Characterization and Functional Analysis of trehalase Related to Chitin Metabolism in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae)
Abstract
Simple Summary
Abstract
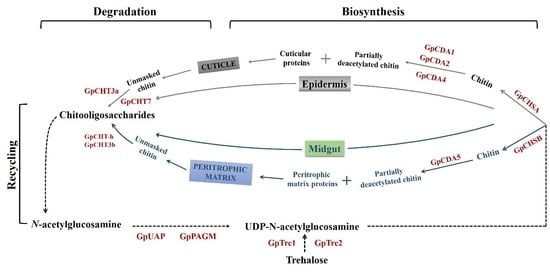
1. Introduction
2. Materials and Methods
2.1. G. pyloalis Raring and Samples Preparation
2.2. Bioinformatics Analysis of GpTre1 and GpTre2
2.3. RNA Isolation and cDNA Synthesis
2.4. RT-qPCR Analysis of GpTre1 and GpTre2 Expression Levels
2.5. dsRNA Synthesis and Injection
2.6. Analysis of the Effect of Validamycin A on GpTre1 and GpTre2 Expressions and Chitin Metabolism
2.7. Determination of Trehalose, Glycogen, and Glucose Content
2.8. Statistical Analyses
3. Results
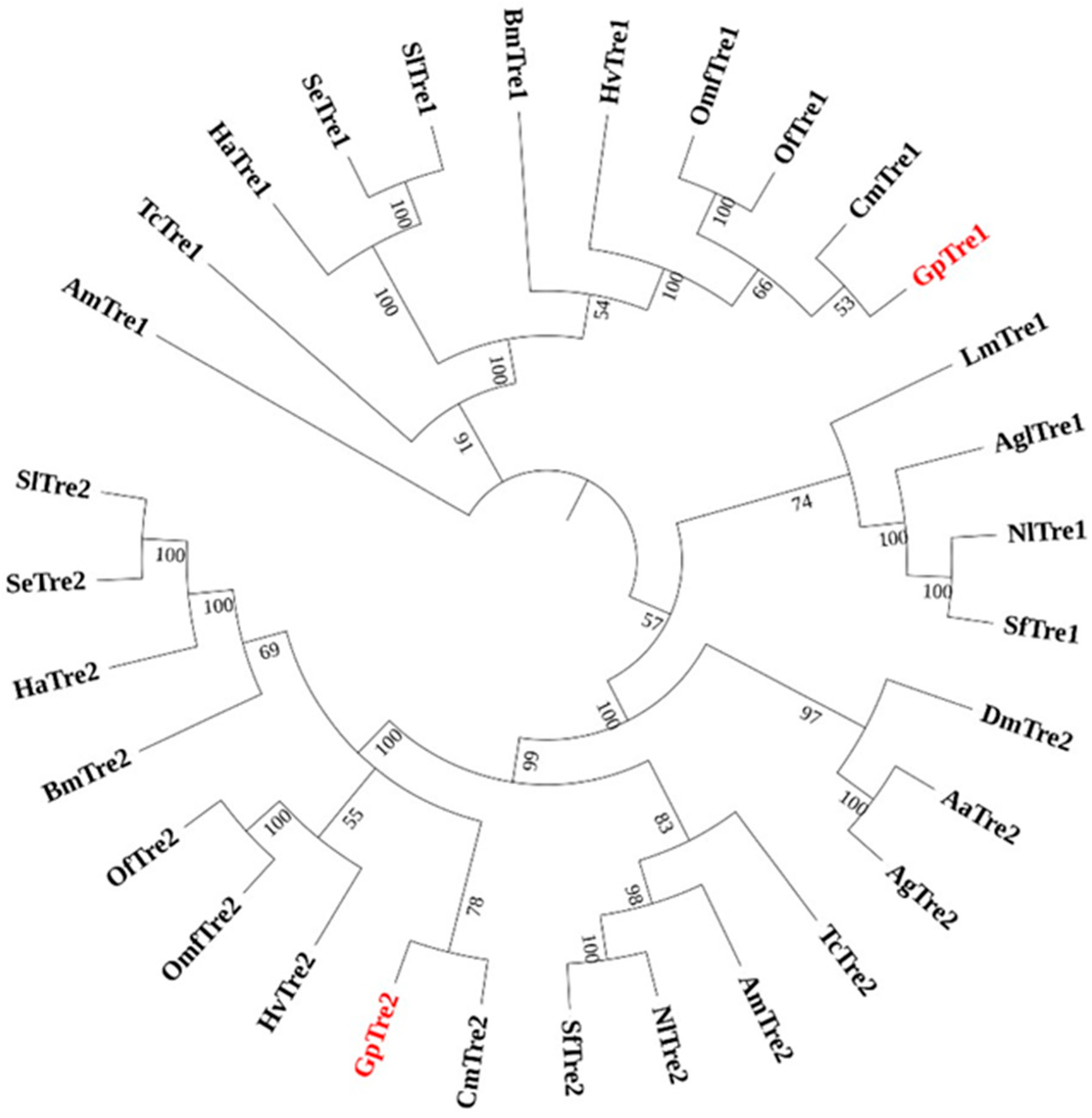
3.1. Gene Identification and Sequence Analyses
3.2. Spatiotemporal Expression Profiles Analysis of GpTre1 and GpTre2
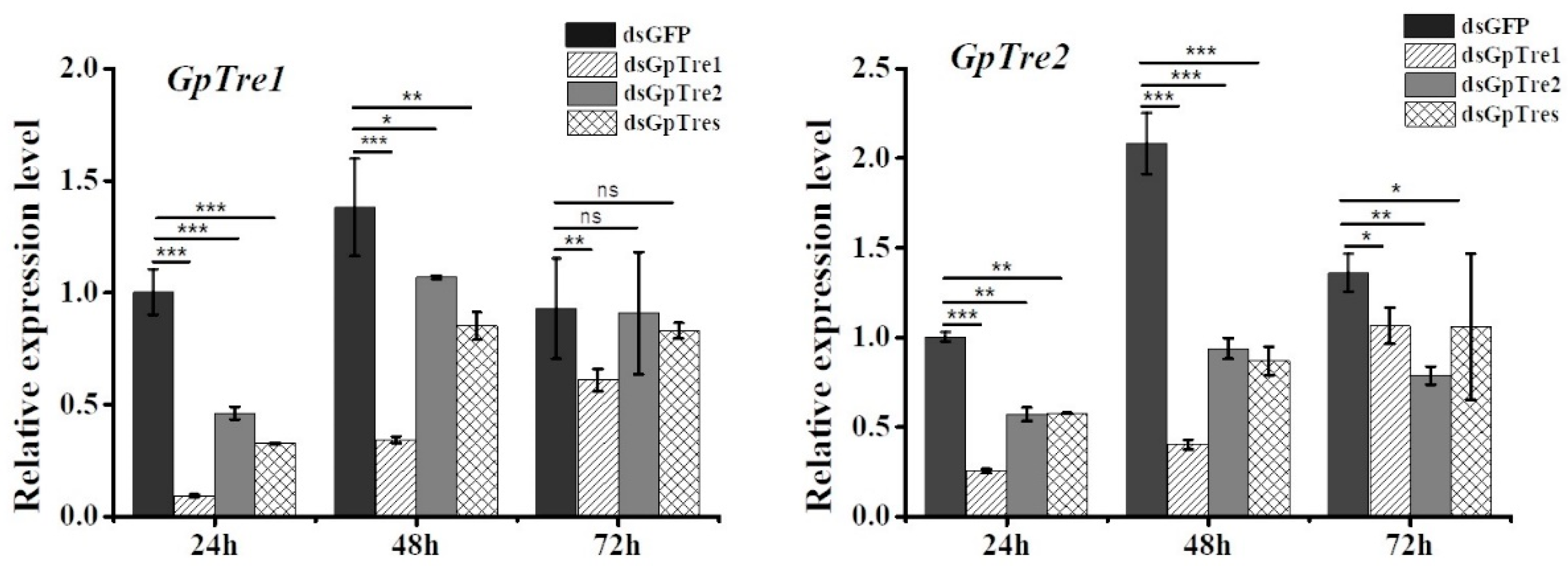
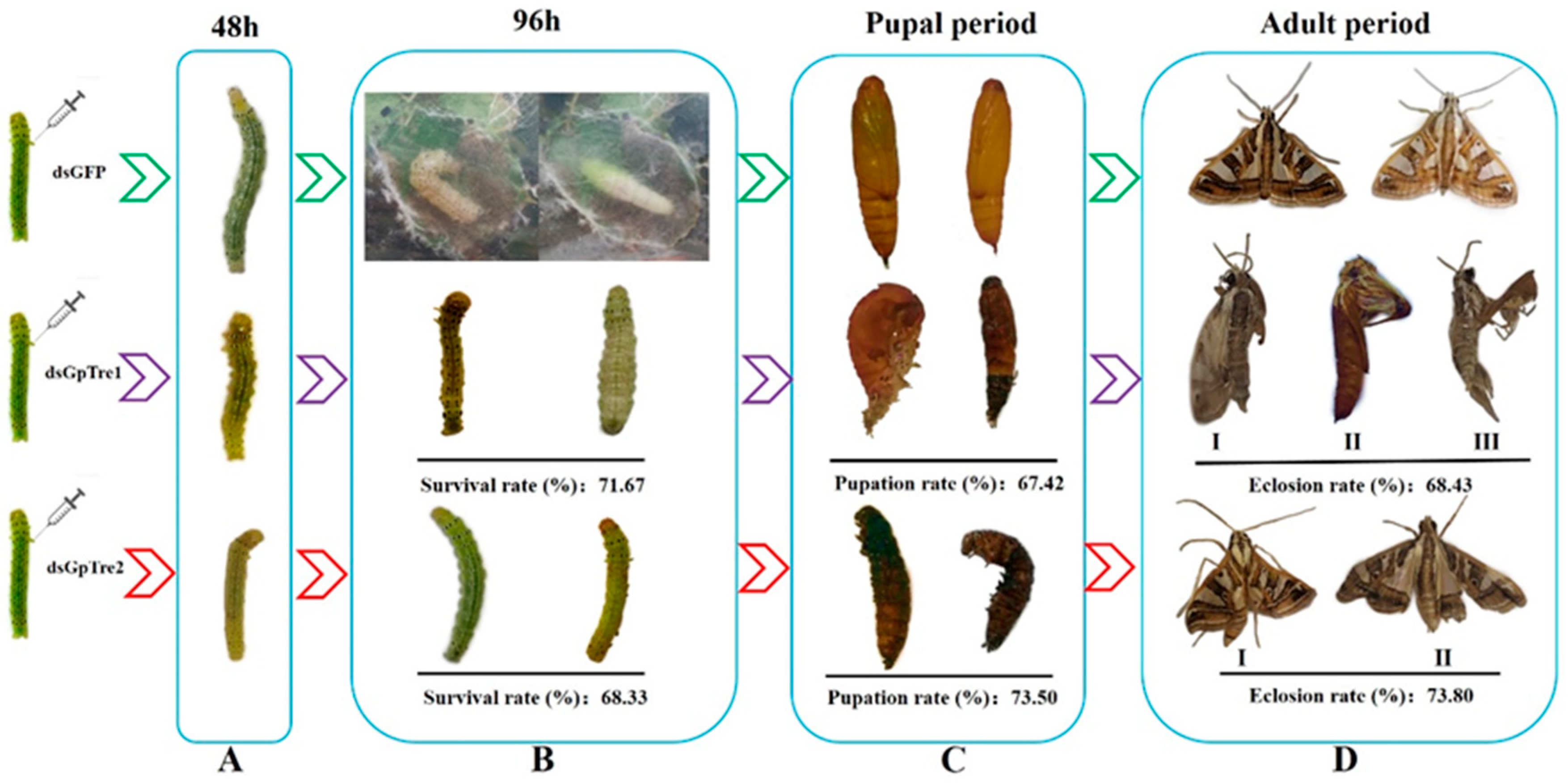
3.3. Analysis of GpTre1 and GpTre2 Expressions and Phenotypes after RNAi
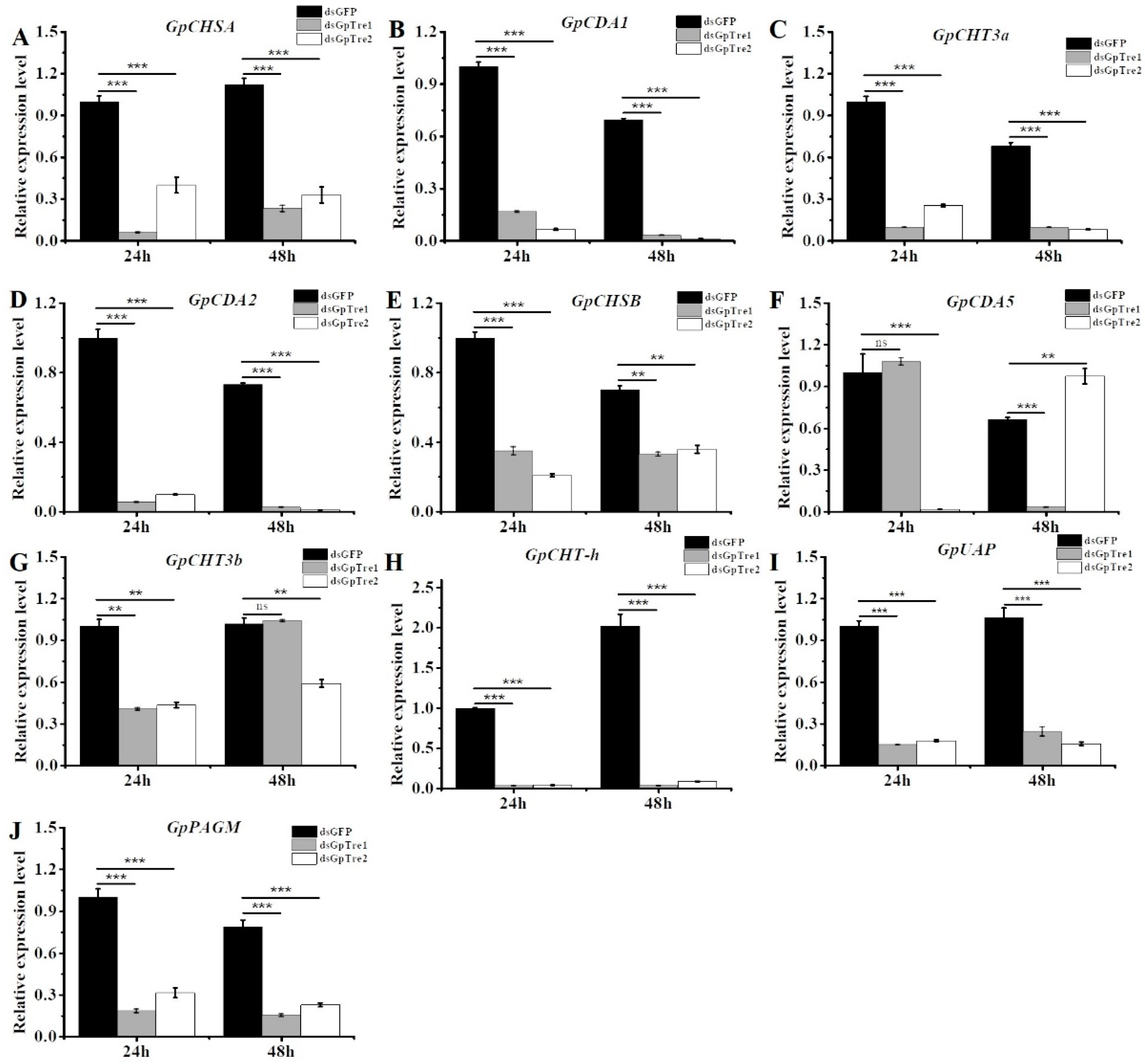
3.4. Analysis of Chitin Metabolism-Related Key Genes after RNAi
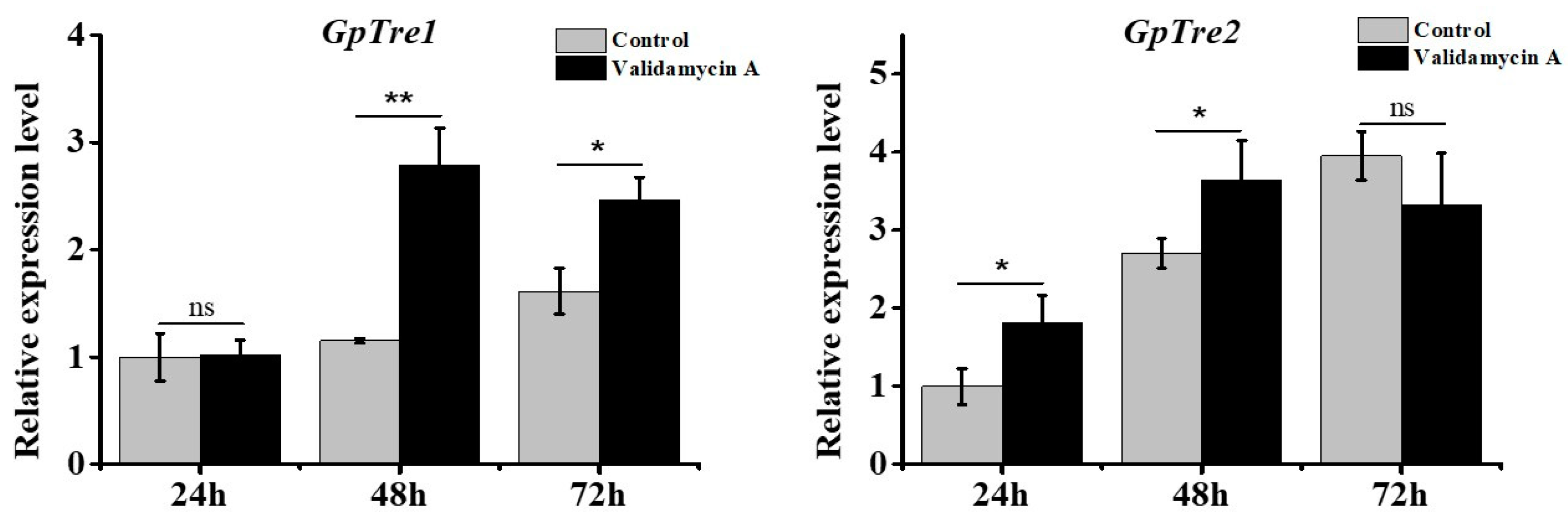
3.5. Analysis of GpTre1 and GpTre2 Expressions after Validamycin A Treatment
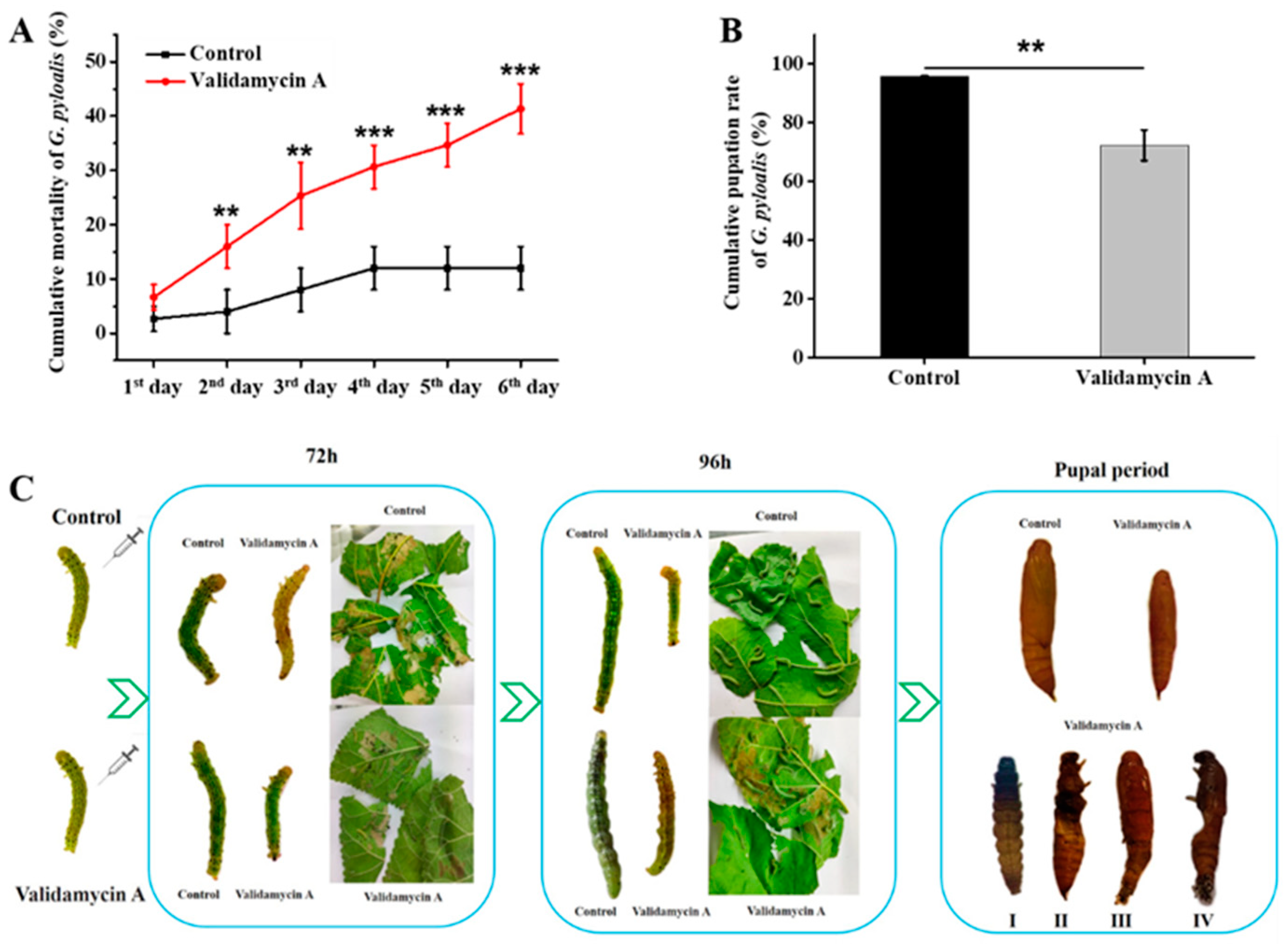
3.6. Analysis of the Effect of Validamycin A on G. pyloalis Development
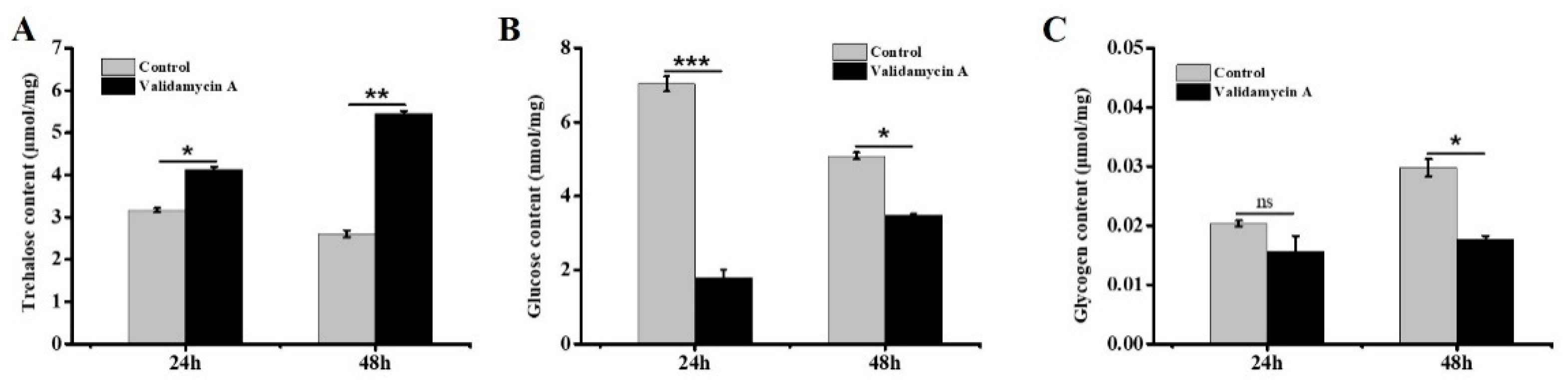
3.7. Analysis of the Effect of Validamycin A on Carbohydrates
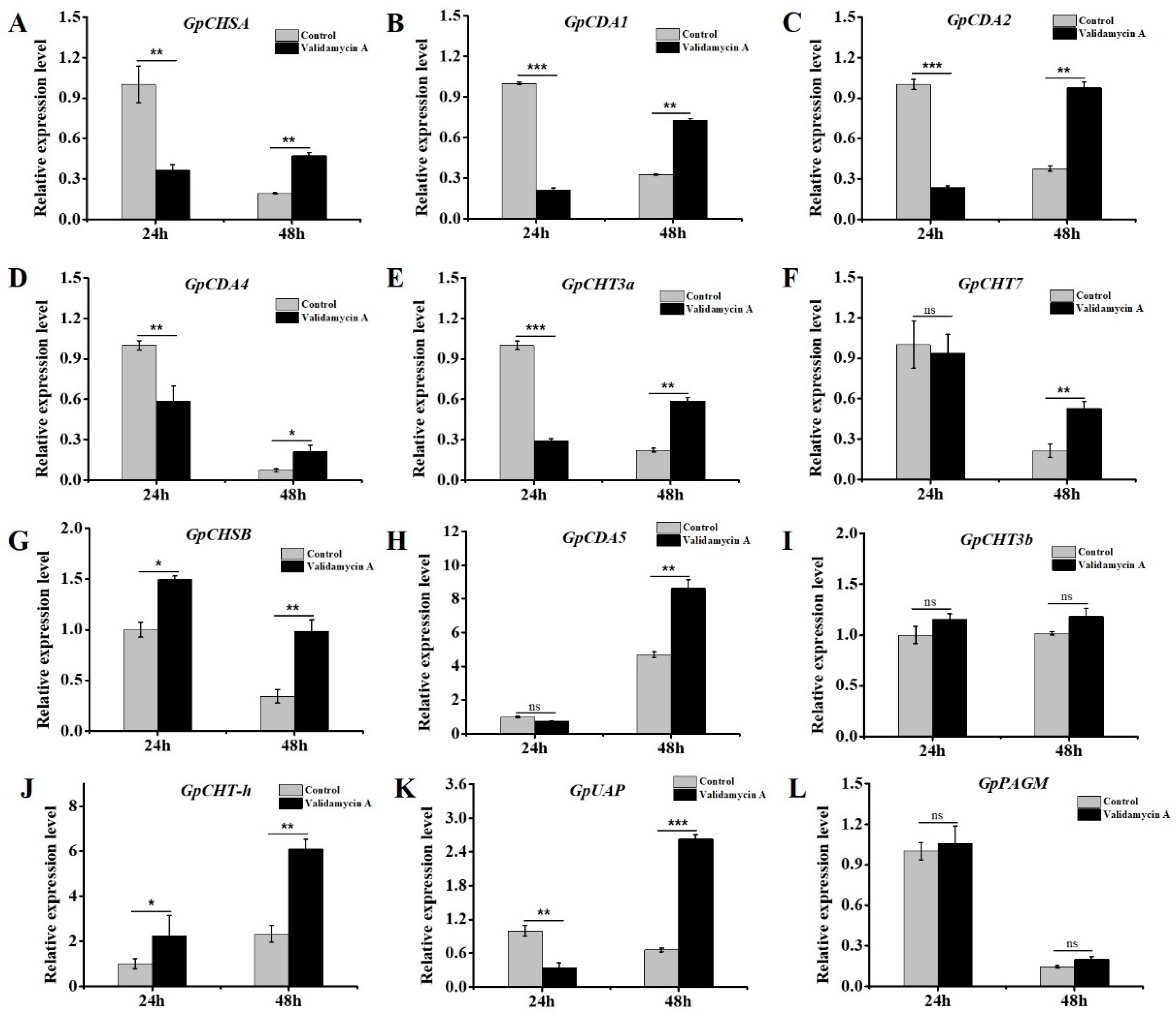
3.8. Validamycin A Treatment Significantly Affected the Expressions of Chitin Metabolism-Related Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Y.C.; Su, H.; Li, R.Q.; Li, X.T.; Xu, Y.S.; Dai, X.P.; Zhou, Y.Y.; Wang, H.B. Comparative transcriptome analysis of Glyphodes pyloalis Walker (Pyralidae: Lepidoptera) reveals novel insights into heat stress tolerance in insects. BMC Genom. 2017, 18, 974. [Google Scholar] [CrossRef]
