What Can the Bacterial Community of Atta sexdens (Linnaeus, 1758) Tell Us about the Habitats in Which This Ant Species Evolves?
Abstract
1. Introduction
2. Materials and Methods
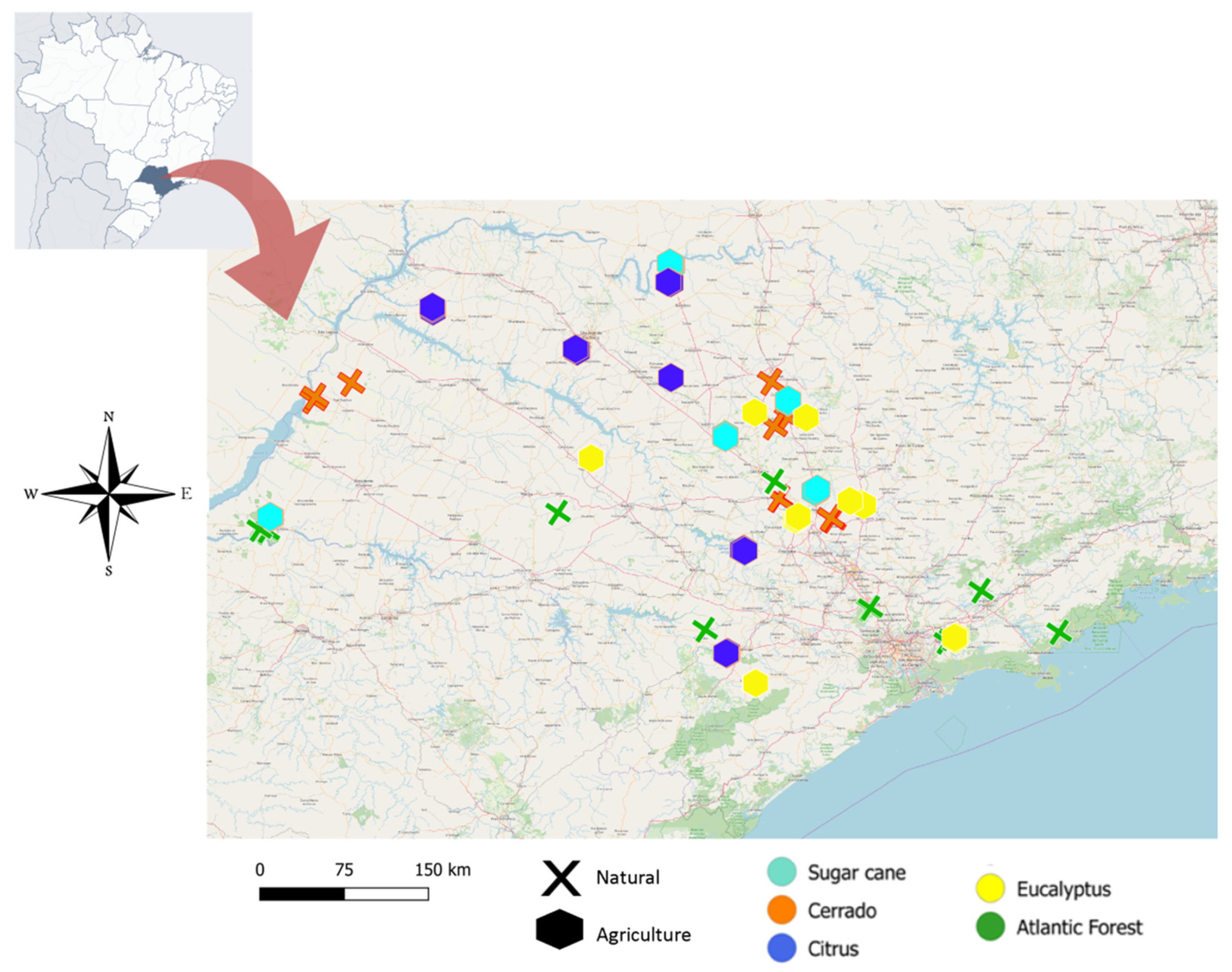
2.1. Sample Collection
2.2. DNA Extraction and Bacterial DNA Sequencing
2.3. Bioinformatic Analysis
2.4. Data Accessibility
3. Results
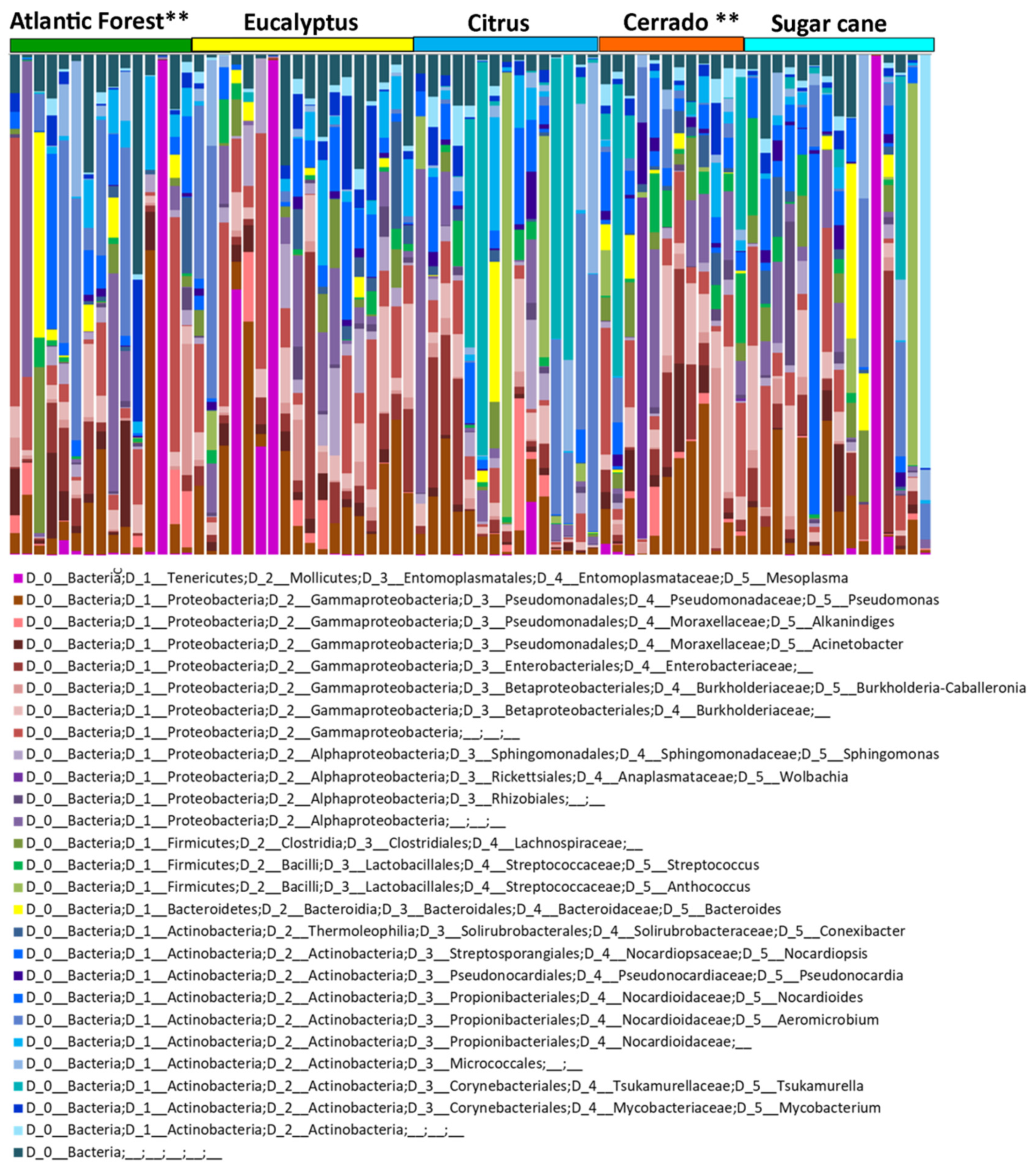
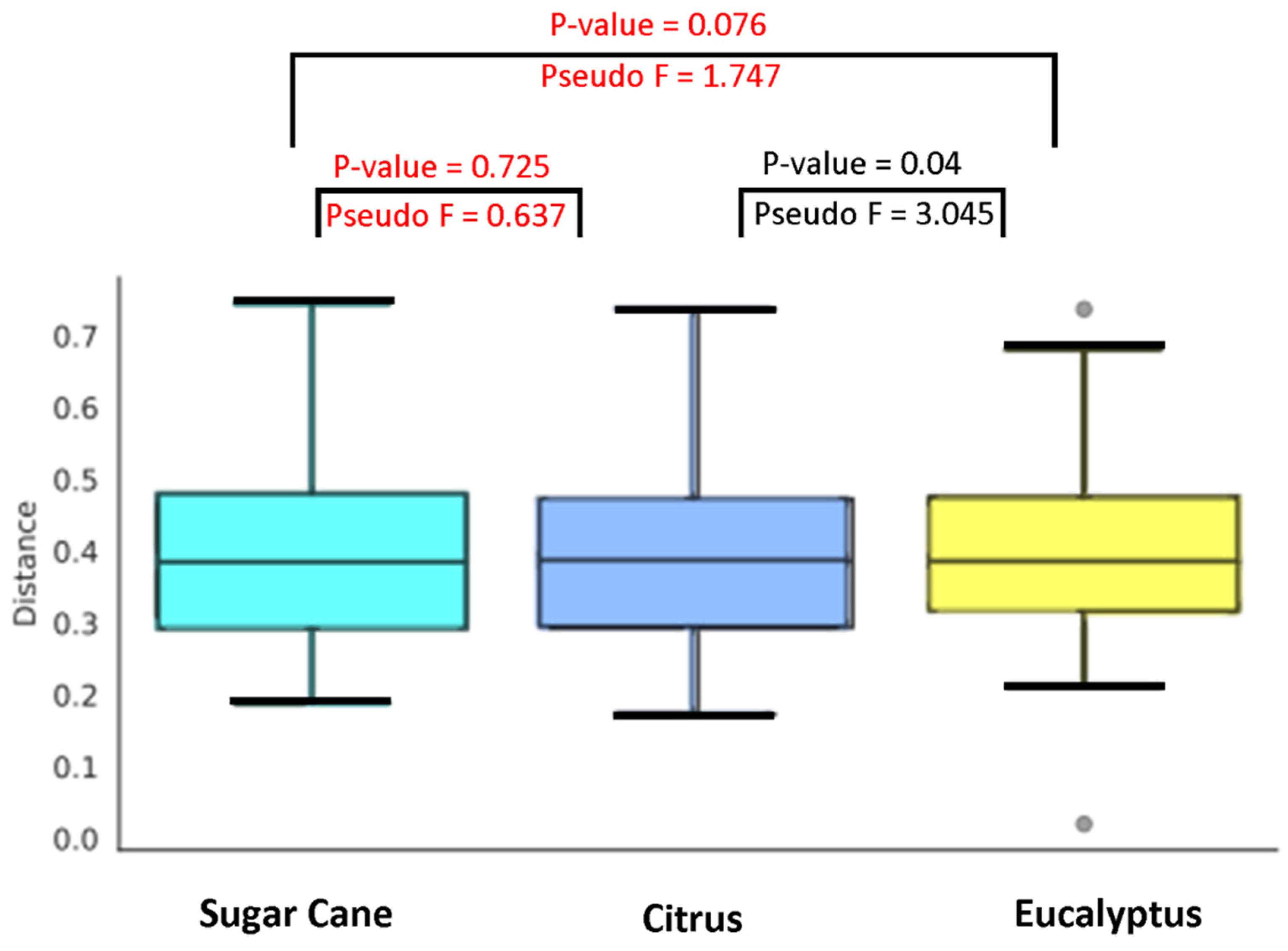
3.1. Beta Diversity and the Influence of Natural and Agricultural Habitats
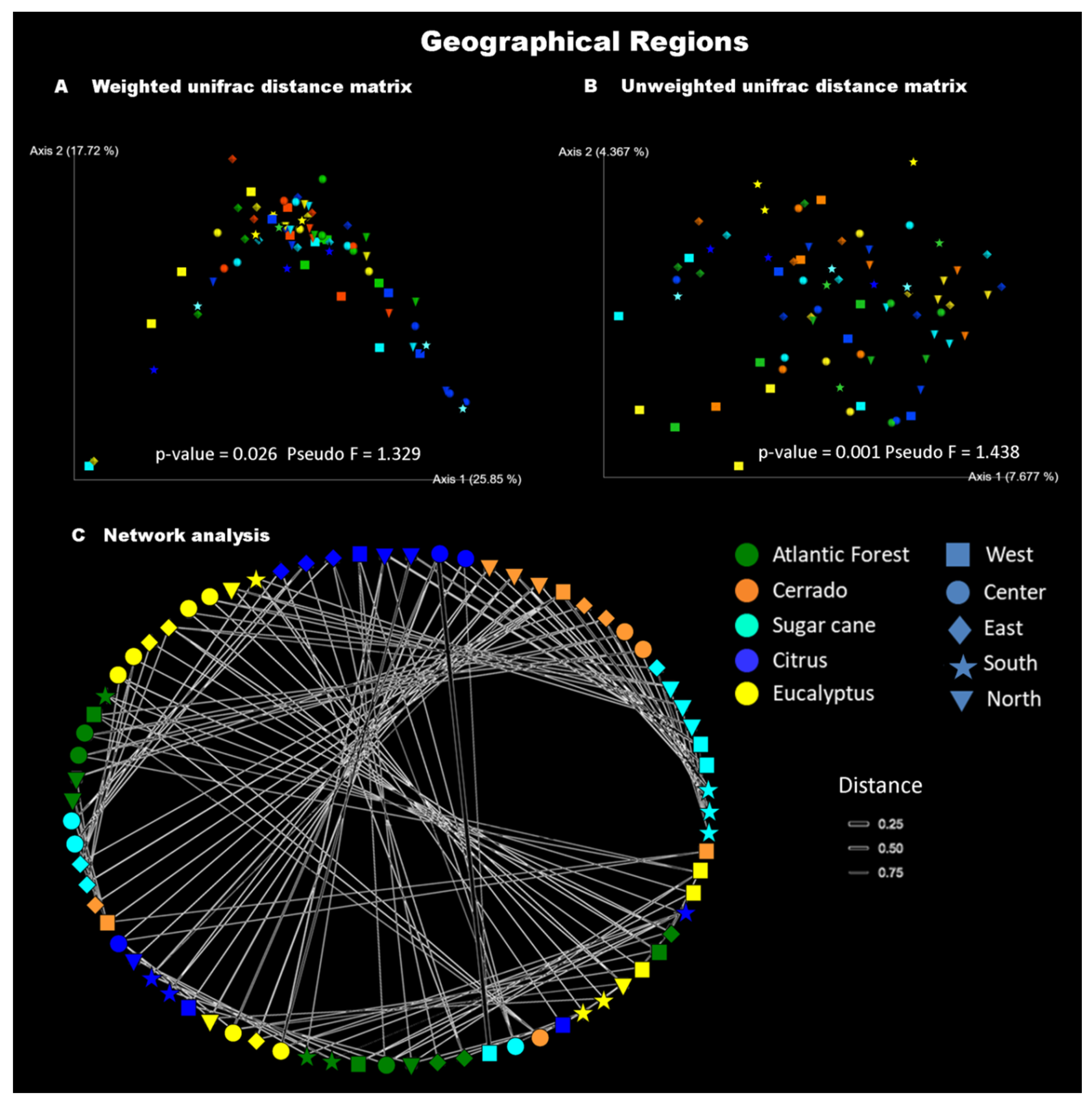
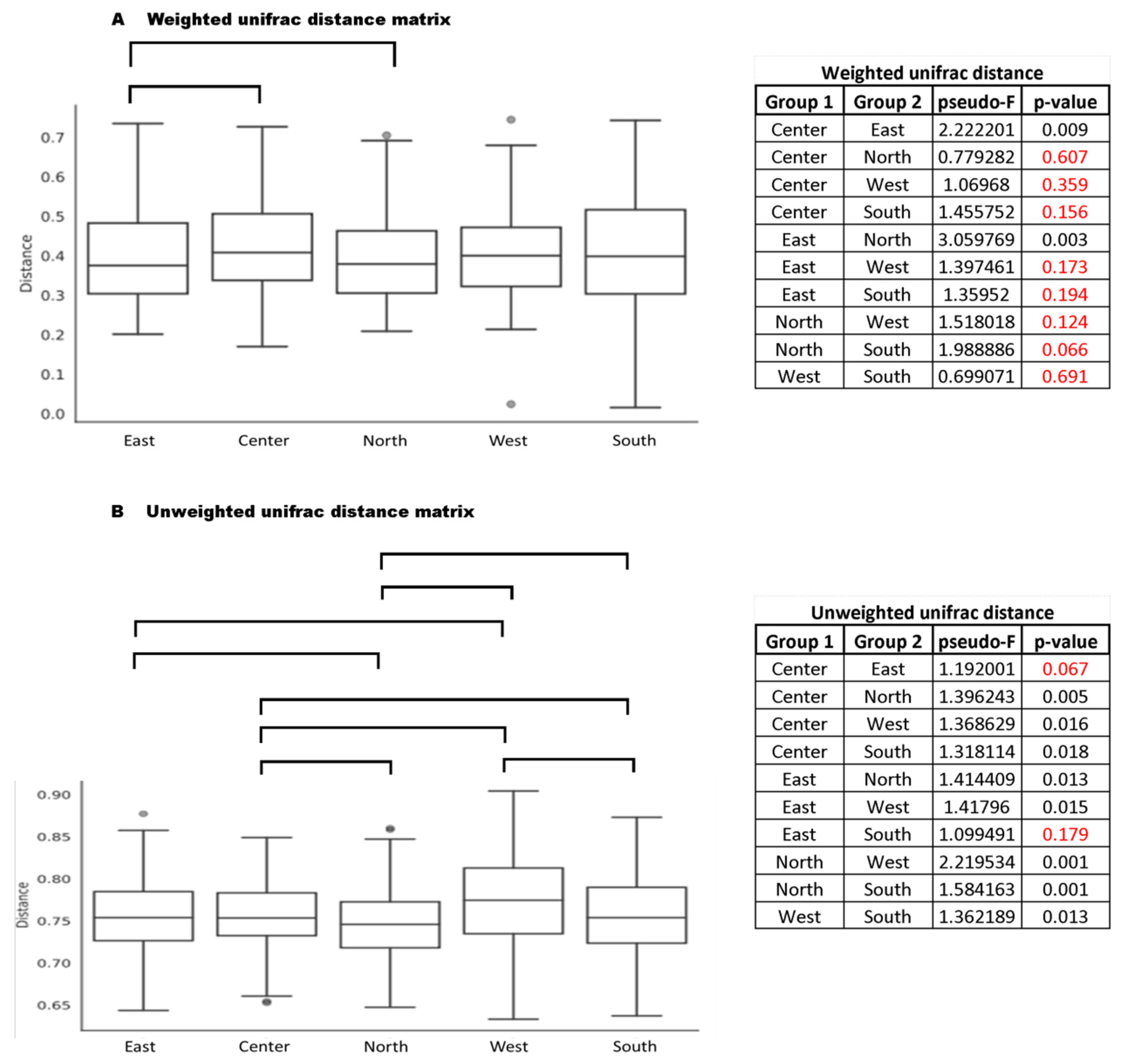
3.2. Beta Diversity and the Influence of Geographic Location
3.3. Alpha Diversity: Different Habitats and Geographical Locations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Hu, Y.; Sanders, J.G.; Łukasik, P.; D’Amelio, C.L.; Millar, J.S.; Vann, D.R.; Lan, Y.; Newton, J.A.; Schotanus, M.; Kronauer, D.J.C.; et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018, 9, 964. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, M.O.; Moreau, C.S.; Bueno, O.C. The Potential Role of Environment in Structuring the Microbiota of Camponotus across Parts of the Body. Adv. Entomol. 2019, 7, 47–70. [Google Scholar] [CrossRef][Green Version]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Species-specific signatures of the microbiome from Camponotus and Colobopsis ants across developmental stages. PLoS ONE 2017, 12, e0187461. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol. Biol. 2017, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Kautz, S.; Wray, B.D.; Moreau, C.S. Dietary specialization in mutualistic acacia-ants affects relative abundance but not identity of host-associated bacteria. Mol. Ecol. 2018, 28, 900–916. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.G.; Łukasik, P.; Frederickson, M.E.; Russell, J.A.; Koga, R.; Knight, R.; Pierce, N.E. Dramatic Differences in Gut Bacterial Densities Correlate with Diet and Habitat in Rainforest Ants. Integr. Comp. Biol. 2017, 57, 705–722. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Duplais, C.; Orivel, J.; Gibson, J.C.; Dejan, A.; Suarez, A.V.; Moreau, C.S. Development but not diet alters microbial communities in the Neotropical arboreal trap jaw ant Daceton armigerum: An exploratory study. Sci. Rep. 2020, 10, 7350. [Google Scholar] [CrossRef]
- Russell, J.A.; Moreau, C.S.; Goldman-Huertas, B.; Fujiwara, M.; Lohman, D.J.; Pierce, N.E. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. USA 2009, 106, 21236–21241. [Google Scholar] [CrossRef]
- Sanders, J.G.; Powell, S.; Kronauer, D.J.C.; Vasconcelos, H.L.; Frederickson, M.E.; Pierce, N.E. Stability and phylogenetic correlation in gut microbiota: Lessons from ants and apes. Mol. Ecol. 2014, 23, 1268–1283. [Google Scholar] [CrossRef]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Webster, G.; Weightman, A.J.; Keith Charnley, A. Diversity of gut microbiota increases with aging and starvation in the desert locust. Antonie Van Leeuwenhoek 2010, 97, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Łukasik, P.; Moreau, C.S.; Russell, J.A. Correlates of gut community composition across an ant species (Cephalotes varians) elucidate causes and consequences of symbiotic variability. Mol. Ecol. 2014, 23, 1284–1300. [Google Scholar] [CrossRef] [PubMed]
