Abstract
The pea aphid is an important pest of vegetables and causes serious losses worldwide. RNA interference (RNAi) is an effective pest control tool, and three sub-pathways have been described: The miRNA pathway, siRNA pathway, and piRNA pathway. A large number of genes in miRNA pathway and piRNA pathway are found to be expanded. To study the roles of these genes, the expression of 25 core RNAi genes was screened in spatiotemporal samples, artificially synthesized dsRNA and miRNA treated samples. The 25 genes were all expressed during different development stages and in different tissues. In dsRNA-treated samples and miRNA-treated samples, the expressions of genes in these three pathways were induced, especially the expanded genes. This suggests a complex network of RNAi core genes in the three sub-pathways. Treatment of miRNA seems to induce gene expression in a dosage-dependent manner. These results increase our knowledge of the siRNA pathway and related factors from RNAi pathway in aphids and promote the use of RNAi for the control of aphid pests.
1. Introduction
RNA interference (RNAi) involves post-transcriptional gene silencing. It silences complementary mRNA through a sequence-specific process [1]. RNAi occurs in all eukaryotes and affects biological activities, including developmental regulation, differentiation, and antiviral responses. The three RNAi pathways are microRNAs (miRNAs), small-interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs). They are classified depending on their region, biogenesis, mechanisms, biological roles, and functions [2]. The siRNA pathway, usually referred to as RNAi, is considered to have the most potential as a pest control strategy [3].
siRNAs are non-coding RNAs with 21–23 nts in length. Generally, siRNAs are produced from exogenous dsRNA (e.g., virus related dsRNA) while miRNA are endogenously transcribed. However, siRNA could also be produced from gonadal and somatic tissues in Drosophila melanogaster [4]. In the cytoplasm, dsRNA is cleaved into siRNA by Dicer2 [5], which is a highly conserved kind of dsRNA-specific endonuclease belonging to the RNase III family [6]. Subsequently, the siRNA interacts with the RNA-induced silencing complex (RISC) with the assistance of RISC-loading complex (RLC). The RLC was consisted of Argonaute2 and a dsRNA-binding partner protein, R2D2 which facilitate siRNA passage from Dicer2 to RISC [7]. The sense strand is sliced by Argonaute2, while the antisense strand remains in RISC [8]. Then, the RISC is guided to its target mRNA by the antisense strand by Argonaute2 which use siRNA recognizes and degrades the complementary mRNA in Drosophila. In comparison to miRNAs, siRNAs are fully complementary to target mRNA and specific genes. The miRNAs, a class of 19–24 nt single-stranded RNAs, are important in biological processes such as cell proliferation, apoptosis, and differentiation [9]. The miRNAs derive from endogenous genes encoding stem-loop primary RNAs (pri-miRNAs), which is typically transcribed from the nucleus into a primary product by RNA polymerase II [10]. In insects, the pri-miRNA is cleaved by the RNase III Drosha and the double-stranded RNA binding protein Pasha into pre-miRNAs (precursor microRNA) with a stem-loop structure of about 60–70 nts [11]. Drosha executes the initiation step of miRNA processing and the Pasha recognizes the substrate pri-miRNA [12]. With the assistance of Exportin-5 (Exp-5), pre-miRNAs are translocated to the cytoplasm. Then, mature miRNA (duplex RNA) is cleaved by another RNase III called Dicer1 and another double-stranded RNA binding protein called Loquacious which interact with Dicer1 and stimulates and directs the specific pre-miRNA processing activity [13]. miRNA is loaded into Ago1 which recognize the target gene to form RNA-induced silencing complex (RISC) where the Argonaute family is a key component. The RISC is guided to a target mRNA and inhibits target mRNA translation [14]. piRNAs are a class of non-coding small RNAs ranging from 24 to 30 nts in length. In the piRNA silencing pathway, Argonaute, Aubergine, and Piwi are the core components [15]. The piRNAs originate from a long single-stranded RNA which is transcribed from repetitive elements or genomic loci. In insects, the piRNAs are mainly derived from the genome of the transposable genetic element or a parasitic element of the host genome [16].