- Oftadeh, M.; Sendi, J.J.; Khosravi, R. Life table parameters of Glyphodes pyloalis Walker (Lep.: Pyralidae) on four varieties of mulberry Morus alba L. (Moraceae). J. Asia-Pac. Entomol. 2015, 18, 315–320. [Google Scholar] [CrossRef]
- Watanabe, H.; Kurihara, Y.; Wang, Y.X.; Shimizu, T. Mulberry pyralid. Glyphodes pyloalis: Habitual host of nonoccluded viruses pathogenic to the silkworm, Bombyx mori. J. Invertebr. Pathol. 1988, 52, 401–408. [Google Scholar] [CrossRef]
- Hassan, F.; Mir, M.A. Effect of Glyphodes pyloalis Walker (Pyralidae: Lepidoptera) infested leaves on the commercial characters of silkworm hybrid (NB4D2 X SH6) B. mori L. J. Entomol. Zool. Stud. 2017, 8, 20–22. [Google Scholar]
- Yazdani, E.; Sendi, J.J.; Aliakbar, A.; Senthil-Nathan, S. Effect of Lavandula angustifolia essential oil against lesser mulberry pyralid Glyphodes pyloalis Walker (Lep: Pyralidae) and identification of its major derivatives. Pestic. Biochem. Physiol. 2013, 107, 250–257. [Google Scholar] [CrossRef]
- Gu, Z.Y.; Li, M.X.; Xia, S.X.; Mao, T.T.; Lu, Z.T.; Chen, J.; Wang, H.; Qu, J.W.; Fang, Y.L.; Li, F.C.; et al. Effects of sublethal phoxim exposure and lower food intake on nutrient metabolism in the midguts of Bombyx mori. Pestic. Biochem. Physiol. 2020, 167, 104593. [Google Scholar] [CrossRef]
- Wang, C.; Feng, M.G. Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol. Control 2014, 68, 129–135. [Google Scholar] [CrossRef]