- Lanan, M.C.; Augusto, P.; Rodrigues, P.; Agellon, A.; Jansma, P.; Wheeler, D.E. A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Lazarus, A.B.; Wernegreen, J.J. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005, 15, 1023–1033. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Vieira, A.S.; Pereira, M.C.; Moreau, C.S.; Bueno, O.C. Transovarian Transmission of Blochmannia and Wolbachia Endosymbionts in the Neotropical Weaver Ant Camponotus textor (Hymenoptera, Formicidae). Curr. Microbiol. 2018, 75, 866–873. [Google Scholar] [CrossRef]
- Wernegreen, J.J.; Kauppinen, S.N.; Brady, S.G.; Ward, P.S. One nutritional symbiosis begat another: Phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol. Biol. 2009, 9, 292. [Google Scholar] [CrossRef]
- AntWeb AntWeb. Available online: https://www.antweb.org (accessed on 24 February 2017).
- Russell, J.A.; Sanders, J.G.; Moreau, C.S. Hotspots for symbiosis: Function, evolution, and specificity of ant-microbe associa-tions from trunk to tips of the ant phylogeny (Hymenoptera: Formicidae). Myrmecol. News 2017, 24, 43–69. [Google Scholar]
- Bolton, B. An Online Catalog of the Ants of the World. Available online: http://www.antcat.org/ (accessed on 20 October 2016).
- Andersen, S.B.; Boye, M.; Nash, D.R.; Boomsma, J.J. Dynamic Wolbachia prevalence in Acromyrmex leaf-cutting ants: Potential for a nutritional symbiosis. J. Evol. Biol. 2012, 25, 1340–1350. [Google Scholar] [CrossRef]
- Vieira, A.S.; Ramalho, M.O.; Martins, C.; Martins, V.G.; Bueno, O.C. Microbial Communities in Different Tissues of Atta sexdens rubropilosa Leaf-cutting Ants. Curr. Microbiol. 2017, 74, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.V.; Dillon, R.J.; Dillon, V.M.; Reynolds, S.E.; Samuels, R.I. Ocurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol. Lett. 2004, 239, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.; Cafaro, M.J.; Erhardt, D.P.; Little, A.E.F.; Gerardo, N.M.; Tebbets, B.; Klein, B.S.; Currie, C.R. Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ. Microbiol. Rep. 2009, 2, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Della Lucia, T.M.; Gandra, L.C.; Guedes, R.N. Managing leaf-cutting ants: Peculiarities, trends and challenges. Pest Manag. Sci. 2014, 70, 14–23. [Google Scholar] [CrossRef]
- Mueller, U.G.; Dash, D.; Rabeling, C.; Rodrigues, A. Coevolution between Attine ants and Actinomycete bacteria: A reevaluation. Evolution 2008, 62, 2894–2912. [Google Scholar] [CrossRef]
- Della Lucia, T. Formigas Cortadeiras: Da Bioecologia ao Manejo; UFV: Viçosa, Brazil, 2011. [Google Scholar]
- Montoya-Lerma, J.; Giraldo-Echeverri, C.; Armbrecht, I.; Farji-Brener, A.; Calle, Z. Leaf-cutting ants revisited: Towards rational management and control. Int. J. Pest Manag. 2012, 58, 225–247. [Google Scholar] [CrossRef]
