Upregulation of Transferrin and Major Royal Jelly Proteins in the Spermathecal Fluid of Mated Honeybee (Apis mellifera) Queens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
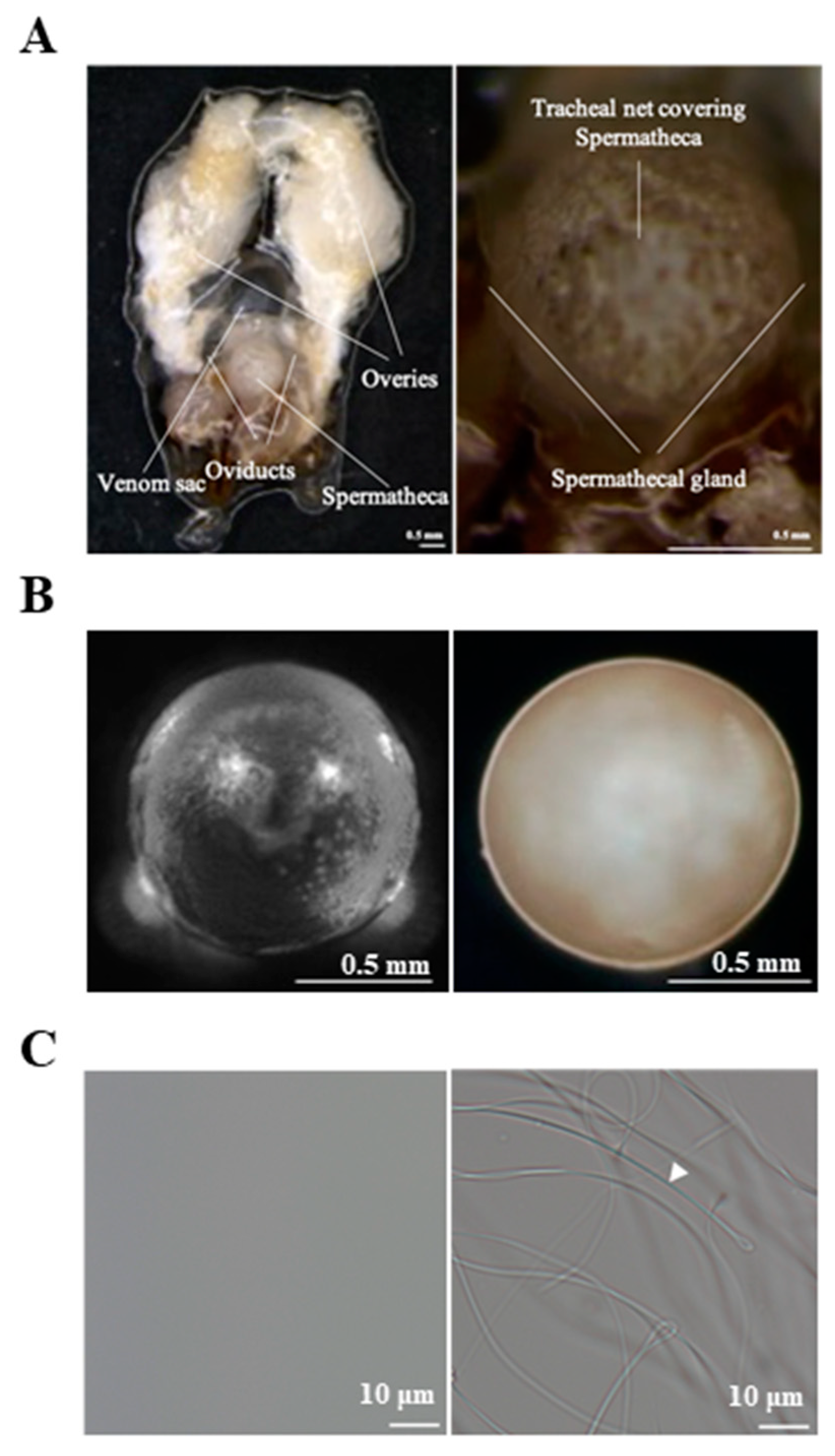
2.1. Sample Preparation
2.2. RNA Sequencing and Data Analysis
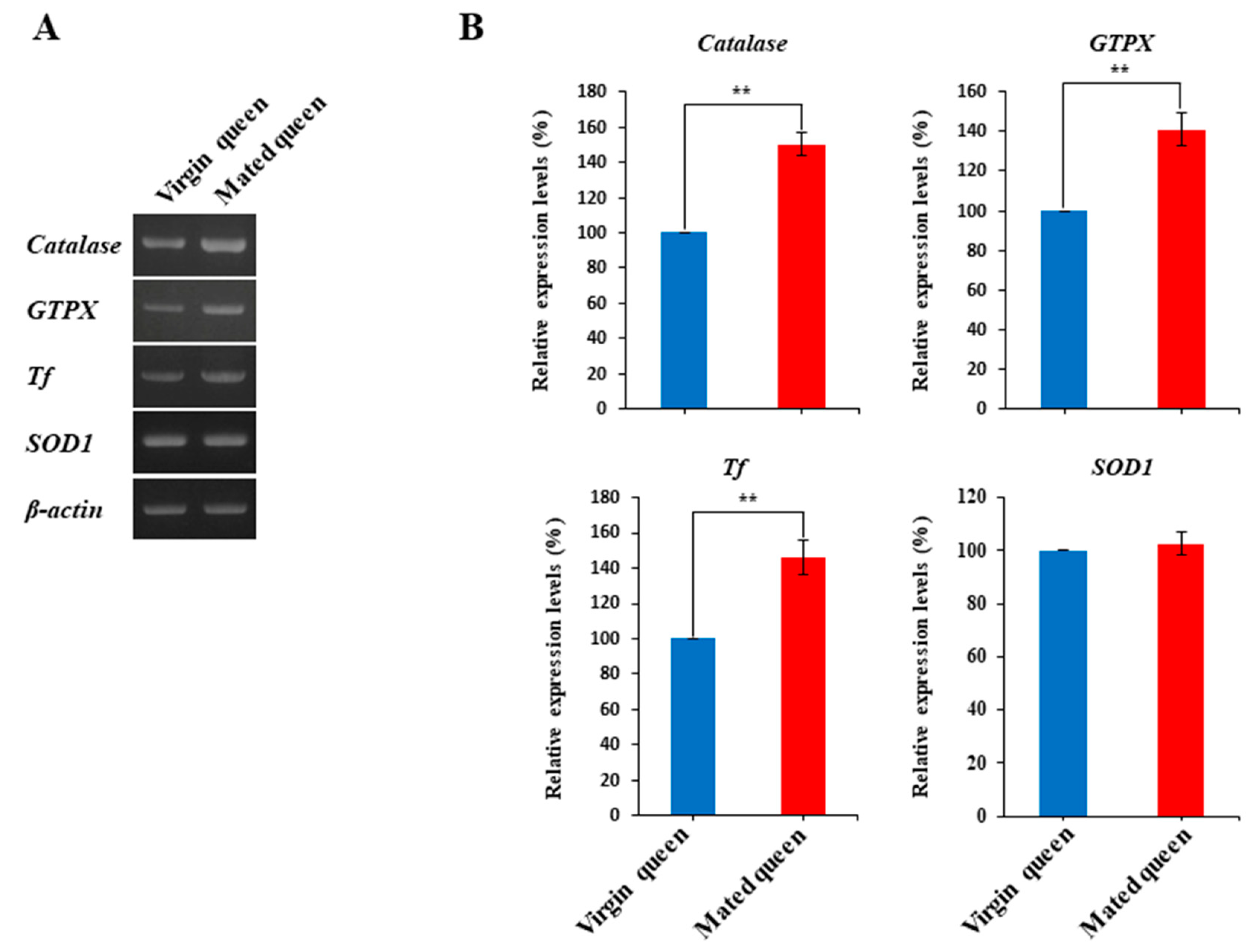
2.3. Reverse Transcription-PCR (RT-PCR)
2.4. Recombinant Protein Expression and Purification
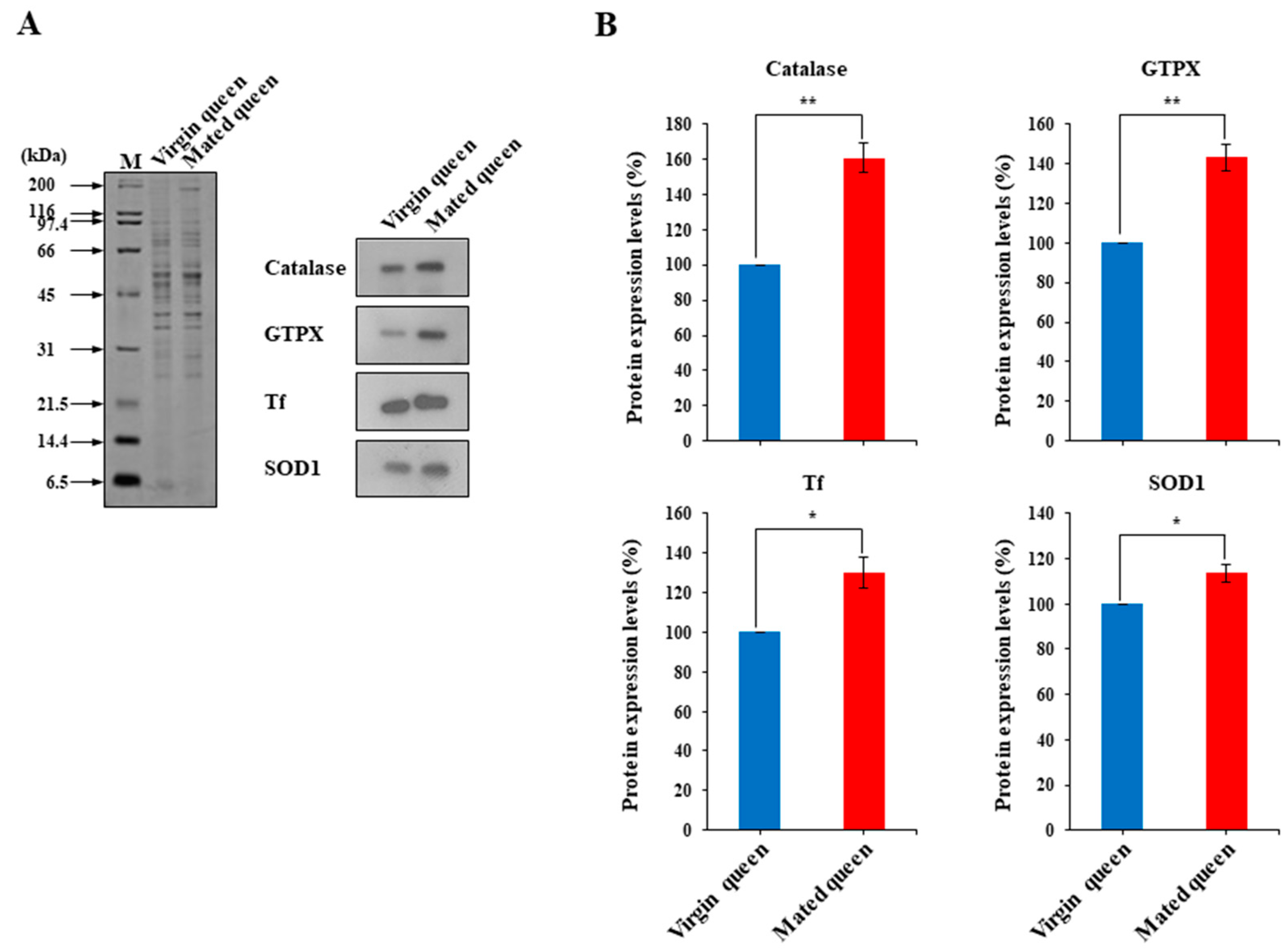
2.5. Polyclonal Antibody Production and Western Blotting
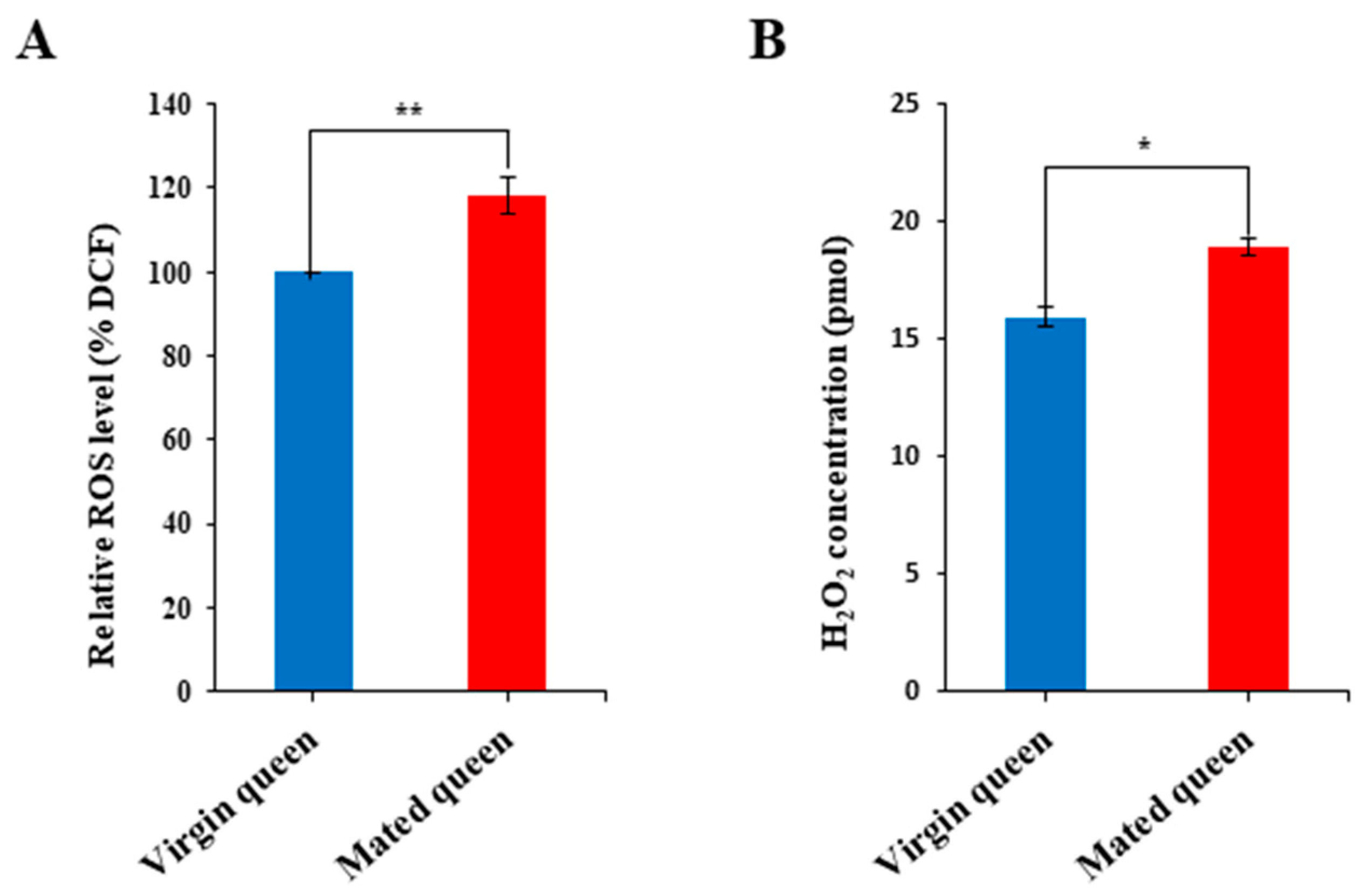
2.6. Measurement of ROS and Iron Levels
2.7. Enzymatic Activity Assay
2.8. Statistical Analysis
3. Results
3.1. Identification of Genes Differentially Expressed between the Mated and Virgin Queens’ Spermathecae
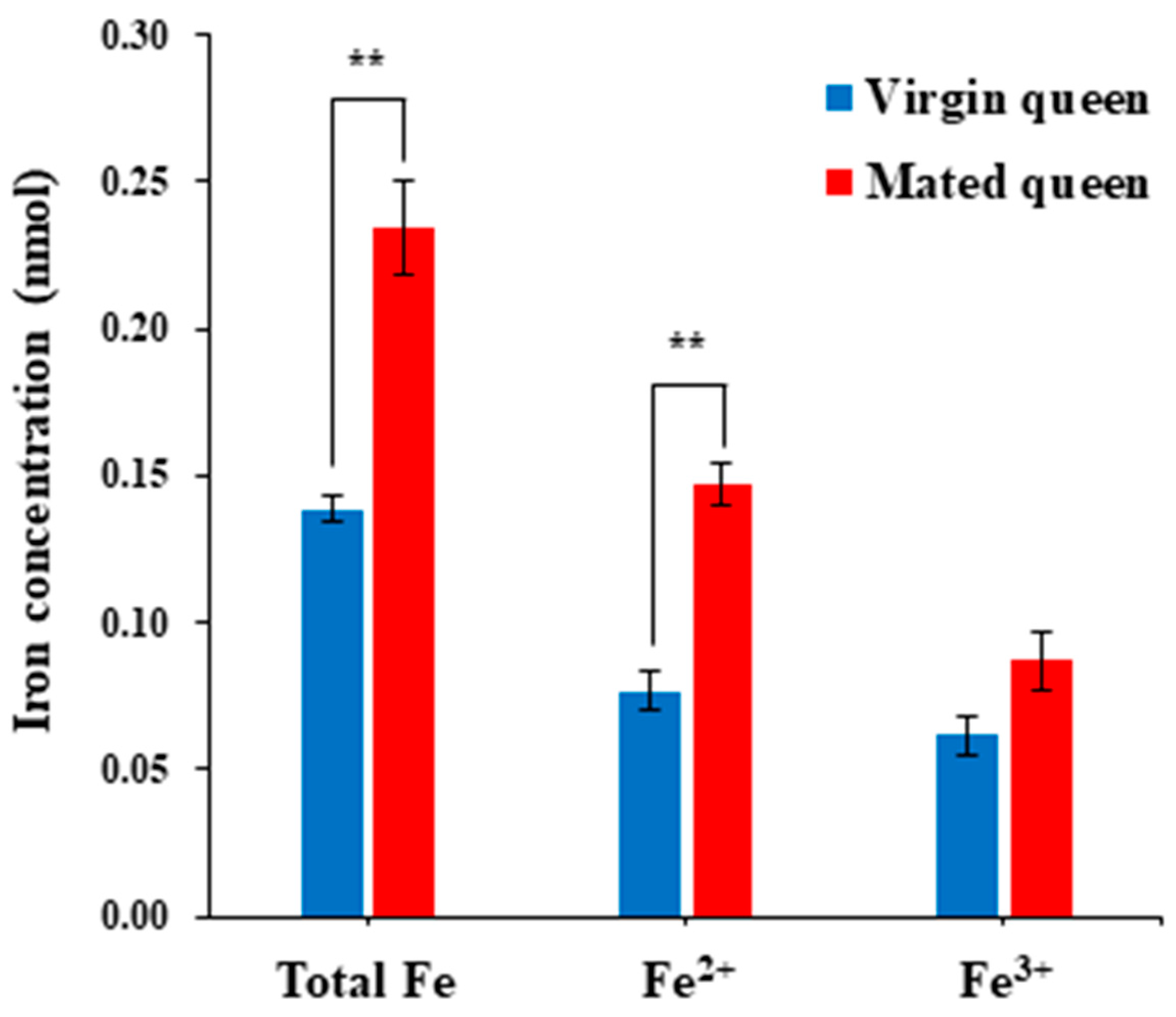
3.2. Antioxidant Defense in the Spermathecae of Mated Queens and Virgin Queens
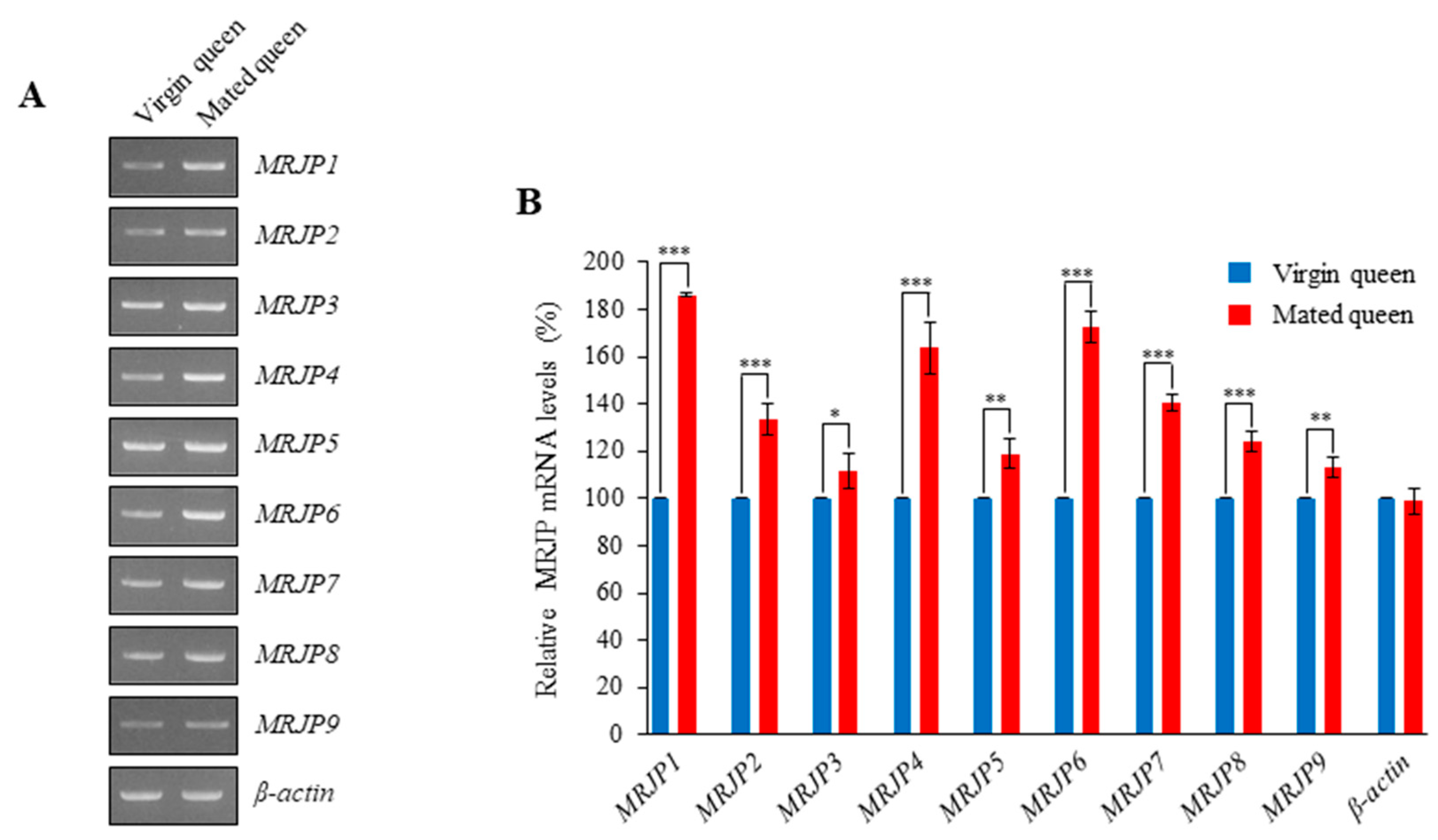
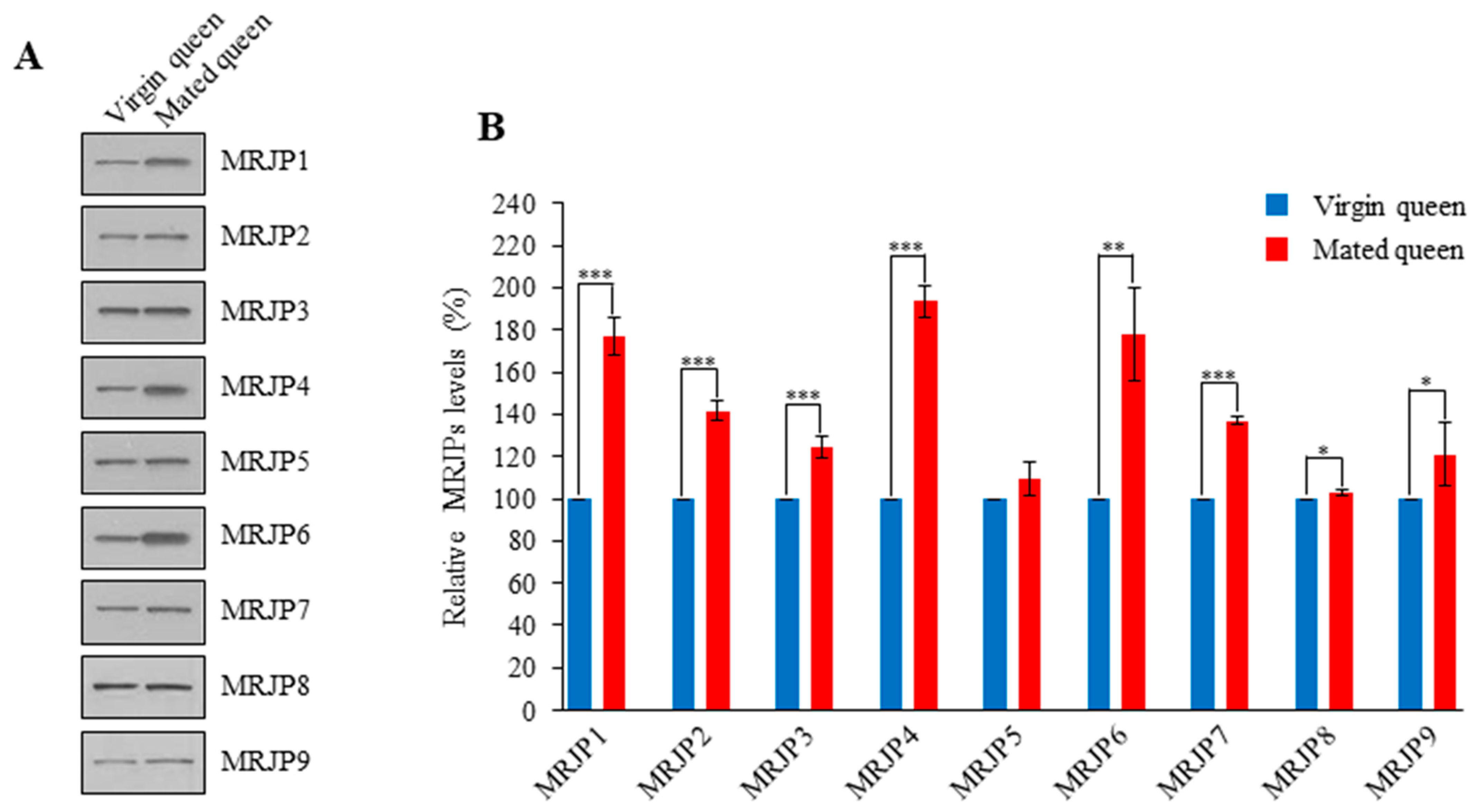
3.3. MRJPs Are Differentially Expressed in the Spermathecae of Mated Queens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Collins, A.M.; Caperna, T.J.; Williams, V.; Garrett, W.M.; Evans, J.D. Proteomic analyses of male contributions to honey bee sperm storage and mating. Insect Mol. Biol. 2006, 15, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Baer, B.; Eubel, H.; Taylor, N.L.; O’Toole, N.; Millar, A.H. Insights into female sperm storage from the spermathecal fluid proteome of the honeybee Apis mellifera. Genome Biol. 2009, 10, R67. [Google Scholar] [CrossRef] [PubMed]