Expansion of portions of the miRNA and piRNA pathways has been found in aphids [17,18]. For siRNA pathway, one copy of Dicer2, Ago2, and R2D2 were already identified in D. melanogaster [19]. However, two Ago2 genes were reported in the Culex pipiens and Musca domestica [20,21] while three Ago2 genes in Glossina morsitans [22]. Specially, Tribolium castaneum has two gene copies of Ago2 and R2D2 [23]. For miRNA pathway, both Aedes aegypti and Diuraphis noxia has two Ago1 genes [21,24]. Besides, the M. domestica has two Dicer1 genes [20]. For piRNA pathway, G. morsitans has three Ago3 genes [25]. Two Aubergine genes which belong to the Piwi family were found in the Nasonia vitripennis and three in the Harpegnathos saltator [19]. However, 5–8 Piwi genes were reported in the A. aegypti, Anopheles gambiae, and D. noxia [21,24,25]. Duplications may lead to subfunctionalization or neofunctionalization in RNAi pathways and could explain different RNAi efficacy in insects. For Ago proteins, Agos belong to a multigene family that is divided into Agonaute and Piwi subgroups [26]. Different types of Ago-type proteins act in different steps of the exo-siRNA or endosiRNA pathway. A study on A. aegypti showed that the expansion of the Piwi protein family might allow Piwi proteins to be functionally specialized in the biogenesis of piRNAs from different origins [27]. The insect species studied so far have only one Pasha, but four Pasha-like genes have been found in the pea aphid, Acyrthosiphon pisum. The difference among the four Pasha proteins in the pea aphid involves a block of 95 AA at the N-terminal end. This block is repeated three times and two times in Pasha1 and Pasha2 while is absent from Pasha3 and Pasha4. However, the sequence of this 95 AA has no homology in the Interpro database and its functional significance is unknown [17]
In D. melanogaster S2 cells, RNAi-mediated downregulation of the GFP reporter included the expression of genes in siRNA, Ago2, and Dicer2. In contrast, silencing Dicer1 and Ago1 inhibited GFP silencing [28]. In Manduca sexta, Dicer2 and Ago2 mRNA levels were elevated following injection with dsRNA in a specific and dose dependent manner [29]. In A. gambiae, some of the piRNA pathway genes are involved in the exogenous dsRNA-induced silencing response [30]. Besides, the mosquitos’ soma which infected with transgenic CHIKV expressing the dsRNA-binding protein B2 suggests that dsRNA molecules are an inducer of the piRNA pathway [31].
Aphis pisum (Hemiptera: Aphididae) is a significant vegetable pest that causes large economic losses. In A. pisum, one copy of Dicer2, Ago2, and R2D2 was found in the siRNA pathway [32]. One copy of Drosha, Exp-5, duplications of Ago1 (Ago1a and Ago1b), Dicer1 (Dicer1a and Dicer1b), and Loquacious (Loquacious1 and Loquacious2), four copies of Pasha (Pasha1, Pasha2, Pasha3, and Pasha4) in the miRNA pathway were reported [17]. As for piRNA pathway, Piwi proteins are divided into Piwi and Aub. Duplication of Ago3 (Ago3a and Ago3b) and eight copies of Piwi/Aub (Piwi1, Piwi2, Piwi3, Piwi4, Piwi5, Piwi6, Piwi7, and Piwi8) of the piRNA pathway have also been reported [18]. The Aubergine belongs to the Piwi protein in pea aphid by BLAST the sequences of D. melanogaster and the study [17]. So we kept its name Piwi. Currently, researches on these genes have focused on bioinformatics. Identification, evolutionary rate, and phylogenetic analysis were studied by bioinformatics approach [17,18]. However, these genes are rarely investigated using experimental approach. There is a need to study three pathways of RNAi activity as a possible aphid control strategy [32,33,34]. To explore the potential role of these expanded genes in three sub-pathways of RNAi, the expressions of 25 core RNAi genes were detected in different development stages, various tissues, dsRNA-treated samples, and miRNA-treated samples. The hypothesis was that there may be crosstalk among miRNA pathway, siRNA pathway and piRNA pathway. These results show a complex network of RNAi core genes in the three sub-pathways.
