Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Identification and Characterization of NvdsRNase
2.3. RNA Isolation, cDNA Synthesis and dsRNA Synthesis
2.4. Developmental Stage-Specific and Tissue-Specific Expression of NvdsRNase and RT-qPCR
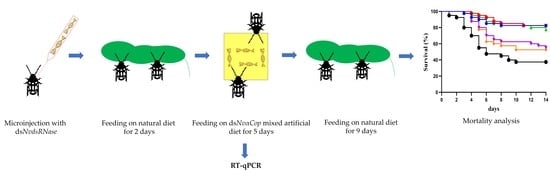
2.5. “RNAi-of-RNAi”– Oral Feeding Bioassay
2.6. “RNAi-of-RNAi”-Ex Vivo Degradation Assay
2.7. Statistical Analysis
3. Results
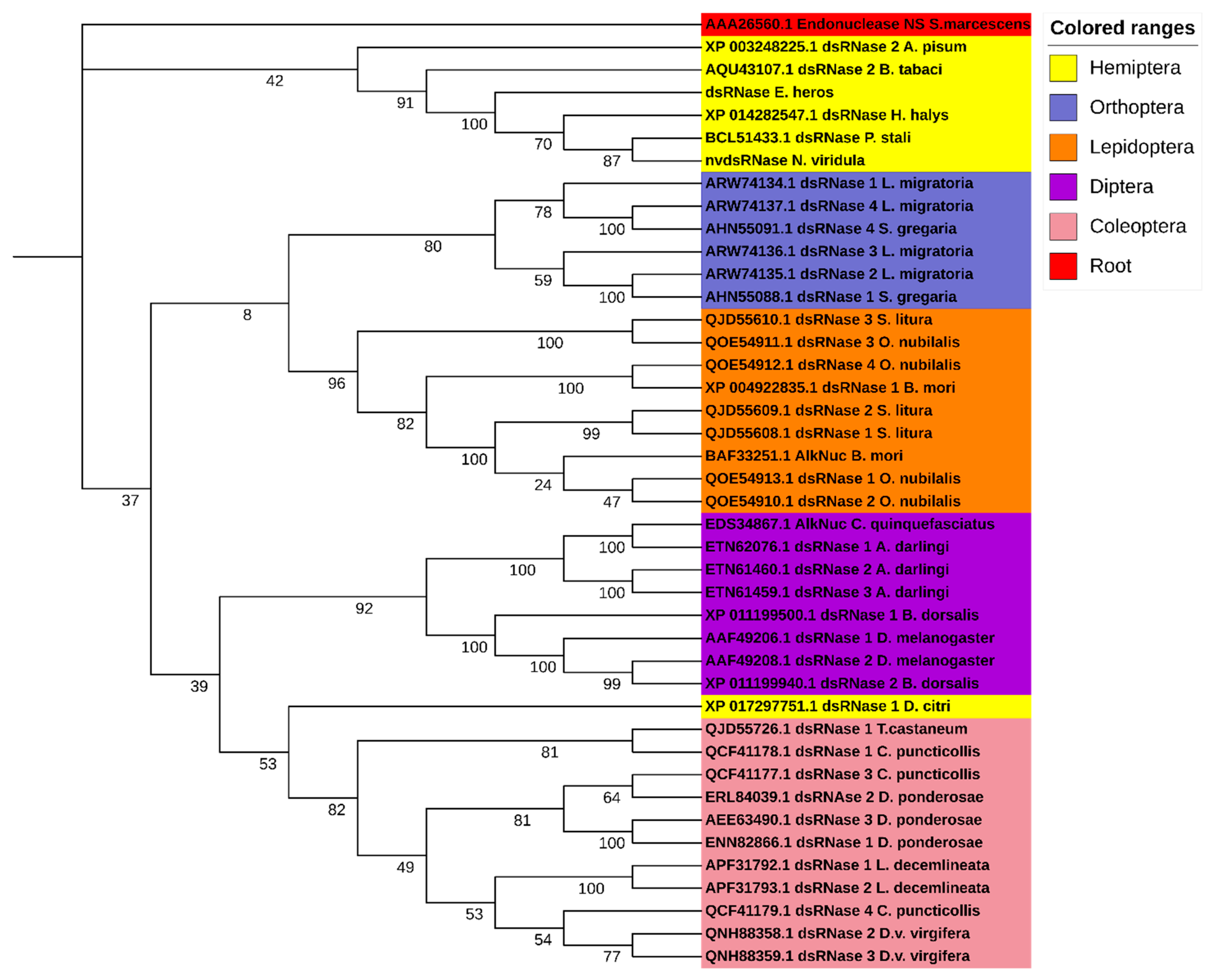
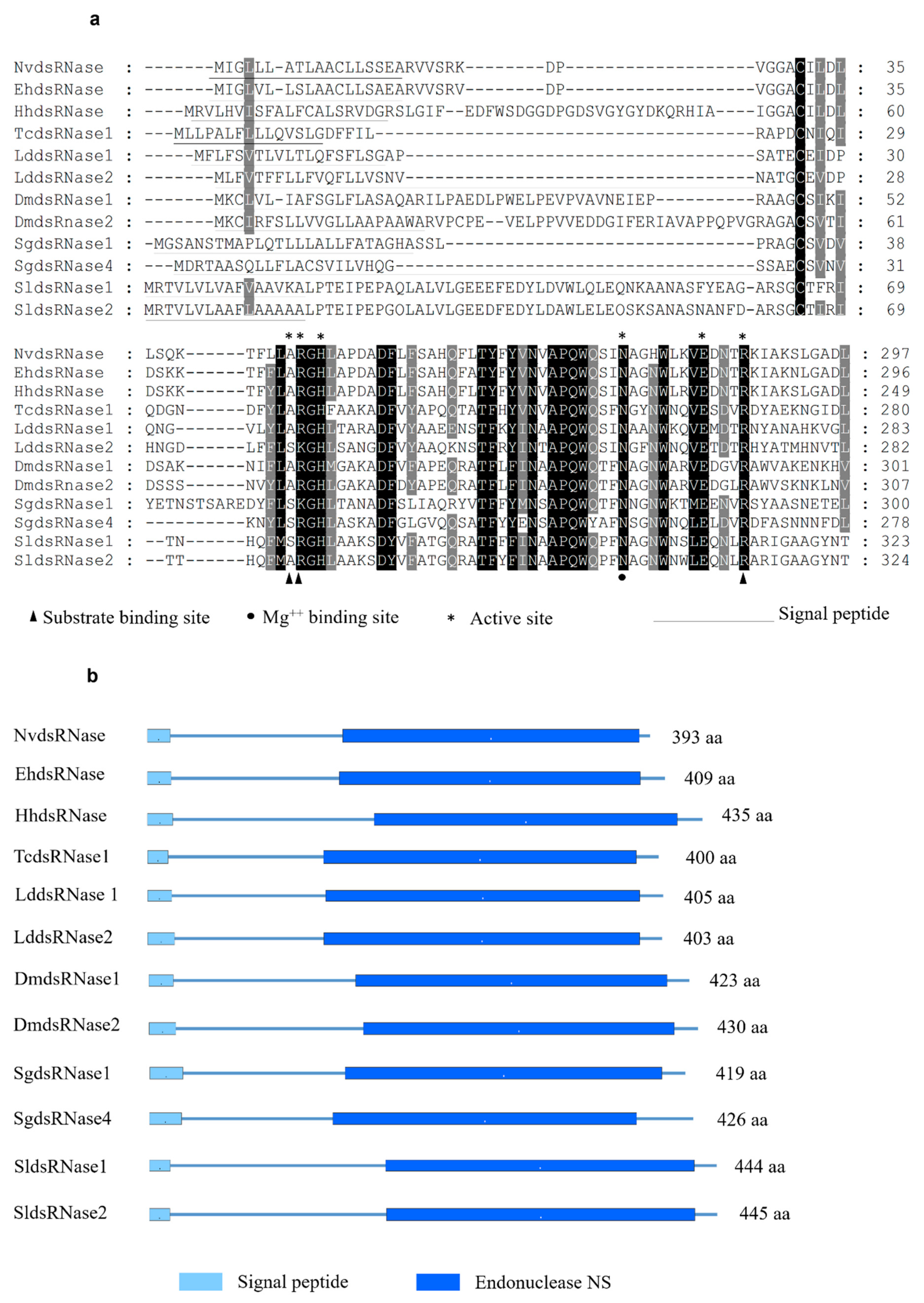
3.1. Identification and Characterization of NvdsRNase
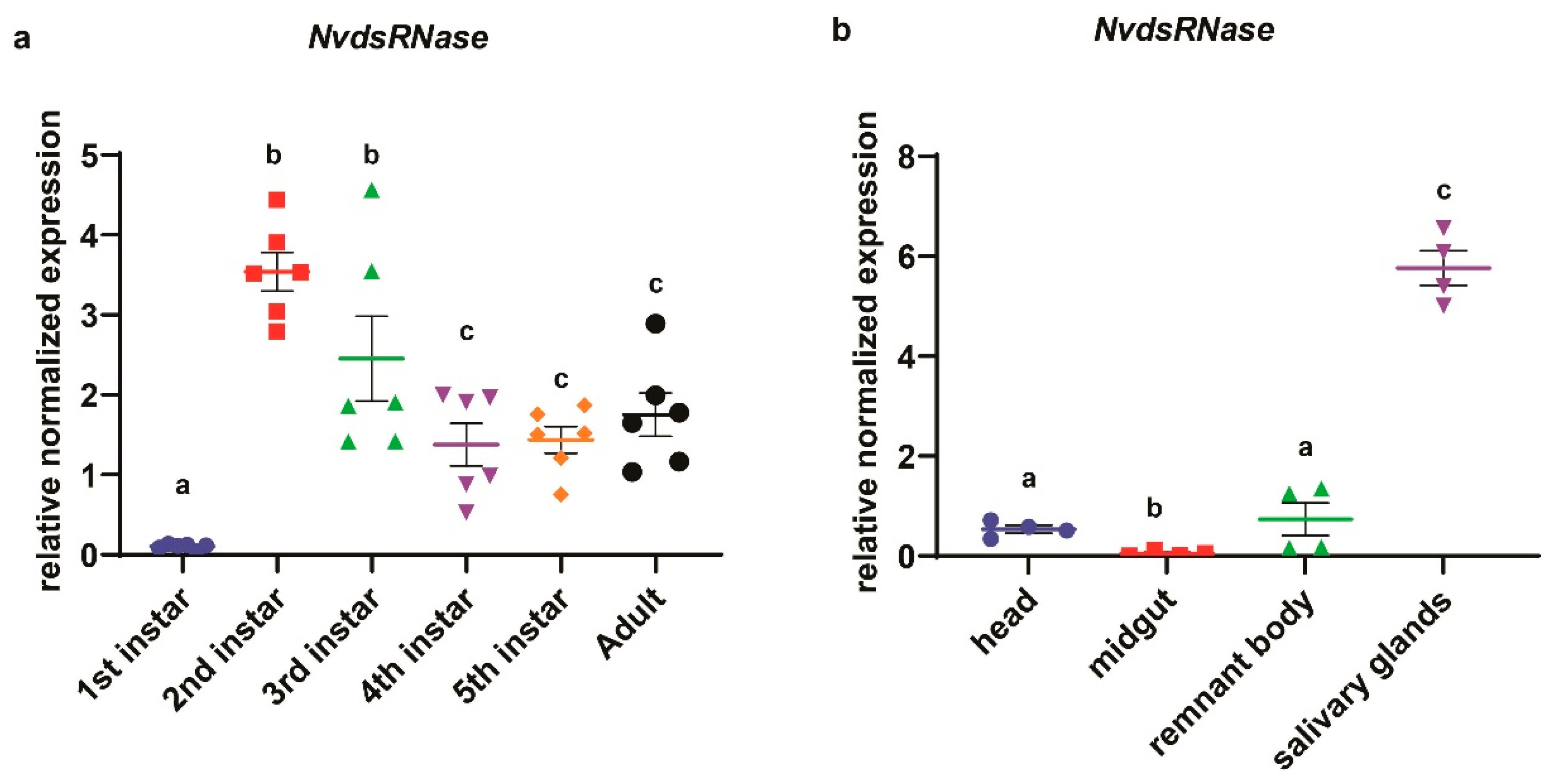
3.2. Developmental Stage-Specific and Tissue-Specific Expression of NvdsRNase
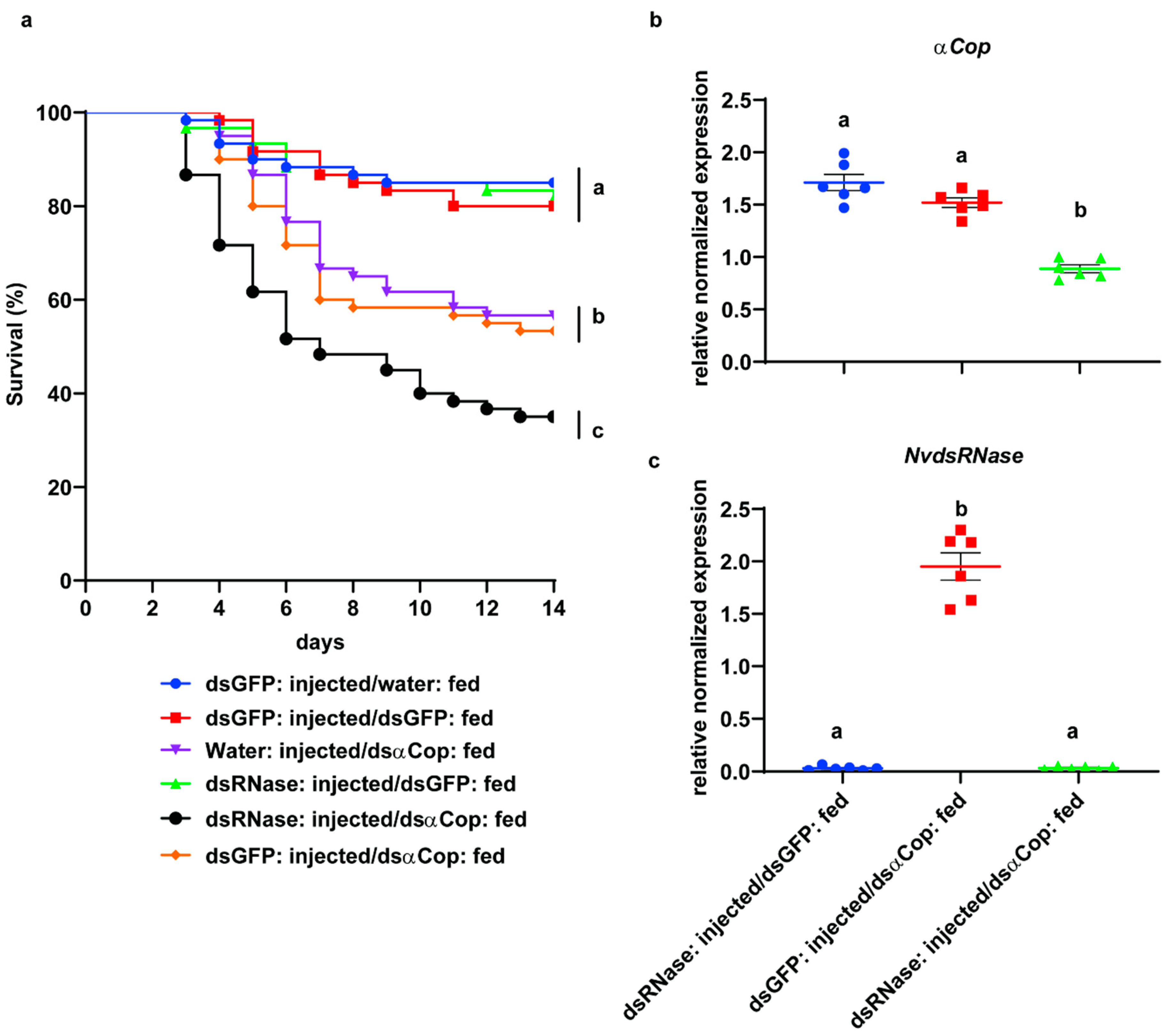
3.3. “RNAi-of-RNAi”– Oral Feeding Bioassay
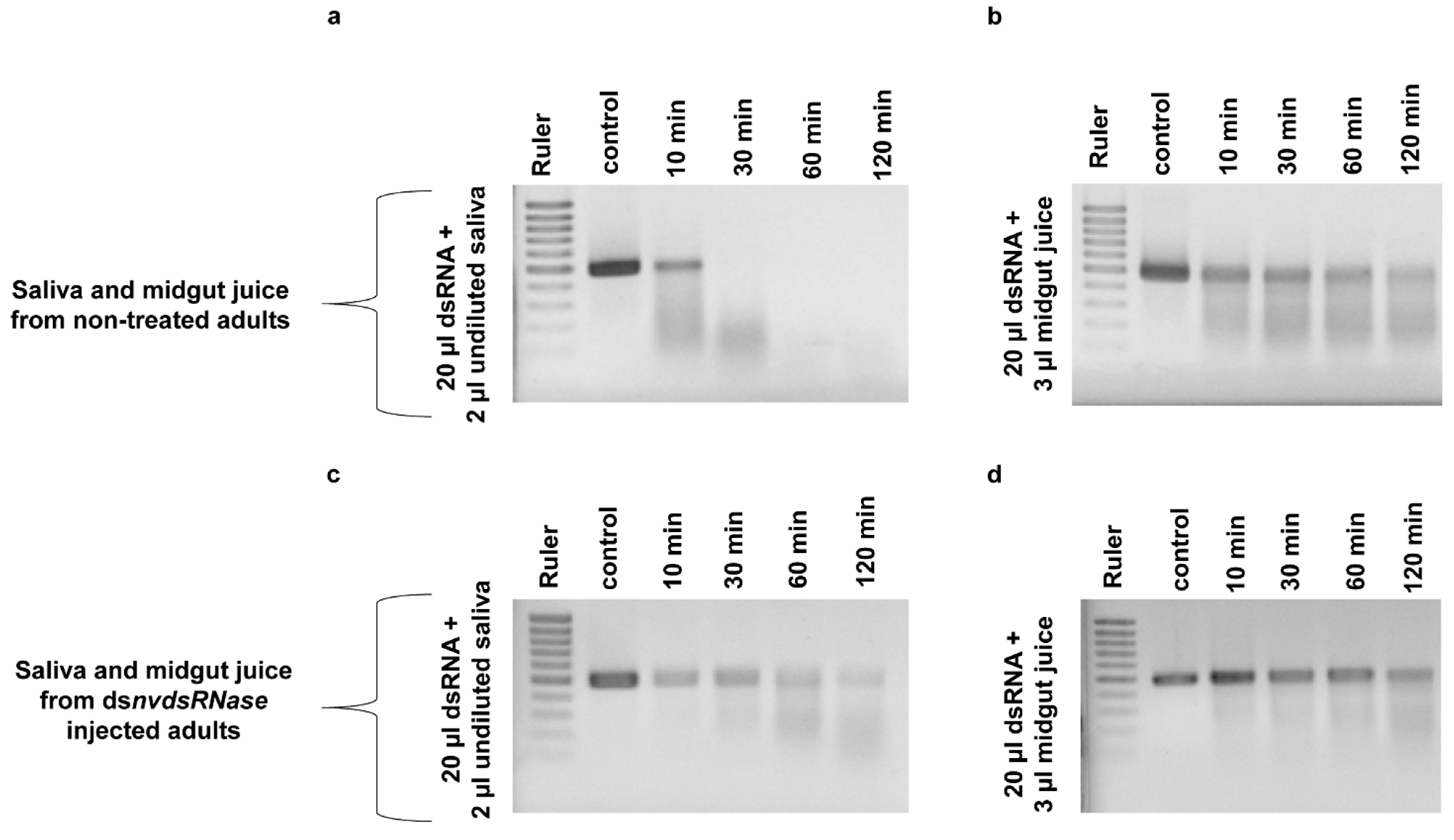
3.4. “RNAi-of-RNAi”-Ex Vivo Degradation Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Werner, B.T.; Gaffar, F.Y.; Schuemann, J.; Biedenkopf, D.; Koch, A.M. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant. Sci. 2020, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [PubMed]
- Sharma, R.; Chandra, K.; Gobu, R.; Loitongbam, B. Post-transcriptional gene silencing and its implication to the asian soybean rust- a review article. Int. J. Agric. Environ. Biotech. 2017, 10, 105–113. [Google Scholar] [CrossRef]