- Shukla, E.; Thorat, L.J.; Nath, B.B.; Gaikwad, S.M. Insect trehalase physiological significance and potential applications. Glycobiology 2014, 25, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, D.J.; van Doorn, J.M.; Beenakkers, A.M.T. Dynamics in the haemolymph trehalose pool during flight of the locust, Locusta migratoria. Insect Biochem. 1978, 8, 413–416. [Google Scholar] [CrossRef]
- Tang, B.; Wei, P.; Zhao, L.; Shi, Z.; Shen, Q.; Yang, M.; Xie, G.; Wang, S. Knockdown of five trehalase genes using RNA interference regulates the gene expression of the chitin biosynthesis pathway in Tribolium castaneum. BMC Biotechnol. 2016, 16, 67. [Google Scholar] [CrossRef]
- Mitsumasu, K.; Azuma, M.; Niimi, T.; Yamashita, O.; Yaginuma, T. Membrane-penetrating trehalase from silkworm Bombyx mori. Molecular cloning and localization in larval midgut. Insect Mol. Biol. 2005, 14, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Mian, M.A.; Mittapalli, O.; Michel, A.P. Molecular characterization and expression analysis of soluble trehalase gene in Aphis glycines, a migratory pest of soybean. Bull. Entomol. Res. 2013, 103, 286–295. [Google Scholar] [CrossRef]
- Tang, B.; Chen, X.; Liu, Y.; Tian, H.; Liu, J.; Hu, J.; Xu, W.; Zhang, W. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Mol. Biol. 2008, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Dai, W.; Li, X.; Zhang, Y.; Zhang, C. Molecular cloning and expression analysis of soluble and membrane-bound trehalase genes in the cotton bollworm, Helicoverpa armigera. J. Asia Pac. Entomol. 2015, 18, 187–195. [Google Scholar] [CrossRef]
- Chen, J.; Tang, B.; Chen, H.; Yao, Q.; Huang, X.; Chen, J.; Zhang, D.; Zhang, W. Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference. PLoS ONE 2010, 5, e10133. [Google Scholar] [CrossRef]