- Klenk, M.; Koeniger, G.; Koeniger, N.; Fasold, H. Proteins in spermathecal gland secretion and spermathecal fluid and the properties of a 29 kDa protein in queens of Apis mellifera. Apidologie 2004, 35, 371–381. [Google Scholar] [CrossRef][Green Version]
- Paynter, E.; Millar, A.H.; Welch, M.; Baer-Imhoof, B.; Cao, D.; Baer, B. Insights into the molecular basis of long-term storage and survival of sperm in the honeybee (Apis mellifera). Sci. Rep. 2017, 7, 40236. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, F.; Li, G.; Chi, X.; Wang, Y.; Wang, H.; Ma, L.; Han, K.; Zhao, G.; Guo, X.; et al. Metabolite support of long-term storage of sperm in the spermatheca of honeybee (Apis mellifera) queens. Front. Physiol. 2020, 11, 574856. [Google Scholar] [CrossRef]
- den Boer, S.P.; Boomsma, J.J.; Baer, B. Seminal fluid enhances sperm viability in the leafcutter ant Atta colombica. Behav. Ecol. Sociobiol. 2008, 62, 1843–1849. [Google Scholar] [CrossRef]
- den Boer, S.P.; Boomsma, J.J.; Baer, B. Honey bee males and queens use glandular secretions to enhance sperm viability before and after storage. J. Insect Physiol. 2009, 55, 538–543. [Google Scholar] [CrossRef]
- Weirich, G.F.; Collins, A.M.; Williams, V.P. Antioxidant enzymes in the honey bee, Apis mellifera. Apidologie 2002, 33, 3–14. [Google Scholar] [CrossRef]
- Collins, A.M.; Williams, V.; Evans, J.D. Sperm storage and antioxidative enzyme expression in the honey bee, Apis mellifera. Insect Mol. Biol. 2004, 13, 141–146. [Google Scholar] [CrossRef]
- Gonzalez, A.N.; Ing, N.; Rangel, J. Upregulation of antioxidant genes in the spermathecae of honey bee (Apis mellifera) queens after mating. Apidologie 2018, 49, 1–11. [Google Scholar] [CrossRef]