- Kusaba, M.; Miyahara, K.; Iida, S.; Fukuoka, H.; Takano, T.; Sassa, H.; Nishimura, M.; Nishio, T. Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 2003, 15, 1455–1467. [Google Scholar] [CrossRef] [Green Version]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Christiaens, O.; Whyard, S.; Velez, A.M.; Smagghe, G. Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef]
- Dubelman, S.; Fischer, J.; Zapata, F.; Huizinga, K.; Jiang, C.; Uffman, J.; Levine, S.; Carson, D. Environmental fate of double-stranded RNA in agricultural soils. PLoS ONE 2014, 9, e93155. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016, 72, 801–809. [Google Scholar]
- Cagliari, D.; Dias, N.P.; Galdeano, D.M.; Dos Santos, E.A.; Smagghe, G.; Zotti, M.J. Management of Pest Insects and Plant Diseases by Non-Transformative RNAi. Front. Plant Sci. 2019, 10, 1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.-S.; Kim, K.; Palli, S.R. Double-stranded RNA in exosomes: Potential systemic RNA interference pathway in the Colorado potato beetle, Leptinotarsa decemlineata. J. Asia Pac. Entomol. 2020, 23, 1160–1164. [Google Scholar] [CrossRef]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar]
- Cooper, A.M.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar]