2. Materials and Methods
2.1. Pea Aphid Strain
The genome strain of pea aphid was originally provided, in 2016, by Professor Guy Smagghe (Ghent University, Belgium). The strain was reared on broad bean (Vicia faba) at 25 ± 1 °C, 75 ± 5% relative humidity, and a 14:10 (L:D) photoperiod.
2.2. Sample Preparation for Different Development Stages and Tissues
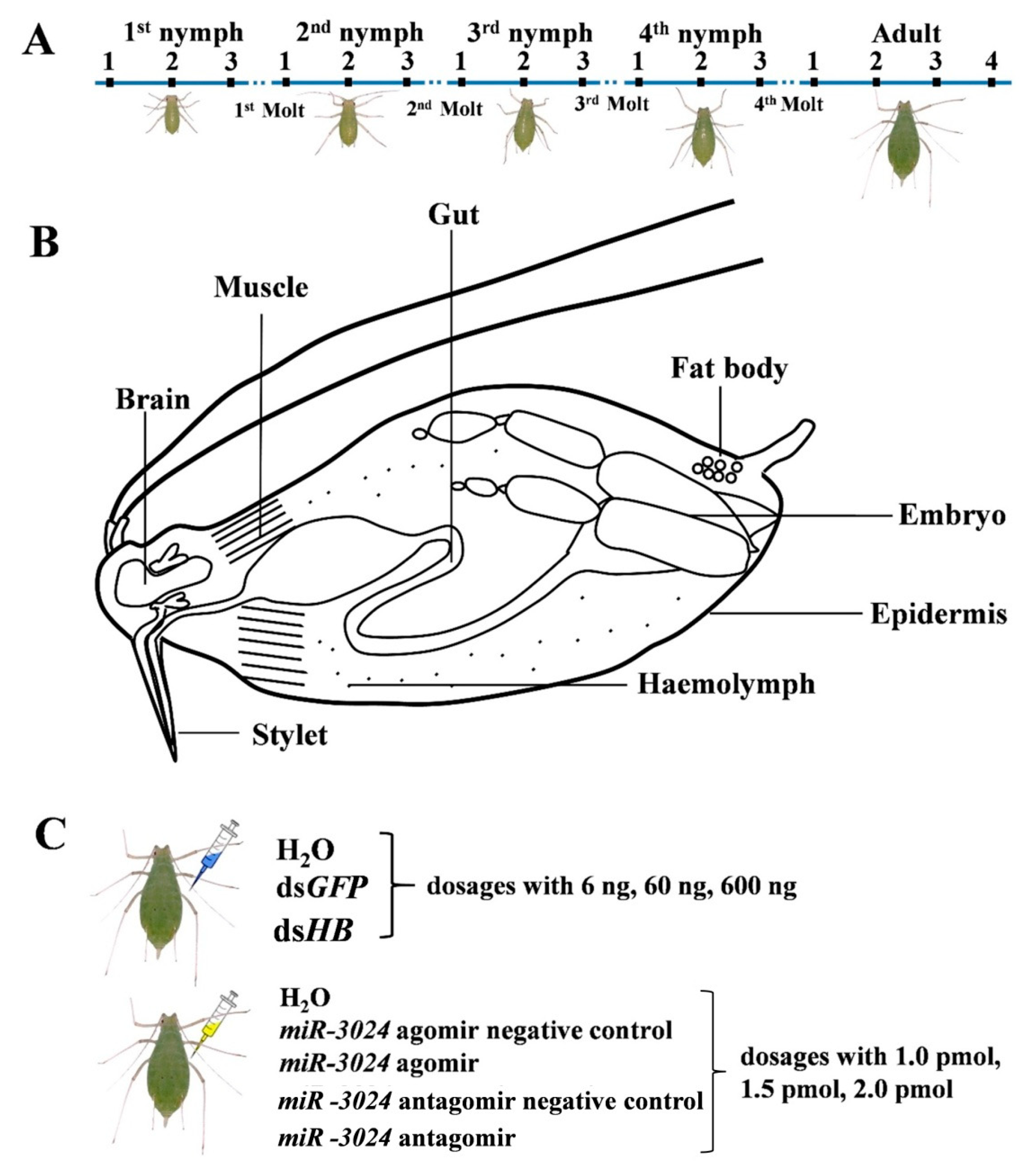
For studies on the different development stages, 500 pea aphid adults were initially cultured on broad bean seedlings, and we collected samples from succeeding generations. The samples of N1st-1, N1st-2, and N1st-3 were collected in the early 4 h, middle time, and the last 4 h before the next molt of the 1st instar nymph, respectively. Samples from other nymph stages were taken using the same pattern. Additionally, the Ad-1, Ad-2, Ad-3, and Ad-4 were collected from 1, 9, 12, and 15 d adults. Several tissues including epidermis, embryo, gut, fat body, brain, stylet, muscle, and hemolymph were isolated from adults within 12 h after molting. Four biological replicates were performed for each sample. The samples were frozen in liquid nitrogen and kept at −80 °C for future use.