- Al-Lawati, H.; Kamp, G.; Bienefeld, K. Characteristics of the spermathecal contents of old and young honeybee queens. J. Insect Physiol. 2009, 55, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Zareie, R.; Eubel, H.; Millar, A.H.; Baer, B. Long-term survival of high quality sperm: Insights into the sperm proteome of the honeybee Apis mellifera. J. Proteome Res. 2013, 12, 5180–5188. [Google Scholar] [CrossRef]
- McAfee, A.; Chapman, A.; Higo, H.; Underwood, R.; Milone, J.; Foster, L.J.; Guarna, M.M.; Tarpy, D.R.; Pettis, J.S. Vulnerability of honey bee queens to heat-induced loss of fertility. Nat. Sustain. 2020, 3, 367–376. [Google Scholar] [CrossRef]
- Rangel, J.; Shepherd, T.F.; Gonzalez, A.N.; Hillhouse, A.; Konganti, K.; Ing, N.H. Transcriptomic analysis of the honey bee (Apis mellifera) queen spermathecae reveals genes that may be involved in sperm storage after mating. PLoS ONE 2021, 16, e0244648. [Google Scholar] [CrossRef] [PubMed]
- Ribou, A.C.; Reinhardt, K. Reduced metabolic rate and oxygen radicals production in stored insect sperm. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2012, 279, 2196–2203. [Google Scholar] [CrossRef]
- Poland, V.; Eubel, H.; King, M.; Solheim, C.; Harvey Millar, A.; Baer, B. Stored sperm differs from ejaculated sperm by proteome alterations associated with energy metabolism in the honeybee Apis mellifera. Mol. Ecol. 2011, 20, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