- Liu, J.; Swevers, L.; Iatrou, K.; Huvenne, H.; Smagghe, G. Bombyx mori DNA/RNA non-specific nuclease: Expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 2012, 58, 1166–1176. [Google Scholar] [CrossRef]
- Prentice, K.; Christiaens, O.; Pertry, I.; Bailey, A.; Niblett, C.; Ghislain, M.; Gheysen, G.; Smagghe, G. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest Manag. Sci. 2017, 73, 44–52. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Christiaens, O.; Berkvens, N.; Casteels, H.; Maes, M.; Smagghe, G. Oral RNAi to control Drosophila suzukii: Laboratory testing against larval and adult stages. J. Pest Sci. 2016, 89, 803–814. [Google Scholar] [CrossRef]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Chen, J.; Wang, J.; Zhang, H.; Ze, L.; Zhu, G.; Zhao, C.; Xiao, H.; Han, Z. Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 2020, 125, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Zhu, G.; Han, Q.; Chen, J.; Elzaki, M.E.A.; Sheng, C.; Zhao, C.; Palli, S.R.; Han, Z. Identification and characterization of multiple dsRNases from a lepidopteran insect, the tobacco cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Pestic Biochem. Physiol. 2020, 162, 86–95. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christiaens, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 2019, 9, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Vanden Broeck, J. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Li, D.; Cooper, A.M.W.; Silver, K.; Li, T.; Liu, X.; Ma, E.; Zhu, K.Y.; Zhang, J. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in Migratory locust. Insect Biochem. Mol. Biol. 2017, 86, 68–80. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Van Eck, J.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef]
- Panizzi, A.R.; Slansky, F. Review of phytophagous Pentatomid (Hemiptera, Pentatomidae) associated with soybean in the Americas. Fla. Entomol. 1985, 68, 184–214. [Google Scholar] [CrossRef]
- Cattelan, A.J.; Dall’Agnol, A. The rapid soybean growth in Brazil. OCL 2018, 25, D102. [Google Scholar] [CrossRef] [Green Version]
- Baur, M.E.; Boethel, D.J.; Boyd, M.L.; Bowers, G.R.; Way, M.O.; Heatherly, L.G.; Rabb, J.; Ashlock, L. Arthropod populations in early soybean production systems in the mid-South. Environ. Entomol. 2000, 29, 312–328. [Google Scholar] [CrossRef]
- Vivan, L.M.; Panizzi, A.R. Geographical distribution of genetically determined types of Nezara viridula (L.) (Heteroptera: Pentatomidae) in Brazil. Neotropical Entomol. 2006, 35, 175–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, L.A.; Rose, H.A.; McDonald, F.J.D. The effects of damage by the green vegetable bug, Nezara viridula (L.) on yield and quality of soybeans. J. Aust. Entomol. Soc. 1978, 16, 421–426. [Google Scholar] [CrossRef]
