Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design, Layout and Environmental Conditions
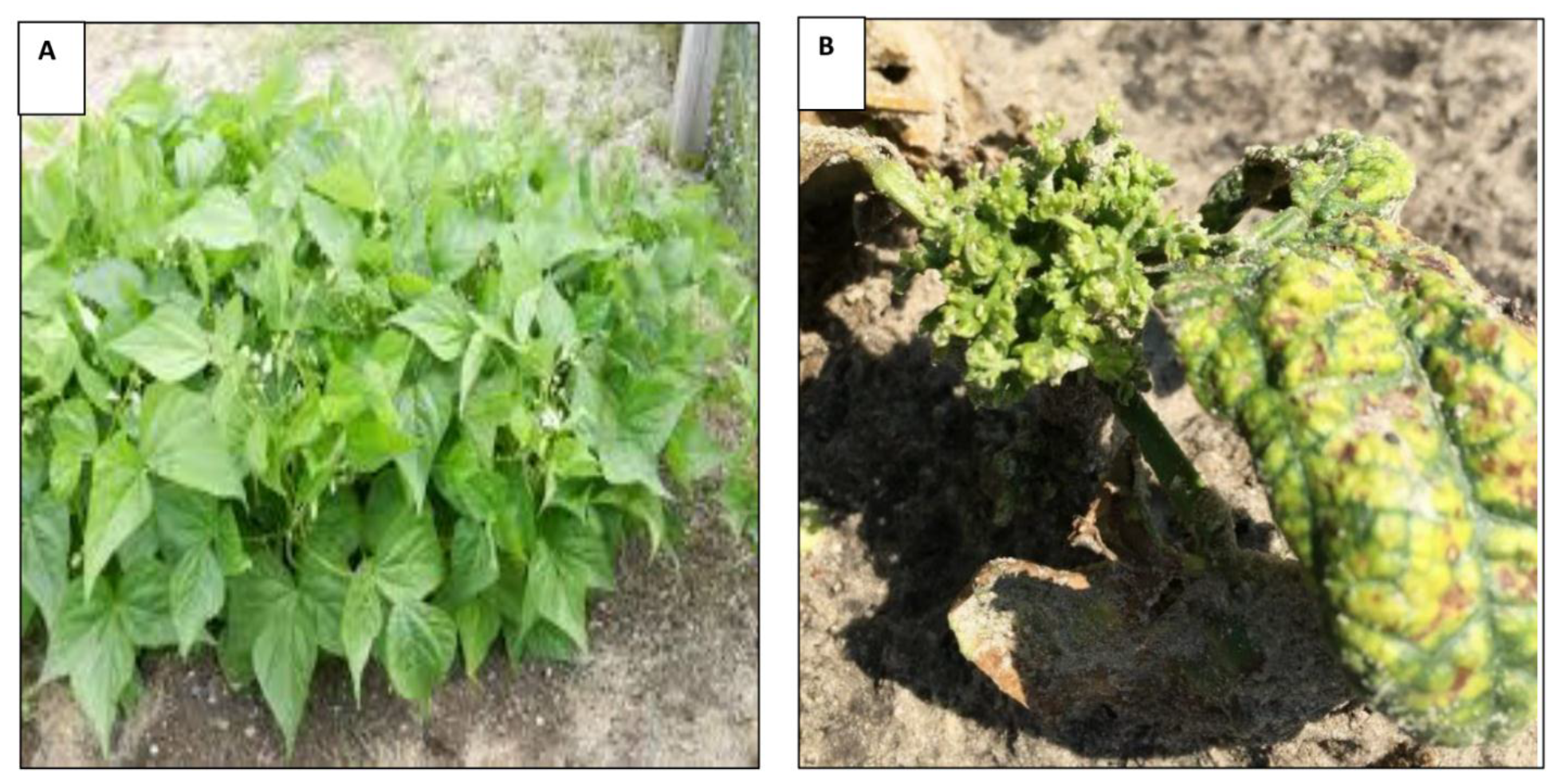
2.3. Response of Phaseolus spp. (Snap Beans and Lima beans) Genotypes to Leaf Crumple Disease in the Field
2.4. Whitefly Count
2.5. DNA Isolation, Library Preparation, Sequencing and Quality Filtering of Raw Data
2.6. Mapping of Filtered Read Data on the Reference Genome and Variant Calling
2.7. Confirmation of Begomoviruses (CuLCrV and SiGMFV) Infection Associated with Leaf Crumple Symptoms in Phaseolus spp.
2.8. Accumulation of CuLCrV and SiGMFV and Leaf Crumple Severity in P. vulgaris Genotypes (Susceptible vs. Resistant; Identified in Field Screen) when Exposed to Viruliferous Whiteflies (Mixed Infected with CuLCrV and SIGMFV) under Greenhouse Conditions