- Crichton, R.R.; Wilmet, S.; Legssyer, R.; Ward, R.J. Molecular and cellular mechanisms of iron homeostasis and toxicity in mammalian cells. J. Inorg. Biochem. 2002, 91, 9–18. [Google Scholar] [CrossRef]
- Fenoglio, I.; Tomatis, M.; Lison, D.; Muller, J.; Fonseca, A.; Nagy, J.B.; Fubini, B. Reactivity of carbon nanotubes: Free radical generation of scavenging activity? Free Radic. Biol. Med. 2006, 40, 1227–1233. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, B.Y.; Kim, H.J.; Seo, S.J.; Yoon, H.J.; Choi, Y.S.; Kim, I.; Han, Y.S.; Je, Y.H.; Lee, S.M.; et al. Transferrin inhibits stress-induced apoptosis in a beetle. Free. Radic. Biol. Med. 2006, 41, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.Y.; Lee, K.S.; Choo, Y.M.; Kim, I.; Je, Y.H.; Woo, S.D.; Lee, S.M.; Park, H.C.; Son, H.D.; Jin, B.R. Insect transferrin functions as an antioxidant protein in a beetle larva. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 150, 161–169. [Google Scholar] [CrossRef]
- Kim, B.Y.; Lee, K.S.; Yoon, H.J.; Kim, I.; Li, J.H.; Sohn, H.D.; Jin, B.R. Expression profile of the iron binding proteins transferrin and ferritin heavy chain subunit in the bumblebee Bombus ignitus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 153, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.L.; Winzerling, J.J. Insect transferrins: Multifunctional proteins. Biochim. Biophys. Acta 2012, 1820, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Harmen, P.H.; John, N.C.; Willem, J.B. Stimulating natural supersedure of honeybee queens, Apis mellifera. Proc. Neth. Entomol. Soc. 2004, 15, 29–33. [Google Scholar]