- Kaiser, W.J.; Vakili, N.G. Insect Transmission of Pathogenic Xanthomonads to Bean and Cowpea in Puerto-Rico. Phytopathology 1978, 68, 1057–1063. [Google Scholar] [CrossRef]
- Ragsdale, D.W.; Larson, A.D.; Newsom, L.D. Microorganisms Associated with Feeding and from Various Organs of Nezara viridula. J. Econ. Entomol. 1979, 72, 725–727. [Google Scholar] [CrossRef]
- Sharma, R.; Christiaens, O.; Taning, C.N.; Smagghe, G. RNAi-mediated mortality in southern green stinkbug Nezara viridula by oral delivery of dsRNA. Pest Manag. Sci. 2021, 77, 77–84. [Google Scholar] [CrossRef]
- Cagliari, D.; Dias, N.P.; Dos Santos, E.A.; Rickes, L.N.; Kremer, F.S.; Farias, J.R.; Lenz, G.; Galdeano, D.M.; Garcia, F.R.M.; Smagghe, G.; et al. First transcriptome of the Neotropical pest Euschistus heros (Hemiptera: Pentatomidae) with dissection of its siRNA machinery. Sci. Rep. 2020, 10, 4856. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.-C.; Shen, H.-B. A New Method for Predicting the Subcellular Localization of Eukaryotic Proteins with Both Single and Multiple Sites: Euk-mPLoc 2.0. PLoS ONE 2010, 5, e9931. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Mittapelly, P.; Chen, Y.; Mamidala, P.; Zhao, C.; Michel, A. Quantitative RT-PCR Gene Evaluation and RNA Interference in the Brown Marmorated Stink Bug. PLoS ONE 2016, 11, e0152730. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bonning, B.C. The Principal Salivary Gland Is the Primary Source of Digestive Enzymes in the Saliva of the Brown Marmorated Stink Bug, Halyomorpha halys. Front. Physiol. 2019, 10, 1255. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Riga, M.; Denecke, S.; Livadaras, I.; Geibel, S.; Nauen, R.; Vontas, J. Development of efficient RNAi in Nezara viridula for use in insecticide target discovery. Arch. Insect Biochem. Physiol. 2020, 103, e21650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis-Vogel, C.; Van Allen, B.; Van Hemert, J.L.; Sethi, A.; Nelson, M.E.; Sashital, D.G. Identification and comparison of key RNA interference machinery from western corn rootworm, fall armyworm, and southern green stink bug. PLoS ONE 2018, 13, e0203160. [Google Scholar] [CrossRef] [Green Version]
- Gurusamy, D.; Howell, J.L.; Chereddy, S.; Koo, J.; Palli, S.R. Transport of orally delivered dsRNA in southern green stink bug, Nezara viridula. Arch. Insect Biochem. Physiol. 2020, 104, e21692. [Google Scholar] [CrossRef]
- Yoon, J.S.; Tian, H.G.; McMullen, J.G., II; Chung, S.H.; Douglas, A.E. Candidate genetic determinants of intraspecific variation in pea aphid susceptibility to RNA interference. Insect Biochem. Mol. Biol. 2020, 123, 103408. [Google Scholar] [CrossRef]