3. Results
3.1. Response of Phaseolus spp. (Snap Beans and Lima Beans) Genotypes to Leaf Crumple Disease in the Field
3.2. Confirmation of Begomoviruses (CuLCrV and/or SiGMFV) Infection in Phaseolus spp.
3.3. Accumulation of CuLCrV and SiGMFV and Leaf Crumple Severity in P. vulgaris Genotypes (Susceptible vs. Resistant; Identified in Field Screen) when Exposed to Viruliferous Whiteflies (Mixed Infected with CuLCrV and SIGMFV) under Greenhouse Conditions
3.4. Whitefly Count
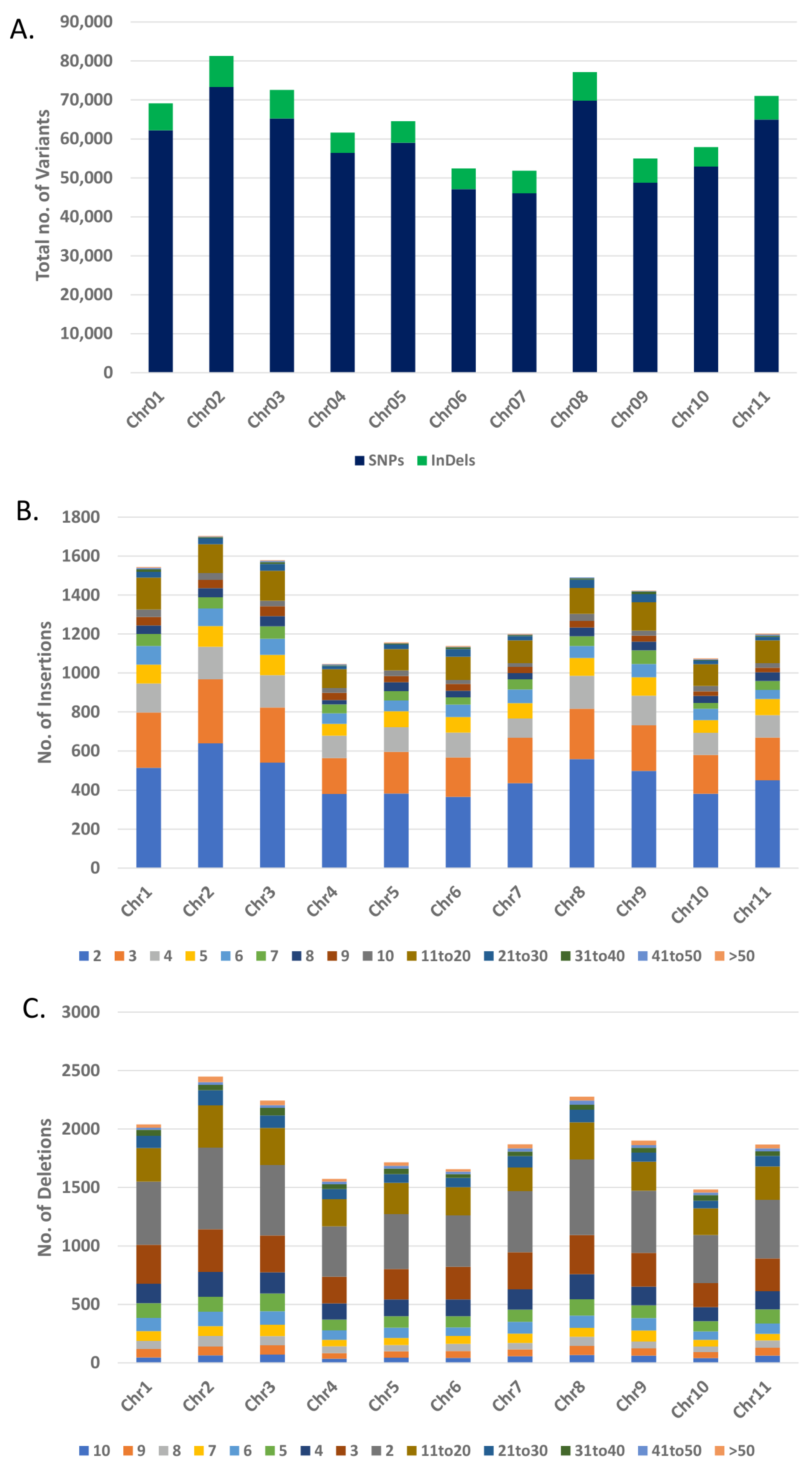