- Schley, P. Einführung in die Technik der Instrumentellen Besamung von Bienenköniginnen, 2nd ed.; Köhler Offset KG: Giessen, Germany, 1987; p. 103. [Google Scholar]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Park, H.G.; Kim, B.Y.; Park, M.J.; Deng, Y.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Antibacterial activity of major royal jelly proteins of the honeybee (Apis mellifera) royal jelly. J. Asia Pac. Entomol. 2019, 22, 737–741. [Google Scholar] [CrossRef]
- Kim, B.Y. Antiapoptotic role of major royal jelly protein 8 of honeybee (Apis mellifera) venom. J. Asia Pac. Entomol. 2021, 24. [Google Scholar] [CrossRef]
- Park, H.G.; Lee, K.S.; Kim, B.Y.; Yoon, H.J.; Choi, Y.S.; Lee, K.Y.; Wan, H.; Li, J.; Jin, B.R. Honeybee (Apis cerana) vitellogenin acts as an antimicrobial and antioxidant agent in the body and venom. Dev. Comp. Immunol. 2018, 85, 51–60. [Google Scholar] [CrossRef]
- Je, Y.H.; Chang, J.H.; Kim, M.H.; Roh, J.Y.; Jin, B.R.; O’Reilly, D.R.; Kang, S.K. A defective viral genome maintained in Escherichia coli for the generation of baculovirus expression vectors. Biotechnol. Lett. 2001, 23, 575–582. [Google Scholar] [CrossRef]
- Liochev, S.I.; Fridovich, I. The effects of superoxide dismutase on H2O2 formation. Free Radic. Biol. Med. 2007, 42, 1465–1469. [Google Scholar] [CrossRef]
- Liochev, S.I.; Fridovich, I. Mechanism of the peroxidase activity of Cu, Zn superoxide dismutase. Free Radic. Biol. Med. 2010, 48, 1565–1569. [Google Scholar] [CrossRef]