- Song, H.; Fan, Y.; Zhang, J.; Cooper, A.M.; Silver, K.; Li, D.; Li, T.; Ma, E.; Zhu, K.Y.; Zhang, J. Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manag. Sci. 2019, 75, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Kiritani, K. The Effect of Colony Size upon the Survival of Larvae of the Southern Green Stink Bug, Nezara viridula. Jpn. J. Appl. Entomol. Zool. 1964, 8, 45–64. [Google Scholar] [CrossRef]
- Pinheiro, D.H.; Taylor, C.E.; Wu, K.; Siegfried, B.D. Delivery of gene-specific dsRNA by microinjection and feeding induces RNAi response in Sri Lanka weevil, Myllocerus undecimpustulatus undatus Marshall. Pest Manag. Sci. 2020, 76, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.; Deikman, J.; Hendrix, B.; Iandolino, A. Barriers to Efficient Foliar Uptake of dsRNA and Molecular Barriers to dsRNA Activity in Plant Cells. Front. Plant Sci. 2020, 11, 816. [Google Scholar] [CrossRef]
- Parsons, K.H.; Mondal, M.H.; McCormick, C.L.; Flynt, A.S. Guanidinium-Functionalized Interpolyelectrolyte Complexes Enabling RNAi in Resistant Insect Pests. Biomacromolecules 2018, 19, 1111–1117. [Google Scholar]