3.5. Data filtering, Mapping and Variants Identification
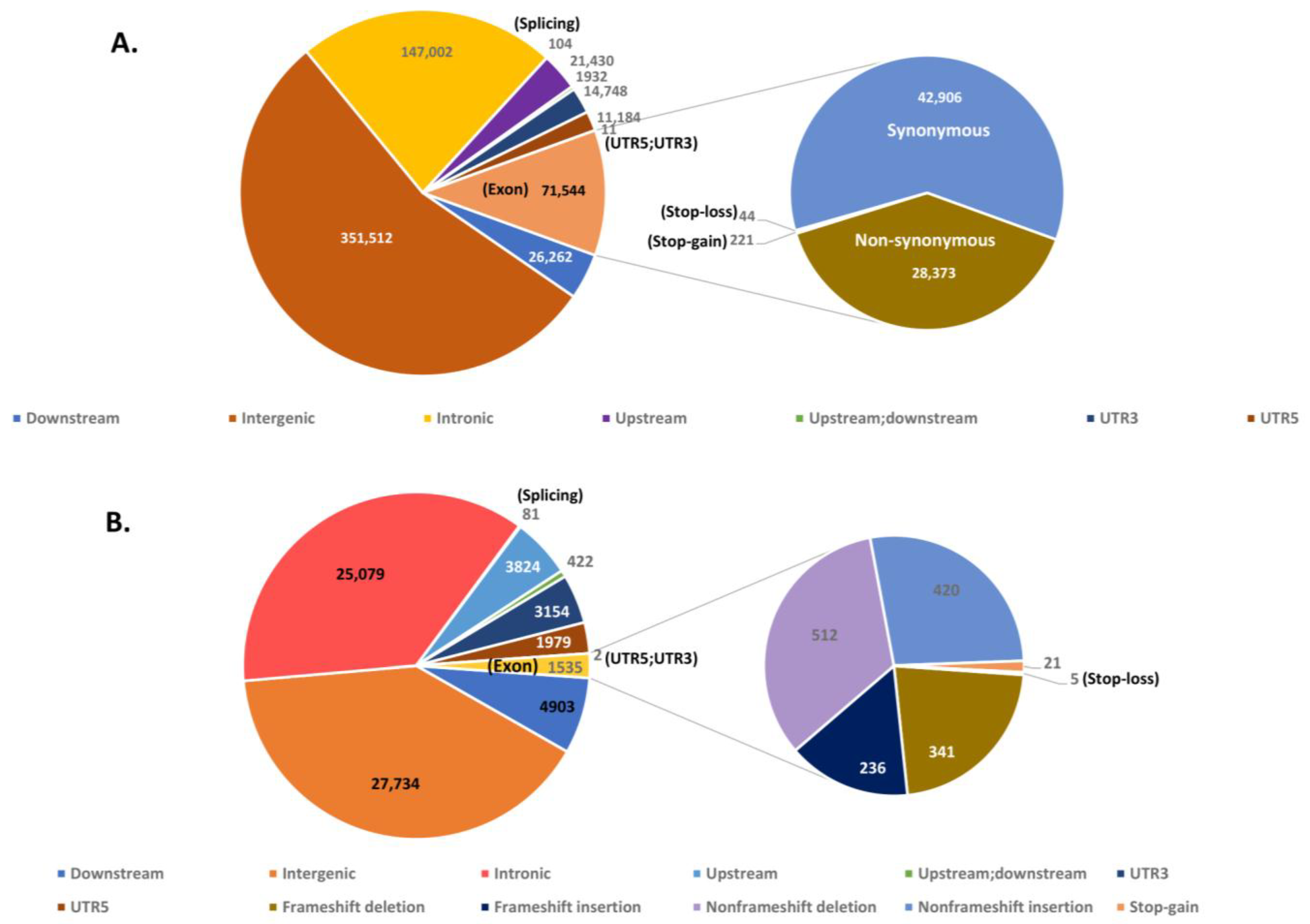
3.6. Analysis and Annotation of SNPs and InDels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- United States Dry Bean Council. Available online: https://www.usdrybeans.com/industry/production-facts/ (accessed on 20 November 2020).
- Georgia Agricultural Commodity Rankings. 2018. Available online: https://caed.uga.edu/content/dam/caes-subsite/caed/publications/annual-reports-farm-gate-value-reports/2018%20Farm%20Gate.pdf (accessed on 22 November 2020).