- Bueno, O.C.; Bueno, F.C. Manejo de Pragas Urbanas. In Controle de Formigas em Áreas Urbanas.; São FEALQ: Paulo, Brazil, 2007; pp. 67–77. [Google Scholar]
- Ortiz, G.; Cintra-Socolowski, P.; Vieira, A.S.; Bueno, O.C. Ação de produtos utilizados no cultivo da cana-de-açúcar sobre as formigas-cortadeiras. In Cana-de-Açúcar e seus Impactos: Uma visão Acadêmica; Fontanetti, C.S., Bueno, O.C., Eds.; Canal6 editora: Bauru, Brazil, 2017; pp. 179–196. [Google Scholar]
- Delabie, J.H.C.; Alves, H.R.; Reuss-Strenzel, G.M.; Carmo, A.D.; Nascimento, I.D. Distribuição das formigas cortadeiras dos gêneros Acromyrmex e Atta no Novo Mundo. In Formigas Cortadeiras: Da Bioecologia ao Manejo; Universidade Federal de Viçosa: Viçosa, Brazil, 2011; pp. 80–101. [Google Scholar]
- Groussin, M.; Mazel, F.; Sanders, J.G.; Smillie, C.S.; Lavergne, S.; Thuiller, W.; Alm, E.J. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 2017, 8, 14319. [Google Scholar] [CrossRef]
- Teseo, S.; van Zweden, J.S.; Pontieri, L.; Kooij, P.W.; Sørensen, S.J.; Wenseleers, T.; Poulsen, M.; Boomsma, J.J.; Sapountzis, P. The scent of symbiosis: Gut bacteria may affect social interactions in leaf-cutting ants. Anim. Behav. 2019, 150, 239–254. [Google Scholar] [CrossRef]
- Meirelles, L.A.; McFrederick, Q.S.; Rodrigues, A.; Mantovani, J.D.; de Melo Rodovalho, C.; Ferreira, H.; Bacci, M.; Mueller, U.G. Bacterial microbiomes from vertically transmitted fungal inocula of the leaf-cutting ant Atta texana. Environ. Microbiol. Rep. 2016, 8, 630–640. [Google Scholar] [CrossRef]
- Zhukova, M.; Sapountzis, P.; Schiøtt, M.; Boomsma, J.J. Diversity and transmission of gut bacteria in Atta and Acromyrmex leaf-cutting ants during development. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Ratter, J. The Brazilian Cerrado Vegetation and Threats to its Biodiversity. Ann. Bot. 1997, 80, 223–230. [Google Scholar] [CrossRef]
- Sano, E.E.; Rosa, R.; Luís, J.; Laerte, S.B.; Ferreira, G. Documentos 190 Mapeamento de Cobertura Vegetal do Bioma Cerrado: Estratégias e resultados Embrapa Cerrados Planaltina, DF 2007 Empresa Brasileira de Pesquisa Agropecuária Embrapa Cerrados Ministério da Agricultura, Pecuária e Abastecimento. 2007. Available online: https://www.infoteca.cnptia.embrapa.br/bitstream/doc/570887/1/doc190.pdf (accessed on 3 November 2019).
- Joly, C.A.; Metzger, J.P.; Tabarelli, M. Experiences from the Brazilian Atlantic Forest: Ecological findings and conservation initiatives. New Phytol. 2014, 204, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.C.; Metzger, J.P.; Martensen, A.C.; Ponzoni, F.J.; Hirota, M.M. The Brazilian Atlantic Forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009, 142, 1141–1153. [Google Scholar] [CrossRef]
- SOS Mata Atlântica. Atlas dos Remanescentes Florestais de Mata Atlântica. Relatório Técnico. 2019. Available online: https://www.sosma.org.br/wp-content/uploads/2019/06/Atlas-mata-atlanticaDIGITAL.pdf (accessed on 3 November 2019).