- Christiaens, O.; Petek, M.; Smagghe, G.; Taning, C.N.T. The Use of Nanocarriers to Improve the Efficiency of RNAi-Based Pesticides in Agriculture. In Nanopesticides: From Research and Development to Mechanisms of Action and Sustainable Use in Agriculture; Fraceto, L.F., de Castro, V.L.S.S., Grillo, R., Ávila, D., Caixeta Oliveira, H., Lima, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 49–68. [Google Scholar]
- Mitchell, P.L.; Cooke, S.B.; Smaniotto, L.F. Probing Behavior of Nezara viridula on Soybean: Characterization and Comparison of Electrical Penetration Graph (EPG) Waveforms on Vegetative and Reproductive Plant Structures. J. Agric. Urban Entomol. 2018, 34, 19–43. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.; Taning, C.N.T.; Smagghe, G.; Christiaens, O. Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula. Insects 2021, 12, 115. https://doi.org/10.3390/insects12020115
Sharma R, Taning CNT, Smagghe G, Christiaens O. Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula. Insects. 2021; 12(2):115. https://doi.org/10.3390/insects12020115
Chicago/Turabian StyleSharma, Rohit, Clauvis Nji Tizi Taning, Guy Smagghe, and Olivier Christiaens. 2021. "Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula" Insects 12, no. 2: 115. https://doi.org/10.3390/insects12020115
APA StyleSharma, R., Taning, C. N. T., Smagghe, G., & Christiaens, O. (2021). Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezara viridula. Insects, 12(2), 115. https://doi.org/10.3390/insects12020115