Invertebr. Pathol. 2012, 110, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Chakroun, M.; Banyuls, N.; Walsh, T.; Downes, S.; James, B.; Ferre, J. Characterization of the resistance to Vip3Aa in Helicoverpa armigera from Australia and the role of midgut processing and receptor binding. Sci. Rep. 2016, 6, 24311. [Google Scholar] [CrossRef]
- Callahan, P.S. Behavior of the Imago of the Corn Earworm, Heliothis Zea (Boddie), with Special Reference to Emergence and Reproduction. Ann. Entomol. Soc. Am. 1958, 51, 271–283. [Google Scholar] [CrossRef]
- Liu, L.; Feng, Q. The study on fusion of testis in lepidoptera insects. J. South China Norm. Univ. 2014, 46, 1–7. [Google Scholar]
- Lu, T. A study on the reproductive system of Agrotis ypsilon ROTT. Acta Entomol. Sin. 1982, 25, 268–274. [Google Scholar]
- Chaudhury, M.F.B.; Raun, E.S. Spermatogenesis and Testicular Development of the European Corn Borer, Ostrinia nubilalis (Lepidoptera: Pyraustidae). Ann. Entomol. Soc. Am. 1966, 59, 1157–1159. [Google Scholar] [CrossRef]
- Chen, D.F.; Niu, B.L.; Weng, H.B. Study on larval sexual character and spermatogenesis of cotton bollworm (Helicoverpa armigera) Acta Agric. Zhejiangensis 2004, 16, 354–357. [Google Scholar]
- Alves, L.; Mancini, K.; Lino-Neto, J.; Dolder, H. Morphology of the male reproductive system and sperm ultrastructure of Leucoptera coffeella (Lepidoptera: Lyonetiidae). Acta Zool. 2010, 87, 131–139. [Google Scholar] [CrossRef]
- Zhang, W.N.; Xiao, H.J.; Liang, G.M.; Guo, Y.Y. Observation on ovarian morphology and oogenesis in the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). Acta Entomol. Sin. 2013, 56, 358–364. [Google Scholar]
- Song, J.; Zhou, S. Post-transcriptional regulation of insect metamorphosis and oogenesis. Cell Mol. Life Sci. 2020, 77, 1–17. [Google Scholar] [CrossRef]
- Tariq, K.; Metzendorf, C.; Peng, W.; Sohail, S.; Zhang, H. miR-8-3p regulates mitoferrin in the testes of Bactrocera dorsalis to ensure normal spermatogenesis. Sci. Rep. 2016, 6, 22565. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Xu, J.; Ling, L.; Yang, D.H.; Chen, S.Q.; Huang, Y.P. MicroRNA-2738 regulates gene expression in the sex determination pathway in Bombyx mori. Insect Sci. 2020, 27, 646–654. [Google Scholar] [CrossRef]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013, 3, 1493–1509. [Google Scholar] [CrossRef]