- Cepea PIB do Agronegócio de São Paulo-Centro de Estudos Avançados em Economia Aplicada-CEPEA-Esalq/USP. Available online: https://www.cepea.esalq.usp.br/br/pib-do-agronegocio-de-sao-paulo.aspx (accessed on 5 September 2019).
- Power, E.F.; Kelly, D.L.; Stout, J.C. Organic Farming and Landscape Structure: Effects on Insect-Pollinated Plant Diversity in Intensively Managed Grasslands. PLoS ONE 2012, 7, e38073. [Google Scholar] [CrossRef]
- Kaluza, B.F.; Wallace, H.M.; Heard, T.A.; Minden, V.; Klein, A.; Leonhardt, S.D. Social bees are fitter in more biodiverse environments. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Dharampal, P.S.; Diaz-Garcia, L.; Haase, M.A.B.; Zalapa, J.; Currie, C.R.; Hittinger, C.T.; Steffan, S.A. Microbial Diversity Associated with the Pollen Stores of Captive-Bred Bumble Bee Colonies. Insects 2020, 11, 250. [Google Scholar] [CrossRef]
- Dharampal, P.S.; Carlson, C.; Currie, C.R.; Steffan, S.A. Pollen-borne microbes shape bee fitness. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182894. [Google Scholar] [CrossRef]
- Hannula, S.E.; Zhu, F.; Heinen, R.; Bezemer, T.M. Foliar-feeding insects acquire microbiomes from the soil rather than the host plant. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Moran, N.A.; Ochman, H.; Hammer, T.J. Evolutionary and Ecological Consequences of Gut Microbial Communities. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 451–475. [Google Scholar] [CrossRef]
- Mackay, W.; Mackay, E. Las hormigas de Colombia: Arrieras del Genero/Atta/(Hymenoptera-Formicidae). Revista Colombiana de Entomología 1986, 12, 23–30. [Google Scholar]
- Moreau, C.S. A practical guide to DNA extraction, PCR, and gene-based DNA sequencing in insects. Halteres 2014, 5, 32–42. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. Gigascience 2012, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Paleontological Statistics Software: Package for Education and Data Analysis. Palaeontol. Electron. 2001, 1–9. [Google Scholar]
- McCune, B.; Grace, J.B.; Urban, D. Analysis of Ecological Communities; MJM Software Design: Gleneden Beach, OR, USA, 2002. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Cham, Switzerland, 2009; ISBN 9780387981406. [Google Scholar]
- Sapountzis, P.; Zhukova, M.; Hansen, L.H.; Sørensen, S.J.; Schiøtt, M.; Boomsma, J.J. Acromyrmex leaf-cutting ants have simple gut microbiota with nitrogen-fixing potential. Appl. Environ. Microbiol. 2015, 81, 5527–5537. [Google Scholar] [CrossRef]
- Kellner, K.; Ishak, H.D.; Linksvayer, T.A.; Mueller, U.G. Bacterial community composition and diversity in an ancestral ant fungus symbiosis. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef]
- Woodhams, D.C.; Brucker, R.M. Disease defence through generations: Leaf-cutter ants and their symbiotic bacteria. Mol. Ecol. 2013, 22, 4141–4143. [Google Scholar] [CrossRef]
- Tseng, S.P.; Wetterer, J.K.; Suarez, A.V.; Lee, C.Y.; Yoshimura, T.; Shoemaker, D.W.; Yang, C.C.S. Genetic Diversity and Wolbachia Infection Patterns in a Globally Distributed Invasive Ant. Front. Genet. 2019, 10, 838. [Google Scholar] [CrossRef]