- Baer, B.; Heazlewood, J.L.; Taylor, N.L.; Eubel, H.; Millar, A.H. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics 2009, 9, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Bantleon, F.I.; McIntyre, M.; Oliert, M.; Spiliner, E. The major royal jelly proteins 8 and 9 (Api m 11) are glycosylated components of Apis mellifera venom with allergenic potential beyond carbohydrate-based reactivity. Clin. Exp. Allergy 2012, 42, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Fratini, F.; Cilia, G.; Mancini, S.; Felicioli, A. Royal jelly: An ancient remedy with remarkable antibacterial properties. Microbiol. Res. 2016, 192, 130–141. [Google Scholar] [CrossRef]
- Tamura, S.; Amano, S.; Kono, T.; Kondoh, J.; Yamaguchi, K.; Kobayashi, S.; Atabe, T.; Moriyama, T. Molecular characteristics and physiological functions of major royal jelly protein 1 oligomer. Proteomics 2009, 9, 5534–5543. [Google Scholar] [CrossRef]
- Ramanathan, A.N.K.G.; Nair, A.J.; Sugunan, V.S. A review on royal jelly proteins and peptides. J. Funct. Foods 2018, 44, 255–264. [Google Scholar] [CrossRef]
- Bílikova, K.; Wub, G.; Šimuth, J. Isolation of a peptide fraction from honeybee royal jelly as a potential antifoulbrood factor. Apidologie 2001, 32, 275–283. [Google Scholar] [CrossRef]
- Kim, B.Y.; Lee, K.S.; Jung, B.; Choi, Y.S.; Kim, H.K.; Yoon, H.J.; Gui, Z.Z.; Lee, J.; Jin, B.R. Honeybee (Apis cerana) major royal jelly protein 4 exhibits antimicrobial activity. J. Asia Pac. Entomol. 2019, 22, 175–182. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, B.Y.; Park, H.G.; Deng, Y.; Yoon, H.J.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Major royal jelly protein 2 acts as an antimicrobial and antioxidant in royal jelly. J. Asia Pac. Entomol. 