- Gautam, S. The Role of Bemisia tabaci in the Transmission of Vegetable Viruses in the Farmscape of Georgia; University of Georgia: Athens, GA, USA, 2019. [Google Scholar]
- Guzman, P.; Sudarshana, M.; Seo, Y.-S.; Rojas, M.; Natwick, E.; Turini, T.; Mayberry, K.; Gilbertson, R. A new bipartite geminivirus (begomovirus) causing leaf curl and crumpling in cucurbits in the Imperial Valley of California. Plant Dis. 2000, 84, 488. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.; Kmiecik, K. First report of Cucurbit leaf crumple virus in snap bean in Georgia. In Proceedings of the Phytopathology, 2010 American Phytopathological Society Annual Meeting, Charlotte, NC, USA, 7–11 August 2010; p. S68. [Google Scholar]
- Durham, T.; Baker, C.; Jones, L.; Snyder, L.U. First report of Sida golden mosaic virus infecting snap bean (Phaseolus vulgaris) in Florida. Plant Dis. 2010, 94, 487. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.; Crossley, M.S.; Dutta, B.; Coolong, T.; Simmons, A.M.; da Silva, A.; Snyder, W.E.; Srinivasan, R. Low Genetic Variability in Bemisia tabaci MEAM1 Populations within Farmscapes of Georgia, USA. Insects 2020, 11, 834. [Google Scholar] [CrossRef]
- Mukeshimana, G.; Butare, L.; Cregan, P.B.; Blair, M.W.; Kelly, J.D. Quantitative trait loci associated with drought tolerance in common bean. Crop Sci. 2014, 54, 923–938. [Google Scholar] [CrossRef]
- Villordo-Pineda, E.; González-Chavira, M.M.; Giraldo-Carbajo, P.; Acosta-Gallegos, J.A.; Caballero-Pérez, J. Identification of novel drought-tolerant-associated SNPs in common bean (Phaseolus vulgaris). Front. Plant Sci. 2015, 6, 546. [Google Scholar] [CrossRef]
- Moghaddam, S.M.; Mamidi, S.; Osorno, J.M.; Lee, R.; Brick, M.; Kelly, J.; Miklas, P.; Urrea, C.; Song, Q.; Cregan, P. Genome-wide association study identifies candidate loci underlying agronomic traits in a Middle American diversity panel of common bean. Plant Genome 2016, 9, 1–21. [Google Scholar] [CrossRef]
- Extension Bulletin. 2007. Available online: https://secure.caes.uga.edu/extension/publications/files/pdf/B%201369_4.PDF (accessed on 30 November 2020).
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Legarrea, S.; Barman, A.; Marchant, W.; Diffie, S.; Srinivasan, R. Temporal effects of a Begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci, and implications for epidemics. PLoS ONE 2015, 10, e0142114. [Google Scholar] [CrossRef]
- Gadhave, K.R.; Gautam, S.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Low Frequency of Horizontal and Vertical Transmission of Cucurbit Leaf Crumple Virus in Whitefly Bemisia tabaci Gennadius. Phytopathology 2020, 110. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.; Gadhave, K.R.; Buck, J.W.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Virus-virus interactions in a plant host and in a hemipteran vector: Implications for vector fitness and virus epidemics. Virus Res. 2020, 286, 198069. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Jhanwar, S.; Priya, P.; Singh, V.K.; Saxena, M.S.; Parida, S.K.; Garg, R.; Tyagi, A.K.; Jain, M. Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers. PLoS ONE 2012, 7, e52443. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Clevenger, J.; Pandey, M.K.; Wang, H.; Shasidhar, Y.; Chu, Y.; Fountain, J.C.; Choudhary, D.; Culbreath, A.K.; Liu, X. High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut. Plant Biotechnol. J. 2018, 16, 1954–1967. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Clevenger, J.; Kale, S.M.; Wang, H.; Pandey, M.K.; Choudhary, D.; Yuan, M.; Wang, X.; Culbreath, A.K.; Holbrook, C.C. A recombination bin-map identified a major QTL for resistance to Tomato Spotted Wilt Virus in peanut (Arachis hypogaea). Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Chavarro, C.; Agarwal, G.; Bertioli, D.J.; Leal-Bertioli, S.C.; Pandey, M.K.; Vaughn, J.; Abernathy, B.; Barkley, N.A. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol. Plant 2017, 10, 309–322. [Google Scholar] [CrossRef]
- Oladzad, A.; Porch, T.; Rosas, J.C.; Moghaddam, S.M.; Beaver, J.; Beebe, S.E.; Burridge, J.; Jochua, C.N.; Miguel, M.A.; Miklas, P.N. Single and multi-trait GWAS identify genetic factors associated with production traits in common bean under abiotic stress environments. G3: GenesGenomesGenet. 2019, 9, 1881–1892. [Google Scholar]
- Lobaton, J.D.; Miller, T.; Gil, J.; Ariza, D.; de la Hoz, J.F.; Soler, A.; Beebe, S.; Duitama, J.; Gepts, P.; Raatz, B. Resequencing of common bean identifies regions of inter–gene pool introgression and provides comprehensive resources for molecular breeding. Plant Genome 2018, 11, 1–21. [Google Scholar] [CrossRef]
- Perseguini, J.M.K.C.; Oblessuc, P.R.; Rosa, J.R.B.F.; Gomes, K.A.; Chiorato, A.F.; Carbonell, S.A.M.; Garcia, A.A.F.; Vianello, R.P.; Benchimol-Reis, L.L. Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLoS ONE 2016, 11, e0150506. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, X.; Yuan, H.; Araki, H.; Wang, J.; Tian, D. The pattern of insertion/deletion polymorphism in Arabidopsis thaliana. Mol. Genet. Genom. 2008, 280, 351–361. [Google Scholar] [CrossRef]
- Jain, M.; Moharana, K.C.; Shankar, R.; Kumari, R.; Garg, R. Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol. J. 2014, 12, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Shankar, R.; Jain, M.; Subudhi, P.K. Genome-wide discovery of DNA polymorphisms by whole genome sequencing differentiates weedy and cultivated rice. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Hyten, D.; Specht, J.; Matukumalli, L.; Song, Q.; Quigley, C.; Lee, M.; Chase, K.; Lark, K.; Reiter, R. A soybean transcript map: Discovery and mapping of single nucleotide polymorphisms in soybean genes. Genetics 2007, 176, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Saxena, R.K.; Upadhyaya, H.D.; Khan, A.W.; Yu, Y.; Kim, C.; Rathore, A.; Kim, D.; Kim, J.; An, S. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 2017, 49, 1082. [Google Scholar] [CrossRef]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.; Li, X.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012, 30, 105–111. [Google Scholar] [CrossRef]