- Schultz, T.R.; Brady, S.G. Major Evolutionary Transitions in Ant Agriculture; National Academy of Sciences: Washington, DC, USA, 2008; Volume 105, pp. 5435–5440. [Google Scholar]
- Chevrette, M.G.; Bratburd, J.R.; Currie, C.R.; Stubbendieck, R.M. Experimental Microbiomes: Models Not to Scale. mSystems 2019, 4, e00175-19. [Google Scholar] [CrossRef]
- Linnenbrink, M.; Wang, J.; Hardouin, E.A.; Künzel, S.; Metzler, D.; Baines, J.F. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol. Ecol. 2013, 22, 1904–1916. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Moreau, C.S. Influence of host phylogeny, geographical location and seed harvesting diet on the bacterial community of globally distributed Pheidole ants. PeerJ 2020, 2020, e8492. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodríguez, M.; Soler, J.J.; Martín-Vivaldi, M.; Martín-Platero, A.M.; Méndez, M.; Peralta-Sánchez, J.M.; Ananou, S.; Valdivia, E.; Martínez-Buenob, M. Environmental factors shape the community of symbionts in the hoopoe uropygial gland more than genetic factors. Appl. Environ. Microbiol. 2014, 80, 6714–6723. [Google Scholar] [CrossRef]
- Song, S.J.; Sanders, J.G.; Delsuc, F.; Metcalf, J.; Amato, K.; Taylor, M.W.; Mazel, F.; Lutz, H.L.; Winker, K.; Graves, G.R.; et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio 2020. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, S.; Luo, J.Y.; Wang, C.Y.; Lv, L.M.; Cui, J.J. Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Lee, A.H.; Husseneder, C.; Hooper-Bùi, L. Culture-independent identification of gut bacteria in fourth-instar red imported fire ant, Solenopsis invicta Buren, larvae. J. Invertebr. Pathol. 2008, 98, 20–33. [Google Scholar] [CrossRef][Green Version]
- Funaro, C.F.; Kronauer, D.J.C.; Moreau, C.S.; Goldman-Huertas, B.; Pierce, N.E.; Russell, J.A. Army ants harbor a host-specific clade of Entomoplasmatales bacteria. Appl. Environ. Microbiol. 2011, 77, 346–350. [Google Scholar] [CrossRef]
- Łukasik, P.; Newton, J.A.; Sanders, J.G.; Hu, Y.; Moreau, C.S.; Kronauer, D.J.C.; O’Donnell, S.; Koga, R.; Russell, J.A. The structured diversity of specialized gut symbionts of the New World army ants. Mol. Ecol. 2017, 26, 3808–3825. [Google Scholar] [CrossRef]
- Sapountzis, P.; Zhukova, M.; Shik, J.Z.; Schiott, M.; Boomsma, J.J. Reconstructing the functions of endosymbiotic mollicutes in fungus-growing ants. Elife 2018, 7, e39209. [Google Scholar] [CrossRef] [PubMed]
- Araújo Teixeira, M.; Soares de Melo, I.; Faria Vieira, R.; Eduardo Carvalho Costa, F.; Harakava, R. Microrganismos Endofíticos de Mandioca de Áreas Comerciais e Etnovariedades em três Estados Brasileiros. Pesquisa Agripecuária Brasileira 2007, 42, 42–49. [Google Scholar]
- De Assumpção, L.C.; Lacava, P.T.; Dias, A.C.F.; de Azevedo, J.L.; Menten, J.O.M. Diversidade e potencial biotecnológico da comunidade bacteriana endofítica de sementes de soja. Pesqui. Agropecuária Bras. 2009, 44, 503–510. [Google Scholar] [CrossRef]