- Adhav, A.S.; Kokane, S.R.; Joshi, R.S. Functional characterization of Helicoverpa armigera trehalase and investigation of physiological effects caused due to its inhibition by Validamycin A formulation. Int. J. Biol. Macromol. 2018, 112, 638–647. [Google Scholar] [CrossRef]
- Tatun, N.; Singtripop, T.; Sakurai, S. Dual control of midgut trehalase activity by 20-hydroxyecdysone and an inhibitory factor in the bamboo borer Omphisa fuscidentalis Hampson. J. Insect Physiol. 2008, 54, 351–357. [Google Scholar] [CrossRef]
- Zhang, L.; Qiu, L.Y.; Yang, H.L.; Wang, H.J.; Zhou, M.; Wang, S.G.; Tang, B. Study on the Effect of Wing Bud Chitin Metabolism and Its Developmental Network Genes in the Brown Planthopper, Nilaparvata lugens, by Knockdown of TRE Gene. Front. Physiol. 2017, 8, 750. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Wang, S.S.; Pan, B.Y.; Wang, S.G.; Wang, S.; Tang, B. Evaluation of the Expression and Function of the TRE2-like and TRE2 Genes in Ecdysis of Harmonia axyridis. Front. Physiol. 2019, 10, 1371. [Google Scholar] [CrossRef]
- Kramer, K.J.; Dziadik-turner, C.; Koga, D. Chitin metabolism in insects. Compr. Insect Physiol. Biochem. Pharmacol. 1985, 3, 75–115. [Google Scholar]
- Liebl, M.; Nelius, V.; Kamp, G.; Ando, O.; Wegener, G. Fate and effects of the trehalase inhibitor trehazolin in the migratory locust (Locusta migratoria). J. Insect Physiol. 2010, 56, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Li, L.; Xu, H.; Minagawa, K.; Yu, Y.; Zhang, Y.; Zhou, X.; Floss, H.G.; Mahmud, T.; Deng, Z. Functional Analysis of the Validamycin Biosynthetic Gene Cluster and Engineered Production of Validoxylamine A. Chem. Biol. 2006, 13, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Takahashi, M.; Matsushita, K.; Nishina, M.; Kameda, Y.; Hori, E. Inhibition of flight in Periplaneta americana (Linn.) by a trehalase inhibitor, validoxylamine A. J. Insect Physiol. 1994, 40, 455–461. [Google Scholar] [CrossRef]