- Mace, E.S.; Tai, S.; Gilding, E.K.; Li, Y.; Prentis, P.J.; Bian, L.; Campbell, B.C.; Hu, W.; Innes, D.J.; Han, X. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Wang, J.; Raskin, L.; Samuels, D.C.; Shyr, Y.; Guo, Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics 2015, 31, 318–323. [Google Scholar] [CrossRef]
- Bai, H.; Cao, Y.; Quan, J.; Dong, L.; Li, Z.; Zhu, Y.; Zhu, L.; Dong, Z.; Li, D. Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of foxtail millet. PLoS ONE 2013, 8, e73514. [Google Scholar] [CrossRef]
- Liu, S.; An, Y.; Tong, W.; Qin, X.; Samarina, L.; Guo, R.; Xia, X.; Wei, C. Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis) and development of InDel markers for genetic research. BMC Genom. 2019, 20, 935. [Google Scholar] [CrossRef]
- Ramakrishna, G.; Kaur, P.; Nigam, D.; Chaduvula, P.K.; Yadav, S.; Talukdar, A.; Singh, N.K.; Gaikwad, K. Genome-wide identification and characterization of InDels and SNPs in Glycine max and Glycine soja for contrasting seed permeability traits. Bmc Plant Biol. 2018, 18, 141. [Google Scholar] [CrossRef] [PubMed]
- Batley, J.; Barker, G.; O’Sullivan, H.; Edwards, K.J.; Edwards, D. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003, 132, 84–91. [Google Scholar] [CrossRef] [PubMed]

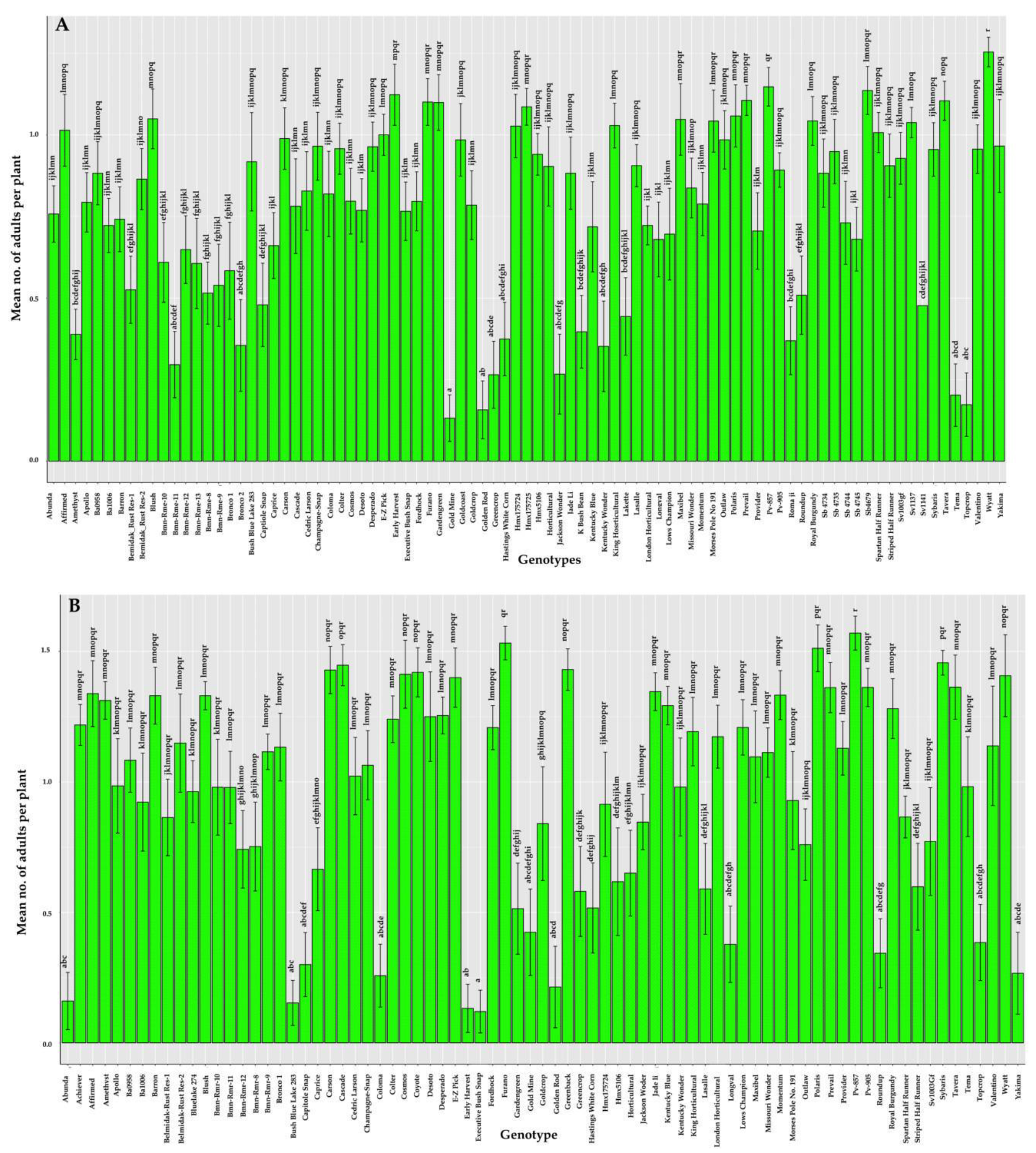

| Serial number | Genotype | Source | Disease Severity (%) | 2019 Mean (%) ± SE | HR/MR/S/HS | |||
|---|---|---|---|---|---|---|---|---|
| 2018 30 DAS | HR a/MR b/S c/HS d | 2019 30 DAS e | 2019 45 DAS | |||||
| 1 | Abunda * | GRIN | 77 f | HS | 100 | 100 | 100 ± 0 | HS |
| 2 | Affirmed * | Seminis | 17 | HR | 53 | 57 | 55 ± 2 | S |
| 3 | Amethyst * | Johhny’s seed | 20 | HR | 33 | 33 | 33 ± 0 | MR |
| 4 | Apollo * | GRIN | 33 | MR | 60 | 60 | 60 ± 0 | S |
| 5 | BA0958 * | Jenna | 53 | S | 70 | 74 | 72 ± 2 | HS |
| 6 | BA1006 * | Jenna | 77 | HS | 65 | 65 | 65 ± 0 | S |
| 7 | Barron * | Harris Moran | 30 | MR | 73 | 27 | 50 ± 23 | MR |
| 8 | Belmidak-Rust Resistant-1 * | GRIN | 73 | HS | 60 | 60 | 60 ± 0 | S |
| 9 | Belmidak-Rust Resistant-2 * | GRIN | 67 | HS | 40 | 37 | 38.5 ± 1.5 | MR |
| 10 | BLUSH * | GRIN | 17 | HR | 47 | 50 | 48.5 ± 1.5 | MR |
| 11 | BMN- RMR- 13 * | GRIN | 52 | S | NS g | NS | _ | _ |
| 12 | BMN-RMR-10 * | GRIN | 53 | S | 63 | 67 | 65 ± 2 | S |
| 13 | BMN-RMR-11 * | GRIN | 73 | HS | 70 | 77 | 73.5 ± 3.5 | HS |
| 14 | BMN-RMR-12 * | GRIN | 80 | HS | 83 | 86 | 84.5 ± 1.5 | HS |
| 15 | BMN-RMR-8 * | GRIN | 50 | MR | 72 | 80 | 76 ± 4 | HS |
| 16 | BMN-RMR-9 * | GRIN | 60 | S | 70 | 70 | 70 ± 0 | HS |
| 17 | Bronco 1 * | Seminis | 80 | HS | 53 | 57 | 55 ± 2 | S |
| 18 | Bronco 2 * | Seminis | 90 | HS | NS | NS | _ | _ |
| 19 | Bush Blue Lake 283 * | Asgrow Seed Co | 80 | HS | 100 | 100 | 100 ± 0 | HS |
| 20 | Capitole Snap * | GRIN | 80 | HS | 70 | 73 | 71.5 ± 1.5 | HS |
| 21 | Caprice * | Harris Moran | 87 | HS | 100 | 100 | 100 ± 0 | HS |
| 22 | Carson * | Syngenta | 22 | MR | 43 | 47 | 45 ± 2 | MR |
| 23 | Cascade * | GRIN | 42 | MR | 57 | 59 | 58 ± 1 | S |
| 24 | Cedric Larson * | GRIN | 27 | MR | 37 | 43 | 40 ± 3 | MR |
| 25 | Champagne * | GRIN | 55 | S | 47 | 59 | 53 ± 6 | S |
| 26 | Coloma * | GRIN | 97 | HS | 93 | 100 | 96.5 ± 3.5 | HS |