- Velazco, L.A.; Fernández, D.B. Study of Oral Acute Toxicity/Pathogenicity of the Tsukamurella Paurometabola Active Agent of the Biological Nematicide C-924 (Hebernem®). Adv. Pharmacoepidemiol. Drug Saf. 2012. [Google Scholar] [CrossRef]
- Ishak, H.D.; Plowes, R.; Sen, R.; Kellner, K.; Meyer, E.; Estrada, D.A.; Dowd, S.E.; Mueller, U.G. Bacterial Diversity in Solenopsis invicta and Solenopsis geminata Ant Colonies Characterized by 16S amplicon 454 Pyrosequencing. Microb. Ecol. 2011, 61, 821–831. [Google Scholar] [CrossRef]
- Reyes, R.D.H.; Cafaro, M.J. Paratrechina longicornis ants in a tropical dry forest harbor specific Actinobacteria diversity. J. Basic Microbiol. 2015, 55, 11–21. [Google Scholar] [CrossRef]
- Andersen, S.B.; Hansen, L.H.; Sapountzis, P.; Sørensen, S.J.; Boomsma, J.J. Specificity and stability of the Acromyrmex-Pseudonocardia symbiosis. Mol. Ecol. 2013, 22, 4307–4321. [Google Scholar] [CrossRef]
- Aylward, F.O.; Burnum, K.E.; Scott, J.J.; Suen, G.; Tringe, S.G.; Adams, S.M.; Barry, K.W.; Nicora, C.D.; Piehowski, P.D.; Purvine, S.O.; et al. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J. 2012, 6, 1688–1701. [Google Scholar] [CrossRef]
- Bartha, R.; Lanzilotta, R.P.; Pramer, D. Stability and effects of some pesticides in soil. Appl. Microbiol. 1967, 15, 67–75. [Google Scholar] [CrossRef]
- Tu, C.M. Influence of five pyrethroid insecticides on microbial populations and activities in soil. Microb. Ecol. 1980, 5, 321–327. [Google Scholar] [CrossRef]
- van den Bosch, T.J.M.; Welte, C.U. Detoxifying symbionts in agriculturally important pest insects. Microb. Biotechnol. 2017, 10, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Saikia, N.; Das, S.K.; Patel, B.K.C.; Niwas, R.; Singh, A.; Gopal, M. Biodegradation of beta-cyfluthrin by Pseudomonas stutzeri strain S1. Biodegradation 2005, 16, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.G.D.; Moraes, L.A.; Trigo, J.R.; Omoto, C.; Cônsoli, F.L. The gut microbiota of insecticide-resistant insects houses insecticide-degrading bacteria: A potential source for biotechnological exploitation. PLoS ONE 2017, 12, e0174754. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Tago, K.; Okubo, T.; Itoh, H.; Kikuchi, Y.; Hori, T.; Sato, Y.; Nagayama, A.; Hayashi, K.; Ikeda, S.; Hayatsu, M. Insecticide-Degrading Burkholderia Symbionts of the Stinkbug Naturally Occupy Various Environments of Sugarcane Fields in a Southeast Island of Japan. Microbes Environ. 2015, 30, 29–36. [Google Scholar] [CrossRef]
- Reeves, D.D.; Price, S.L.; Ramalho, M.O.; Moreau, C.S. The Diversity and Distribution of Wolbachia, Rhizobiales, and Ophiocordyceps Within the Widespread Neotropical Turtle Ant, Cephalotes atratus (Hymenoptera: Formicidae). Neotrop. Entomol. 2020, 49, 52–60. [Google Scholar] [CrossRef]
- Kelly, M.; Price, S.L.; de Oliveira Ramalho, M.; Moreau, C.S. Diversity of Wolbachia Associated with the Giant Turtle Ant, Cephalotes atratus. Curr. Microbiol. 2019, 76, 1330–1337. [Google Scholar] [CrossRef]
- Gadagkar, R. The birth of ant genomics. Proc. Natl. Acad. Sci. USA 2011, 108, 5477–5478. [Google Scholar] [CrossRef]