- Kono, Y.; Takeda, S.; Kameda, Y. Lethal Activity of a Trehalase Inhibitor, validoxylamine A, aginst Mamestra brassicas and Spodoptera littura. J. Pestic. Sci. 1994, 19, 39–42. [Google Scholar] [CrossRef][Green Version]
- Kono, Y.; Takeda, S.; Kameda, Y.; Takahashi, M. Lethal activity of a trehalase inhibitor, validoxylamine A, and its influence on the blood sugar level in Bombyx mori (Lepidoptera: Bombycidae). Appl. Entomol. Zool. 1993, 28, 379–386. [Google Scholar] [CrossRef]
- Marten, A.D.; Stothard, A.I.; Kalera, K.; Swarts, B.M.; Conway, M.J. Validamycin A Delays Development and Prevents Flight in Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 2020, 57, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.M.; Li, Y.J.; Zhang, X.R.; Chu, J.; Ma, J.H.; Liu, Z.X.; Wang, J.; Sheng, S.; Wu, F.A. Identification and Functional Study of Chitin Metabolism and Detoxification-Related Genes in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) Based on Transcriptome Analysis. Int. J. Mol. Sci. 2020, 21, 1904. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Gao, Y.; Liu, Y.; Li, X.; Liang, Y.; Dai, X.; Xu, Y.; Zhou, Y.; Wang, H. Comparative transcriptome profiling reveals candidate genes related to insecticide resistance of Glyphodes pyloalis. Bull. Entomol. Res. 2020, 110, 57–67. [Google Scholar] [CrossRef]