| 27 | Colter * | Harris Moran | 27 | MR | 57 | 60 | 58.5 ± 1.5 | S |
| 28 | Cosmos * | Johnny’s seed | 60 | S | 53 | 63 | 58 ± 5 | S |
| 29 | Desoto * | Harris Moran | 20 | HR | 40 | 50 | 45 ± 5 | MR |
| 30 | Desperado * | Burpee | 20 | HR | 47 | 47 | 47 ± 0 | MR |
| 31 | Early Harvest * | GRIN | 80 | HS | 60 | 100 | 80 ± 20 | HS |
| 32 | Executive Bush Snap * | GRIN | 87 | HS | 100 | 100 | 100 ± 0 | HS |
| 33 | E-Z pick * | Johhny’s seed | 82 | HS | 97 | 99 | 98 ± 1 | HS |
| 34 | Fordhook * | Seedway | 23 | MR | 25 | 18 | 21.5 ± 3.5 | MR |
| 35 | Furano * | Syngenta | 22 | MR | 32 | 40 | 36 ± 4 | MR |
| 36 | Gardengreen * | GRIN | 33 | MR | 67 | 83 | 75 ± 8 | HS |
| 37 | Gold Mine * | Seminis | 87 | HS | 100 | 100 | 100 ± 0 | HS |
| 38 | Goldcoast * | GRIN | 67 | HS | 100 | 100 | 100 ± 0 | HS |
| 39 | Goldcrop * | GRIN | 17 | HR | 47 | 47 | 47 ± 0 | MR |
| 40 | Greencrop * | Seedway | 80 | HS | 88 | 67 | 77.5 ± 10.5 | HS |
| 41 | Hastings White Cornfield * | GRIN | 35 | MR | 45 | 45 | 45 ± 0 | MR |
| 42 | Hmx175724 * | Harris Moran | 27 | MR | 50 | 47 | 48.5 ± 1.5 | MR |
| 43 | Hmx5106 * | Harris Moran | 12 | HR | 47 | 46 | 46.5 ± 0.5 | MR |
| 44 | Horticultural * | Seedway | 93 | HS | 81 | 90 | 85.5 ± 4.5 | HS |
| 45 | Jackson Wonder * | GRIN | 5 | HR | 23 | 12 | 17.5 ± 5.5 | HR |
| 46 | Jade II * | Harris Moran | 40 | MR | 57 | 57 | 57 ± 0 | S |
| 47 | Kentucky Blue * | Sieger | 50 | MR | 70 | 72 | 71 ± 1 | HS |
| 48 | Kentucky Wonder * | Seedway | 35 | MR | 67 | 68 | 67.5 ± 0.5 | HS |
| 49 | King Horticultural * | GRIN | 40 | MR | 60 | 65 | 62.5 ± 2.5 | S |
| 50 | Lakatte * | GRIN | 93 | HS | NS | NS | _ | _ |
| 51 | Lasalle * | Harris Moran | 80 | HS | 95 | 100 | 97.5 ± 2.5 | HS |
| 52 | London Horticultural * | GRIN | 33 | MR | 50 | 57 | 53.5 ± 3.5 | S |
| 53 | Longval * | GRIN | 87 | HS | 77 | 95 | 86 ± 9 | HS |
| 54 | Lows Champion * | GRIN | 17 | HR | 67 | 43 | 55 ± 12 | S |
| 55 | Maxibel * | Johhny’s seed | 57 | S | 53 | 57 | 55 ± 2 | S |
| 56 | Missouri Wonder * | GRIN | 57 | S | 60 | 80 | 70 ± 10 | HS |
| 57 | Momentum * | Syngenta | 20 | HR | 37 | 40 | 38.5 ± 1.5 | MR |
| 58 | Morses Pole No 191 * | GRIN | 53 | S | 53 | 53 | 53 ± 0 | S |
| 59 | Outlaw * | Stokes seeds | 73 | HS | 47 | 63 | 55 ± 8 | S |
| 60 | Polaris * | GRIN | 40 | MR | 66 | 70 | 68 ± 2 | HS |
| 61 | Prevail * | Syngenta | 13 | HR | 45 | 47 | 46 ± 1 | MR |
| 62 | Provider * | Seedway | 93 | HS | 95 | 97 | 96 ± 1 | HS |
| 63 | PV-857 * | Seedway | 20 | HR | 35 | 37 | 36 ± 1 | MR |
| 64 | PV-905 * | PopVriend | 27 | MR | 53 | 53 | 53 ± 0 | S |
| 65 | Roma II * | Seedway | 93 | HS | 80 | 100 | 90 ± 10 | HS |
| 66 | Roundup * | GRIN | 80 | HS | 80 | 87 | 83.5 ± 3.5 | HS |
| 67 | Royal Burgundy * | Johhny’s seed | 23 | MR | 60 | 43 | 51.5 ± 8.5 | S |
| 68 | SB4679 * | GRIN | 17 | HR | NS | NS | _ | _ |
| 69 | SB4734 * | GRIN | 20 | HR | NS | NS | _ | _ |
| 70 | SB4735 * | GRIN | 50 | MR | NS | NS | _ | _ |
| 71 | SB4744 * | GRIN | 37 | MR | NS | NS | _ | _ |
| 72 | Spartan Half Runner * | GRIN | 53 | S | 40 | 40 | 40 ± 0 | MR |
| 73 | Striped Half Runner * | GRIN | 33 | MR | 79 | 39 | 59 ± 20 | S |