- Gadau, J.; Helmkampf, M.; Nygaard, S.; Roux, J.; Simola, D.F.; Smith, C.R.; Suen, G.; Wurm, Y.; Smith, C.D. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 2012, 28, 14–21. [Google Scholar] [CrossRef]
- Libbrecht, R.; Oxley, P.R.; Kronauer, D.J.; Keller, L. Ant genomics sheds light on the molecular regulation of social organization. Genome Biol. 2013, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.R.; Moreau, C.S. ARTICLE Comparative genomics reveals convergent rates of evolution in ant–plant mutualisms. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, N.D. Dissecting ant recognition systems in the age of genomics. Biol. Lett. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Funaro, C.F.; Giraldo, Y.M.; Goldman-Huertas, B.; Suh, D.; Kronauer, D.J.C.; Moreau, C.S.; Pierce, N.E. A Veritable Menagerie of Heritable Bacteria from Ants, Butterflies, and Beyond: Broad Molecular Surveys and a Systematic Review. PLoS ONE 2012, 7, e51027. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Strategies for Enhanced Crop Resistance to Insect Pests. Annu. Rev. Plant Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef] [PubMed]

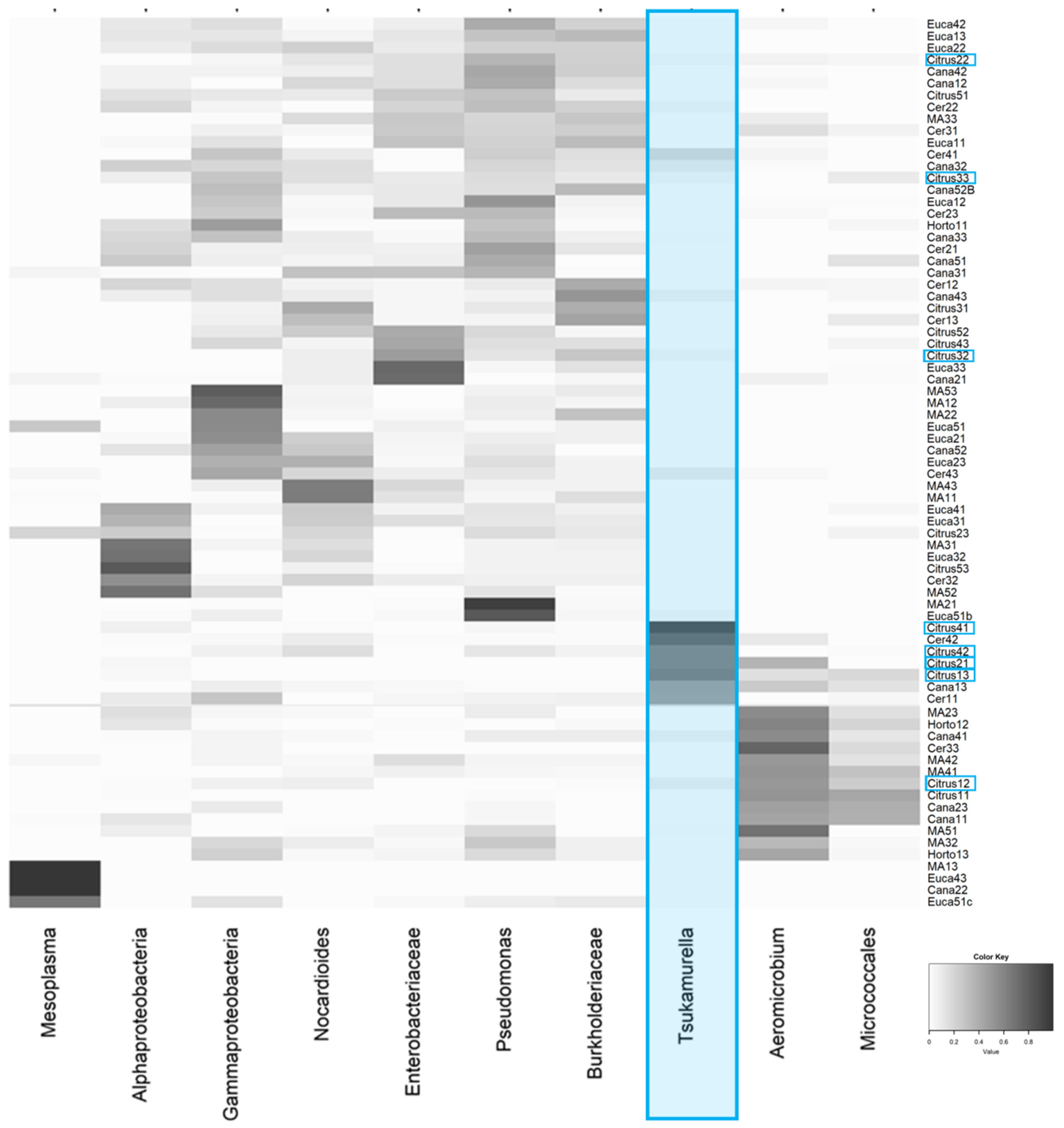
| Habitats | Overall Average Dissimilarity | Most Influential ASV/Taxonomy | Percent Contribuition to Difference | Cumulative % |
|---|---|---|---|---|
| Citrus vs. Eucalyptus | 85.00% | Tsukamurella | 12.05 | 12.05 |
| Aeromicrobium | 7.07 | 19.12 | ||
| Anthococcus | 5.89 | 25.01 | ||
| Mesoplasma | 5.11 | 30.12 | ||
| Micrococcales | 4.3 | 34.42 | ||
| Alphaproteobacteria | 3.91 | 38.32 | ||
| Enterobacteriaceae | 2.84 | 41.17 | ||
| Nocardioides | 2.66 | 43.83 | ||
| Pseudomonas | 2.53 | 46.36 | ||
| Gammaproteobacteria | 2.09 | 48.45 | ||
| Burkholderiaceae | 2.04 | 50.49 |
| Regions | Permanova Weighted Unifrac Distance | Permanova Unweighted Unifrac Distance | ||
|---|---|---|---|---|
| p-Value | Pseudo F | p-Value | Pseudo F | |
| Center | 0.025 | 1.940 | 0.001 | 1.162 |
| East | 0.254 | 1.127 | 0.018 | 1.212 |
| North | 0.494 | 0.977 | 0.001 | 1.192 |
| West | 0.075 | 1.43 | 0.014 | 1.280 |
| South | 0.376 | 1.092 | 0.016 | 1.198 |
| Regions | Overall Average Dissimilarity | Most Influential ASV/Taxonomy | Percent Contribution to Difference | Cumulative % |
|---|---|---|---|---|
| Center vs. East | 74.77% | Alphaproteobacteria | 10.63 | 10.63 |
| Aeromicrobium | 8.38 | 19.01 | ||
| Gammaproteobacteria | 7.64 | 26.65 | ||
| Mesoplasma | 6.925 | 33.58 | ||
| Pseudomonas | 6.403 | 39.98 | ||
| Tsukamurella | 5.928 | 45.91 | ||
| Micrococcales | 5.739 | 51.65 | ||
| East vs. North | 75.89% | Alphaproteobacteria | 8.30 | 8.30 |
| Gammaproteobacteria | 8.14 | 16.44 | ||
| Aeromicrobium | 7.96 | 24.40 | ||
| Tsukamurella | 7.43 | 31.83 | ||
| Mesoplasma | 7.30 | 39.13 | ||
| Nocardioides | 6.23 | 45.36 | ||
| Pseudomonas | 4.92 | 50.28 | ||
| Center vs. North | 73.01% | Aeromicrobium | 12.93 | 12.93 |
| Tsukamurella | 11.34 | 24.27 | ||
| Micrococcales | 6.81 | 31.08 | ||
| Gammaproteobacteria | 6.52 | 37.60 | ||