2.3. RNA Extraction
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA using manufacturer’s instructions. The concentration and the purity of total RNA were evaluated at OD260/280 and OD260/230 using a NanoDrop One (Thermo Scientific, Wilmington, DE, USA). RNA integrity was confirmed by 1% agarose gel electrophoresis.
2.4. First-Strand cDNA Synthesis for mRNA
One μg total RNA was treated with RQ1 RNase-Free DNaseI (Promega, Madison, WI, USA) to remove the genomic DNA. A PrimerScript RT Reagent Kit (Takara, Dalian, China) was used to synthesize the first strand cDNA. Then, the synthesized cDNA was stored at −20 °C until use.
2.5. dsRNA Synthesis and Delivery
The primers used for the dsRNA synthesis were designed using Primer 3.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and are listed in Table S1. According to our previous study, Hunchback (HB) was used as the indicator gene to calculate gene silencing efficiency. This gene is functionally involved in abdominal identity suppression and germband growth [35]. DsRNA of HB and Green fluorescent protein (dsGFP) were synthesized in vitro using a Transcript Aid T7 High Yield Transcription Kit (Thermo Scientific, Wilmington, DE, USA) basing on manufacturer protocol. The final concentration of dsHB (dsGFP) was diluted into 60 ng/μL, 600 ng/μL, and 6000 ng/μL by nuclease-free water. 10 nL has 0.6 ng, 6 ng, and 60 ng for 60 ng/μL, 600 ng/μL, and 6000 ng/μL was injected into dorsal part of adults’ abdomens using a M3301 micromanipulator (World Precision Instruments, Sarasota, FL, United States). After injection, aphids were put on broad bean seedlings and 36 h, the aphids were collected for RNA extraction. We choose 36 h to collect samples based on our previous study [36]. The RNAi efficiency of the targeted gene was optimal at this time point. Four biological replicates were performed for each treatment.
2.6. microRNA Agomir/Antagomir Synthesis and Delivery
An aphid specific expressed miRNA, miR-3024, was used as a miRNA indicator. The function of miR-3024 is still unknown while it was indicated to be involved in aphid-Buchera interactions [37]. The miR-3024 agomir/antagomir and agomir/antagomir negative controls were synthesized by Ribobio (Guangzhou, China). A 10 nL amount of each agomir/antagomir was injected into adults. The final dosages of miR-3024 agomir/antagomir were 1 pmol (agomir 15.4 ng; antgomir 8 ng), 1.5 pmol (agomir 23.1 ng; antgomir 12 ng) and 2 pmol (agomir 30.8 ng; antgomir 16 ng). After injection, aphids were put on broad bean seedlings and 36 h later, the aphids were collected for RNA extraction. Four biological replicates were performed in each treatment.