- Liu, W.; Huang, H.; Xing, C.; Li, C.; Tan, F.; Liang, S. Identification and characterization of the expression profile of microRNAs in Anopheles anthropophagus. Parasites Vectors 2014, 7, 159. [Google Scholar] [CrossRef][Green Version]
- Winter, F.; Edaye, S.; Huttenhofer, A.; Brunel, C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007, 35, 6953–6962. [Google Scholar] [CrossRef]
- Quah, S.; Breuker, C.J.; Holland, P.W. A diversity of conserved and novel ovarian microRNAs in the speckled wood (Pararge aegeria). PLoS ONE 2015, 10, e0142243. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Yang, M.; Zheng, K.; Wang, Z.; Gu, S.; Tong, J.; Liu, J.; Shah, N.A.; Nie, L. Comparison of adult testis and ovary microRNA expression profiles in Reeves’ Pond Turtles (Mauremys reevesii) with temperature-dependent sex determination. Front. Genet. 2020, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; He, P.; Zhang, X.; Li, W.; Zhang, L.; Guan, J.; Chen, X.; Lin, Y.; Zhuo, X.; Li, Q.; et al. Identification and characterization of microRNAs in the gonads of Crassostrea hongkongensis using high-throughput sequencing. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100606. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 32–42. [Google Scholar] [CrossRef]
- Traut, W.; Vogel, H.; Glockner, G.; Hartmann, E.; Heckel, D.G. High-throughput sequencing of a single chromosome: A moth W chromosome. Chromosome Res. 2013, 21, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Zhang, Y.; Zhang, M.; Huang, J.; Li, C.; Ni, X.; Li, X. Characterization of the first W-specific protein-coding gene for sex identification in Helicoverpa armigera. Front. Genet. 2020, 11, 649. [Google Scholar] [CrossRef]
- Kiuchi, T.; Koga, H.; Kawamoto, M.; Shoji, K.; Sakai, H.; Arai, Y.; Ishihara, G.; Kawaoka, S.; Sugano, S.; Shimada, T.; et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014, 509, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Hetie, P.; Matunis, E.L. Niche signaling promotes stem cell survival in the Drosophila testis via the JAK-STAT target DIAP1. Dev. Biol. 2015, 404, 27–39. [Google Scholar] [CrossRef]
- Al Baki, M.A.; Lee, D.W.; Jung, J.K.; Kim, Y. Insulin signaling mediates previtellogenic development and enhances juvenile hormone-mediated vitellogenesis in a lepidopteran insect, Maruca vitrata. BMC Dev. Biol. 2019, 19, 14. [Google Scholar] [CrossRef] [PubMed]


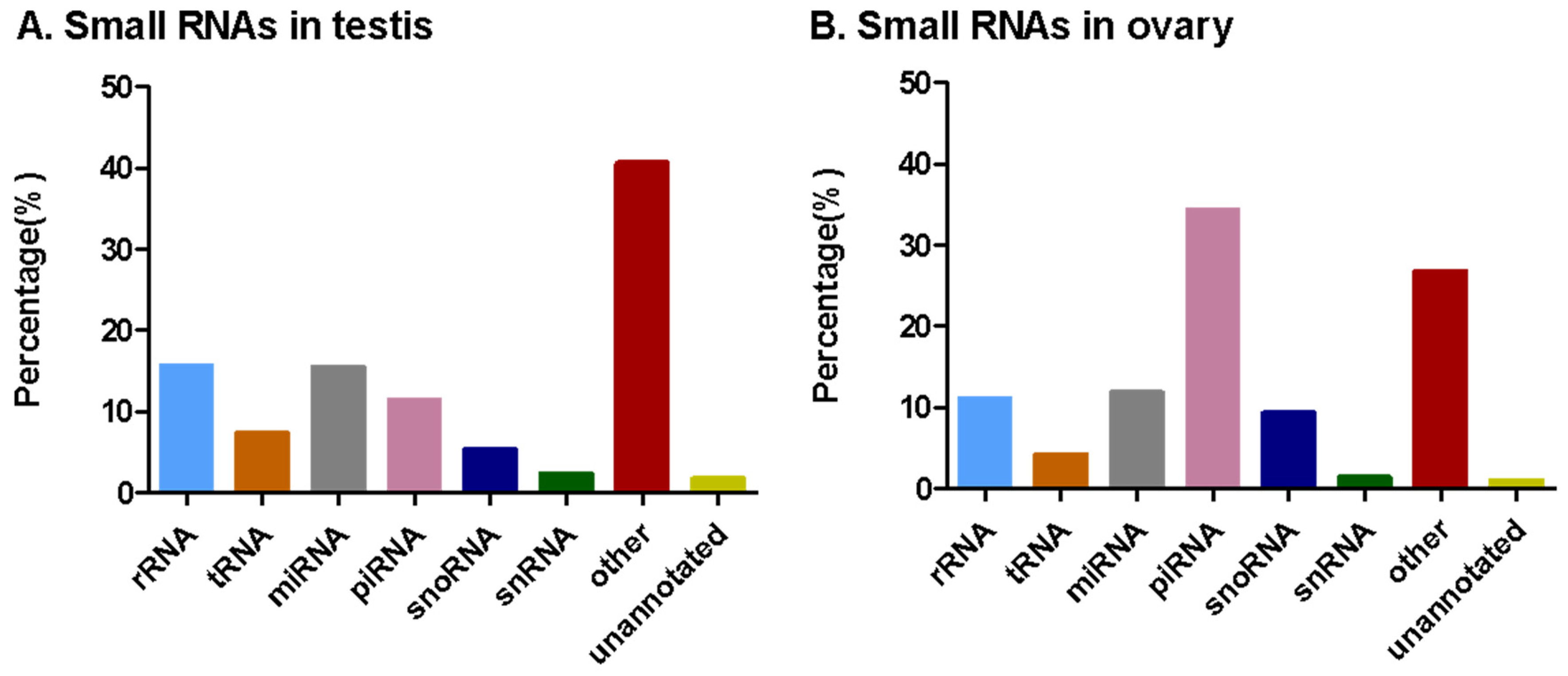


| Ovary | Testis | |||
|---|---|---|---|---|
| Type | Read Count | Percentage (%) | Read Count | Percentage (%) |