- Shao, Z.M.; Li, Y.J.; Ding, J.H.; Liu, Z.X.; Zhang, X.R.; Wang, J.; Sheng, S.; Wu, F.A. Identification, Characterization, and Functional Analysis of Chitin Synthase Genes in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). Int. J. Mol. Sci. 2020, 21, 4656. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Quintana, A.; Sánchez, M.; Rodríguez, E.N.; Cremata, J.; Sánchez, J.C. Rapid and sensitive anthrone-sulfuric acid assay in microplate format to quantify carbohydrate in biopharmaceutical products: Method development and validation. Biologicals 2008, 36, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.Q.; Jin, L.Q.; Zheng, Y.G. The enzymatic properties of trehalase and its exploitation as a target of new pesticides. Chem. Bioeng. 2009, 26, 7–11. [Google Scholar]
- Zhao, L.; Yang, M.; Shen, Q.; Liu, X.; Shi, Z.; Wang, S.; Tang, B. Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference. Sci. Rep. 2016, 6, 27841. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Shao, Y.; Zhang, C.; Liu, Z.; Zhang, Y. Characterization of putative soluble and membrane-bound trehalases in a hemipteran insect, Nilaparvata lugens. J. Insect Physiol. 2009, 55, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.F.; Wang, Y.; Dou, W.; Wang, J.J. Characterization of the cDNA encoding membrane-bound trehalase, its expression and enzyme activity in Bactrocera dorsalis (Diptera: Tephritidae). Fla. Entomol. 2013, 96, 1233–1242. [Google Scholar] [CrossRef]
- Zou, Q.; Wei, P.; Xu, Q.; Zheng, H.Z.; Tang, B.; Wang, S.G. cDNA cloning and characterization of two trehalases from Spodoptera litura (Lepidoptera; Noctuidade). Genet Mol. Res. 2013, 12, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Mitsumasu, K.; Azuma, M.; Niimi, T.; Yamashita, O.; Yaginuma, T. Changes in the expression of soluble and integral-membrane trehalases in the midgut during metamorphosis in Bombyx mori. Zool. Sci. 2008, 25, 693–698. [Google Scholar] [CrossRef]