| Nocardioides | 5.188 | 42.79 | ||
| Alphaproteobacteria | 4.634 | 47.42 | ||
| Pseudomonas | 4.315 | 51.74 | ||
| Center vs. West | 82.17% | Tsukamurella | 11.01 | 11.01 |
| Aeromicrobium | 9.871 | 20.88 | ||
| Enterobacteriaceae | 7.052 | 27.93 | ||
| Mesoplasma | 6.649 | 34.58 | ||
| Micrococcales | 6.587 | 41.17 | ||
| Alphaproteobacteria | 6.284 | 47.45 | ||
| Bacteria | 5.37 | 52.82 | ||
| Center vs. South | 79.77% | Aeromicrobium | 8.51 | 8.51 |
| Mesoplasma | 8.15 | 16.66 | ||
| Anthococcus | 8.05 | 24.70 | ||
| Tsukamurella | 7.87 | 32.57 | ||
| Micrococcales | 5.74 | 38.31 | ||
| Burkholderiaceae | 5.10 | 43.41 | ||
| Pseudomonas | 4.73 | 48.14 | ||
| Alphaproteobacteria | 4.57 | 52.71 | ||
| East vs. West | 82.48% | Mesoplasma | 12.77 | 12.77 |
| Alphaproteobacteria | 10.09 | 22.86 | ||
| Enterobacteriaceae | 7.458 | 30.32 | ||
| Tsukamurella | 6.881 | 37.20 | ||
| Gammaproteobacteria | 6.798 | 44.00 | ||
| Pseudomonas | 5.869 | 49.87 | ||
| Bacteria | 4.881 | 54.75 | ||
| North vs. West | 81.97% | Tsukamurella | 11.83 | 11.83 |
| Aeromicrobium | 9.297 | 21.13 | ||
| Mesoplasma | 6.928 | 28.06 | ||
| Nocardioides | 5.837 | 33.89 | ||
| Gammaproteobacteria | 5.473 | 39.37 | ||
| Enterobacteriaceae | 5.438 | 44.8 | ||
| Bacteria | 4.399 | 49.2 | ||
| Nocardiopsis | 4.18 | 53.39 | ||
| North vs. South | 80.48% | Anthococcus | 9.13 | 9.13 |
| Tsukamurella | 8.84 | 17.98 | ||
| Mesoplasma | 8.41 | 26.39 | ||
| Aeromicrobium | 8.16 | 34.55 | ||
| Nocardioides | 5.86 | 40.41 | ||
| Gammaproteobacteria | 5.39 | 45.79 | ||
| Burkholderiaceae | 4.52 | 50.31 | ||
| West vs. South | 86.00% | Mesoplasma | 13.33 | 13.33 |
| Tsukamurella | 8.571 | 21.90 | ||
| Anthococcus | 8.282 | 30.18 | ||
| Enterobacteriaceae | 5.651 | 35.83 | ||
| Aeromicrobium | 5.253 | 41.08 | ||
| Burkholderiaceae | 4.228 | 45.31 | ||
| Bacteria | 4.195 | 49.51 | ||
| Pseudomonas | 4.104 | 53.61 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramalho, M.d.O.; Martins, C.; Morini, M.S.C.; Bueno, O.C. What Can the Bacterial Community of Atta sexdens (Linnaeus, 1758) Tell Us about the Habitats in Which This Ant Species Evolves? Insects 2020, 11, 332. https://doi.org/10.3390/insects11060332
Ramalho MdO, Martins C, Morini MSC, Bueno OC. What Can the Bacterial Community of Atta sexdens (Linnaeus, 1758) Tell Us about the Habitats in Which This Ant Species Evolves? Insects. 2020; 11(6):332. https://doi.org/10.3390/insects11060332
Chicago/Turabian StyleRamalho, Manuela de Oliveira, Cintia Martins, Maria Santina Castro Morini, and Odair Correa Bueno. 2020. "What Can the Bacterial Community of Atta sexdens (Linnaeus, 1758) Tell Us about the Habitats in Which This Ant Species Evolves?" Insects 11, no. 6: 332. https://doi.org/10.3390/insects11060332
APA StyleRamalho, M. d. O., Martins, C., Morini, M. S. C., & Bueno, O. C. (2020). What Can the Bacterial Community of Atta sexdens (Linnaeus, 1758) Tell Us about the Habitats in Which This Ant Species Evolves? Insects, 11(6), 332. https://doi.org/10.3390/insects11060332