2019, 22, 684–689. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, B.Y.; Deng, Y.; Park, H.G.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Antioxidant capacity of major royal jelly proteins of honeybee (Apis mellifera) royal jelly. J. Asia Pac. Entomol. 2020, 23, 445–448. [Google Scholar] [CrossRef]

| Gene | Primer Sequence | Product Length (bp) |
|---|---|---|
| Catalase | forward 5ʹ-ATGACTGAAATAAAACGA-3ʹ | 1542 |
| reverse 5ʹ-TCACAATCTAGCTGTCTTGCCATAA-3ʹ | ||
| GTPX | forward 5′-ATGAGTGGAAACGACAAC-3′ | 507 |
| reverse 5′-TTAAAAATATTTTTCAAGATGATTT-3′ | ||
| Tf | forward 5′-ATGATGCTCCGATGCAAT-3′ | 2139 |
| reverse 5′-TTAAGCAGCACCGTAATTATTT-3′ | ||
| SOD1 | forward 5′-ATGACTAAAGCAGTGTGC-3′ | 459 |
| reverse 5′-TTAGACTTTTGTAATTCCAA-3′ | ||
| MRJP1 | forward 5′-ATGACAAGATTGTTTATG-3′ | 1299 |
| reverse 5′-TTACAAATGGATTGAAATTTTGAAA-3′ | ||
| MRJP2 | forward 5′-ATGACAAGGTGGTTGTTC-3′ | 1359 |
| reverse 5′-TTAATTATCATTCTGATTGTTATTC-3′ | ||
| MRJP3 | forward 5′-ATGACAAAGTGGTTGTTG-3′ | 1635 |
| reverse 5′-TTAATGTAATTTTGAAGAATGATGA-3′ | ||
| MRJP4 | forward 5′-ATGACAAAATGGTTGCTG-3′ | 1395 |
| reverse 5′-TTAATCGTTATTGTTATGCCGATTG-3′ | ||
| MRJP5 | forward 5′-ATGACAACTTGGTTGTTG-3′ | 1797 |
| reverse 5′-TTAATTATTATGCTTATTTTGATTG-3′ | ||
| MRJP6 | forward 5′-ATGACAAATTGGTTACTG-3′ | 1314 |
| reverse 5′-CTAATCTAAATGAGCTTGATTCTTA-3′ | ||
| MRJP7 | forward 5′-ATGACAAGGTGGTTGTTT-3′ | 1332 |
| reverse 5′-CTAATTAAATGAAGCGTCTATTGTG-3′ | ||
| MRJP8 | forward 5′-ATGATAAGATGGTTGCTG-3′ | 1251 |
| reverse 5′-TCAAGGAGATATGCAACGAGTATTC-3′ | ||
| MRJP9 | forward 5′-ATGTCTTTCAATATCTGG-3′ | 1272 |