- Ge, L.Q.; Zhao, K.F.; Huang, L.J.; Wu, J.C. The effects of triazophos on the trehalose content, trehalase activity and their gene expression in the brown planthopper Nilaparvata lugens (Stål) (Hemiptera: Delphacidae). Pestic. Biochem. Physiol. 2011, 100, 172–181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, M.M.; Zhao, L.N.; Shen, Q.D.; Xie, G.Q.; Wang, S.G. Knockdown of two trehalose-6-phosphate synthases severely affects chitin metabolism gene expression in the brown planthopper Nilaparvata lugens. Pest Manag. Sci. 2017, 73, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lu, D.H.; Pu, J.; Wu, M.; Han, Z.J. Cloning and RNA interference effects of trehalase genes in Laodelphax striatellus (Homoptera: Delphacidae). Acta Entomol. Sin. 2012, 55, 911–920. [Google Scholar]
- Liu, X.J.; Sun, Y.W.; Cui, M.; Ma, E.B.; Zhang, J.Z. Molecular characteristics and functional analysis of trehalase genes in Locusta migratoria. Sci. Agric. Sin. 2016, 49, 4375–4386. [Google Scholar]
- Chen, X.; Quan, Y.; Wang, H.; Luo, H. Trehalase Regulates Neuroepithelial Stem Cell Maintenance and Differentiation in the Drosophila Optic Lobe. PLoS ONE 2014, 9, e101433. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shi, J.F.; Xu, Q.Y.; Sun, Q.K.; Meng, Q.W.; Mu, L.L.; Guo, W.C.; Li, G.Q. Physiological roles of trehalose in Leptinotarsa larvae revealed by RNA interference of trehalose-6-phosphate synthase and trehalase genes. Insect Biochem. Mol. Biol. 2016, 77, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Okuda, T.; Hasegawa, E.; Kono, Y. Suppression of oocyte development by a trehalase inhibitor, Validoxylamine A, through inhibition of juvenile hormone biosynthesis and vitellogenesis in the migratory locust, Locusta migratoria L. Entomol. Sci. 1998, 1, 313–320. [Google Scholar]