| Total | 7,592,150 | 100 | 8,815,237 | 100 |
| Unannotated | 76,990 | 1.01 | 152,387 | 1.73 |
| rRNA | 842,672 | 11.10 | 1,379,525 | 15.65 |
| snRNA | 106,536 | 1.40 | 205,099 | 2.33 |
| snoRNA | 706,253 | 9.30 | 469,250 | 5.32 |
| tRNA | 312,634 | 4.18 | 649,231 | 7.36 |
| miRNA | 899,621 | 11.85 | 1,362,386 | 15.45 |
| piRNA | 2,611,968 | 34.40 | 1,012,824 | 11.49 |
| Other | 2,035,476 | 26.81 | 3,584,535 | 40.66 |
| miRNA | Number of Putative Target Transcripts |
|---|---|
| miR-998 | 4978 |
| miR-989a | 10,670 |
| miR-34 | 12,041 |
| miR-2c | 3194 |
| miR-2765 | 2239 |
| miR-2763 | 324 |
| miR-252a-5p | 2134 |
| miR-263b-5p | 5474 |
| Total | 41,054 |
| Shared by 2 or more miRNA | 10,882 |
| Total—shared | 30,172 |
| KEGG_A_Class | KEGG_B_Class | Pathway | Transcript Number | Pathway ID |
|---|---|---|---|---|
| Cellular Processes | Cell growth and death | Oocyte meiosis | 411 | ko04114 |
| Environmental Information Processing | Signal transduction | mTOR signaling pathway | 653 | ko04150 |
| JAK-STAT signaling pathway | 166 | ko04630 | ||
| Metabolism | Lipid metabolism | Steroid hormone biosynthesis | 285 | ko00140 |
| Metabolism of terpenoids and polyketides | Insect hormone biosynthesis | 62 | ko00981 | |
| Organismal Systems | Endocrine system | Insulin signaling pathway | 711 | ko04910 |
| GnRH signaling pathway | 385 | ko04912 | ||
| Ovarian steroidogenesis | 100 | ko04913 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Wang, S.; Huang, K.; Zhang, Y.; Li, Y.; Zhang, M.; Huang, J.; Deng, Z.; Ni, X.; Li, X. Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae). Insects 2021, 12, 749. https://doi.org/10.3390/insects12080749
Li L, Wang S, Huang K, Zhang Y, Li Y, Zhang M, Huang J, Deng Z, Ni X, Li X. Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae). Insects. 2021; 12(8):749. https://doi.org/10.3390/insects12080749
Chicago/Turabian StyleLi, Leyao, Shan Wang, Kaiyuan Huang, Yuting Zhang, Yalu Li, Min Zhang, Jinyong Huang, Zhongyuan Deng, Xinzhi Ni, and Xianchun Li. 2021. "Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae)" Insects 12, no. 8: 749. https://doi.org/10.3390/insects12080749
APA StyleLi, L., Wang, S., Huang, K., Zhang, Y., Li, Y., Zhang, M., Huang, J., Deng, Z., Ni, X., & Li, X. (2021). Identification and Characterization of MicroRNAs in Gonads of Helicoverpa armigera (Lepidoptera: Noctuidae). Insects, 12(8), 749. https://doi.org/10.3390/insects12080749