2.7. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) for mRNA
The primers for quantitative reverse transcription PCR (RT-qPCR) were designed using Primer 3.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and listed in Table S1. All primers were evaluated using a serial dilution of one cDNA sample to assure amplification efficiency and specificity. RT-qPCR was performed on a BIO-RAD CFX Connect Real-Time System (Bio-Rad, Hercules, CA) using the NovoStar SYBR RT-qPCR SuperMix (Novoprotein Scientific, Shanghai, China). The reactions were performed in a 10 μL volume of a mixture containing 5 μL SYBR Green mix, 0.5 μL cDNA template, 0.5 μL of each primer and 3.5 μL nuclease-free water. The amplication program was a two-step method and its program was as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 1.5 s, 60 °C for 30 s. A melt curve analysis from 60 °C to 95 °C was conducted to ensure specificity. The elongation factor-1 alpha (EF1α) and ribosomal protein S2-like (RPS2) were used as reference genes [38,39]. Four biological replicates were performed for each sample and the relative expression levels were analyzed using qBase software based on the 2ΔΔ-CT method [40].
2.8. First Strand cDNA Synthesis of miRNA
Two μg total RNA was purified using phenol chloroform (phenol water: chloroform (v/v) = 1:1). Before the first-strand cDNA of miRNA was synthesized, the miRNA was added to the poly(A) using a miRNA cDNA synthesis kit (Tiangen, Beijing, China) based on the manufacturer’s instructions. The poly(A) reaction contained 2 μg purified RNA, 2.5 μL 1×poly(A) polymerase buffer, 2.5 μL ATP, 0.5 μL E. coli poly(A) polymerase and RNase-free water up to 25 μL. The reaction condition was 37 °C for 15 min. Then, the first cDNA of miRNA was synthesized in a 20 μL volume of a mixture containing 1 μL ultrapure dNTP mix, 3 μL RT primers, 4 μL 5×superRT buffer, 0.5 μL superRT, 7.5 μL RNase-free water and 4 μL poly(A) reaction fluid. The reaction program was 42 °C for 50 min and 85 °C for 5 min. The cDNA was stored at −20 °C until use.
2.9. RT-qPCR for miRNA
RT-qPCR was performed on a BIO-RAD CFX Connect Real-Time System using a miRNA RT-qPCR assay kit (Tiangen, Beijing, China). The mature miR-3024 gene sequence was used as the forward primer and the U6 snRNA was used as a reference [41] (Table S1). The reactions were performed in a 20 μL volume of a mixture containing 1 μL 2×miRNA RT-qPCR mix, 1 μL cDNA template, 0.4 μL of each primer, and 8.2 μL of nuclease-free water. The program was as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 1.5 s, 60 °C for 1 min, and 60 °C for 1 min. A melt curve analysis from 60 °C to 95 °C was conducted to ensure specificity for all reactions. Four biological replicates were performed for each sample. The relative expression levels were analyzed using qBase software based on the 2ΔΔ-CT method.
2.10. Statistical Analysis
All statistical analyses were performed with SPSS 16.0 for Windows (SPSS Inc., Chicago, IL, USA). Significant differences in transcript levels of different issues, development stages, dsRNA and miRNA agomir/antagomir were tested by one-way analysis of variance (ANOVA), followed by Tukey’s honestly significant difference multiple comparison test (p < 0.01). The Significant differences of dsRNA and miRNA agomir/antagomir was analyzed in each dosage, independently.