- Forcella, M.; Cardona, F.; Goti, A.; Parmeggiani, C.; Parenti, P. A membrane-bound trehalase from Chironomus riparius larvae: Purification and sensitivity to inhibition. Glycobiology 2010, 20, 1186–1195. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, Z.-m.; Ding, J.-h.; Jiang, D.-l.; Liu, Z.-x.; Li, Y.-j.; Wang, J.; Wang, J.; Sheng, S.; Wu, F.-a. Characterization and Functional Analysis of trehalase Related to Chitin Metabolism in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). Insects 2021, 12, 370. https://doi.org/10.3390/insects12040370
Shao Z-m, Ding J-h, Jiang D-l, Liu Z-x, Li Y-j, Wang J, Wang J, Sheng S, Wu F-a. Characterization and Functional Analysis of trehalase Related to Chitin Metabolism in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). Insects. 2021; 12(4):370. https://doi.org/10.3390/insects12040370
Chicago/Turabian StyleShao, Zuo-min, Jian-hao Ding, De-lei Jiang, Zhi-xiang Liu, Yi-jiangcheng Li, Jiao Wang, Jun Wang, Sheng Sheng, and Fu-an Wu. 2021. "Characterization and Functional Analysis of trehalase Related to Chitin Metabolism in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae)" Insects 12, no. 4: 370. https://doi.org/10.3390/insects12040370
APA StyleShao, Z.-m., Ding, J.-h., Jiang, D.-l., Liu, Z.-x., Li, Y.-j., Wang, J., Wang, J., Sheng, S., & Wu, F.-a. (2021). Characterization and Functional Analysis of trehalase Related to Chitin Metabolism in Glyphodes pyloalis Walker (Lepidoptera: Pyralidae). Insects, 12(4), 370. https://doi.org/10.3390/insects12040370