| reverse 5′-TCAAAGGAAAATTGAGAAAAAATTT-3′ | ||
| β–Actin | forward 5′-ATGTCTGACGAAGAAGTT-3′ | 1131 |
| reverse 5′-TTAGAAGCACTTCCTGTGGA-3′ |
| Protein | Gene ID | Fold Change |
|---|---|---|
| Mated Queen/Virgin Queen | ||
| Tf | GB50226 | 2.51 |
| Catalase | GB41427 | 2.77 |
| GTPX | GB47478 | 2.07 |
| MRJP1 | GB55205 | 2.78 |
| MRJP4 | GB55206 | 3.00 |
| MRJP6 | GB55207 | 4.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.-G.; Kim, B.-Y.; Kim, J.-M.; Choi, Y.-S.; Yoon, H.-J.; Lee, K.-S.; Jin, B.-R. Upregulation of Transferrin and Major Royal Jelly Proteins in the Spermathecal Fluid of Mated Honeybee (Apis mellifera) Queens. Insects 2021, 12, 690. https://doi.org/10.3390/insects12080690
Park H-G, Kim B-Y, Kim J-M, Choi Y-S, Yoon H-J, Lee K-S, Jin B-R. Upregulation of Transferrin and Major Royal Jelly Proteins in the Spermathecal Fluid of Mated Honeybee (Apis mellifera) Queens. Insects. 2021; 12(8):690. https://doi.org/10.3390/insects12080690
Chicago/Turabian StylePark, Hee-Geun, Bo-Yeon Kim, Jin-Myung Kim, Yong-Soo Choi, Hyung-Joo Yoon, Kwang-Sik Lee, and Byung-Rae Jin. 2021. "Upregulation of Transferrin and Major Royal Jelly Proteins in the Spermathecal Fluid of Mated Honeybee (Apis mellifera) Queens" Insects 12, no. 8: 690. https://doi.org/10.3390/insects12080690
APA StylePark, H.-G., Kim, B.-Y., Kim, J.-M., Choi, Y.-S., Yoon, H.-J., Lee, K.-S., & Jin, B.-R. (2021). Upregulation of Transferrin and Major Royal Jelly Proteins in the Spermathecal Fluid of Mated Honeybee (Apis mellifera) Queens. Insects, 12(8), 690. https://doi.org/10.3390/insects12080690