| 74 | SV1003GF * | Stokes seed | 20 | HR | 70 | 40 | 55 ± 15 | S |
| 75 | SV1137 * | GRIN | 63 | S | NS | NS | _ | _ |
| 76 | Sybaris * | Seminis | 13 | HR | 35 | 37 | 36 ± 1 | MR |
| 77 | Tavera * | Johhny seed | 53 | S | 43 | 53 | 48 ± 5 | MR |
| 78 | Tema * | Semins | 5 | HR | 47 | 50 | 48.5 ± 1.5 | MR |
| 79 | Topcrop * | Seedway | 100 | HS | 77 | 83 | 80 ± 3 | HS |
| 80 | Valentino * | Stokes seed | 17 | HR | 66 | 45 | 55.5 ± 10.5 | S |
| 81 | Wyatt * | Harris Moran | 37 | MR | 37 | 40 | 38.5 ± 1.5 | MR |
| 82 | Yakima * | GRIN | 20 | HR | 53 | 57 | 55 ± 2 | S |
| 83 | Achiever | Dave’s garden | NS | _ | 53 | 57 | 55 ± 2 | S |
| 84 | Bluelake 274 | Ferry Morse | NS | _ | 75 | 75 | 75 ± 0 | HS |
| 85 | Coyote | Syngenta | NS | _ | 45 | 47 | 46 ± 1 | MR |
| 86 | Golden Rod | Seminis | 77 | HS | 100 | 100 | 100 ± 0 | HS |
| 87 | Greenback | Seedway | NS | _ | 40 | 40 | 40 ± 0 | MR |
| 88 | K Bush Bean | GRIN | 83 | HS | 97 | 100 | 98.5 ± 1.5 | HS |
| Total Raw Reads | Filtered Clean Reads | Filtered Data (Gb) | Total Reads Mapped | Av Reads Mapped (%) |
|---|---|---|---|---|
| 6,033,783,354 | 6,026,076,892 | 903.6 | 5,204,929,327 | 88.59 |
| Chromosome No. | Size (Mb) | No. of SNPs | No. of Insertions | No. of Deletions | No. of InDels |
|---|---|---|---|---|---|
| Chr01 | 51.43 | 62,199 | 3156 | 3766 | 6922 |
| Chr02 | 49.67 | 73,326 | 3522 | 4429 | 7951 |
| Chr03 | 53.44 | 65,222 | 3241 | 4094 | 7335 |
| Chr04 | 48.05 | 56,422 | 2249 | 2928 | 5177 |
| Chr05 | 40.92 | 59,007 | 2383 | 3146 | 5529 |
| Chr06 | 31.24 | 47,079 | 2339 | 3014 | 5353 |
| Chr07 | 40.04 | 46,028 | 2500 | 3301 | 5801 |
| Chr08 | 63.05 | 69,823 | 3144 | 4177 | 7321 |
| Chr09 | 38.25 | 48,746 | 2830 | 3414 | 6244 |
| Chr10 | 44.30 | 52,900 | 2226 | 2793 | 5019 |
| Chr11 | 53.58 | 64,977 | 2575 | 3488 | 6063 |
| Total | 513.97 | 645,729 | 30,165 | 38,550 | 68,715 |
| Substitution Type | Substitution | Count |
|---|---|---|
| Transversions (Tv) | C/G | 46,390 |
| G/T | 59,644 | |
| A/C | 59,095 | |
| A/T | 73,275 | |
| Transitions (Ts) | A/G | 204,568 |
| C/T | 202,757 | |
| Ratio | Ts | 407,325 |
| Tv | 238,404 | |
| Ts/Tv | 1.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, G.; Kavalappara, S.R.; Gautam, S.; Silva, A.d.; Simmons, A.; Srinivasan, R.; Dutta, B. Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA. Insects 2021, 12, 49. https://doi.org/10.3390/insects12010049
Agarwal G, Kavalappara SR, Gautam S, Silva Ad, Simmons A, Srinivasan R, Dutta B. Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA. Insects. 2021; 12(1):49. https://doi.org/10.3390/insects12010049
Chicago/Turabian StyleAgarwal, Gaurav, Saritha Raman Kavalappara, Saurabh Gautam, Andre da Silva, Alvin Simmons, Rajagopalbabu Srinivasan, and Bhabesh Dutta. 2021. "Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA" Insects 12, no. 1: 49. https://doi.org/10.3390/insects12010049
APA StyleAgarwal, G., Kavalappara, S. R., Gautam, S., Silva, A. d., Simmons, A., Srinivasan, R., & Dutta, B. (2021). Field Screen and Genotyping of Phaseolus vulgaris against Two Begomoviruses in Georgia, USA. Insects, 12(1), 49. https://doi.org/10.3390/insects12010049