3. Results
3.1. Spatiotemporal Expression Profiles of Core Genes of Three RNAi Pathways in Different Developmental Stages and Different Tissues of the Pea Aphid
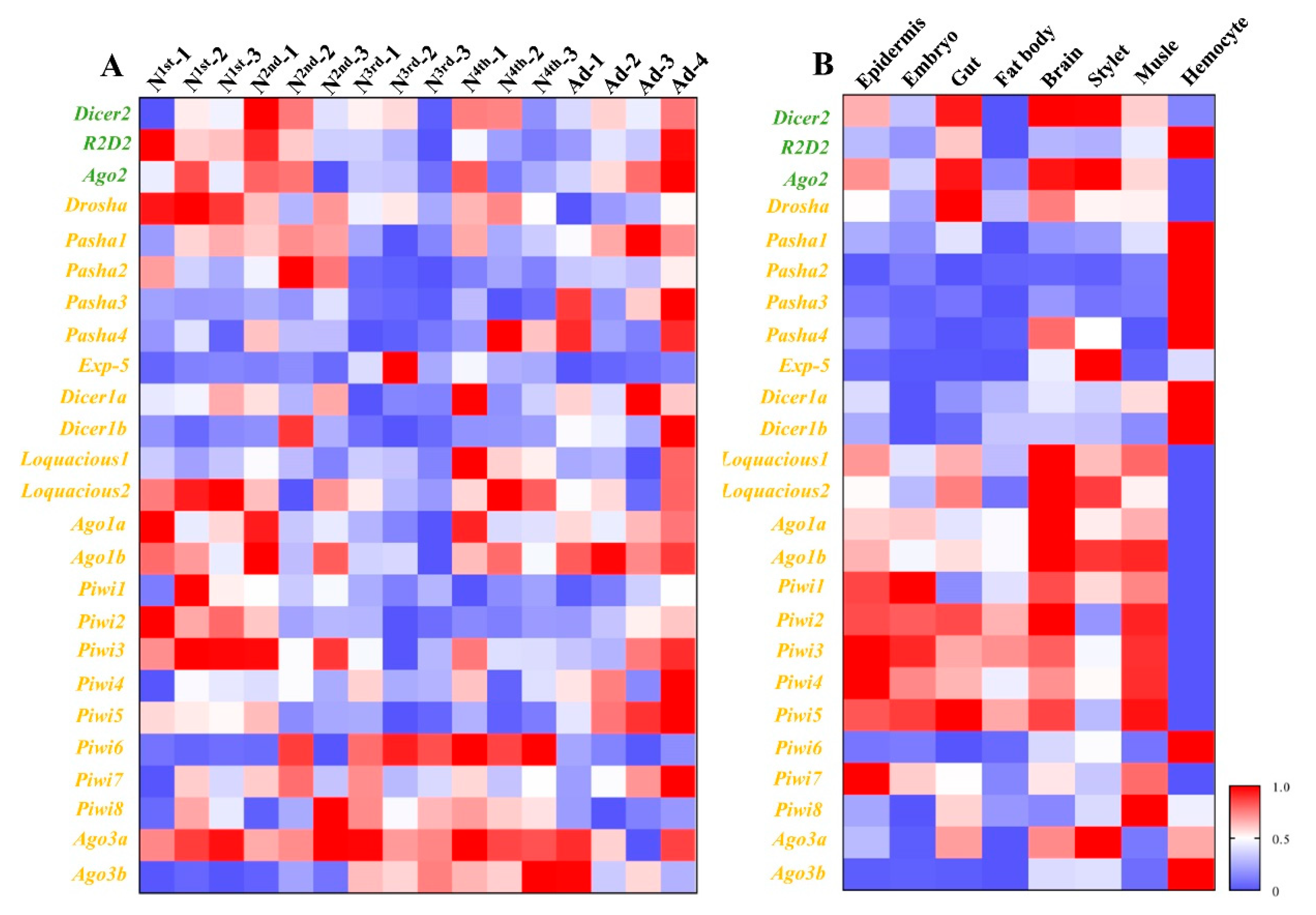
Different developmental stages and different tissues of pea aphid adults were collected (Figure 1), and the transcriptions of 25 genes from three RNAi pathways, miRNA, siRNA, and piRNA were measured by RT-qPCR (Figure 2). The results showed most genes were all expressed in all developmental stages and different tissues. Exp-5 only exhibited high expression in the middle of third instar nymphs, and Piwi6 showed high expression in third and fourth instar nymphs. The transcript levels of Ago1a, Ago1b, Dicer1a, Dicer1b, Pasha1, Pasha2, Pasha3, Pasha4, Ago2, R2D2, Piwi1, Piwi2, Piwi5, and Piwi7 were lower in the old nymphs, but higher in the young nymphs and adults. However, Exp-5, Ago3b, Piwi6, and Piwi8 possessed contrasting expression patterns. The expression levels of the remaining genes were not different among developmental stages (Figure 2). The relative expressions of most genes were below 1.2 while Pasha3, Pasha4, Exp-5, Piwi4, Piwi6, and Ago3b were greater than 1.2 in fourth instar nymphs and adults. Expression of Exp-5 and Piwi6 exceeded 3.
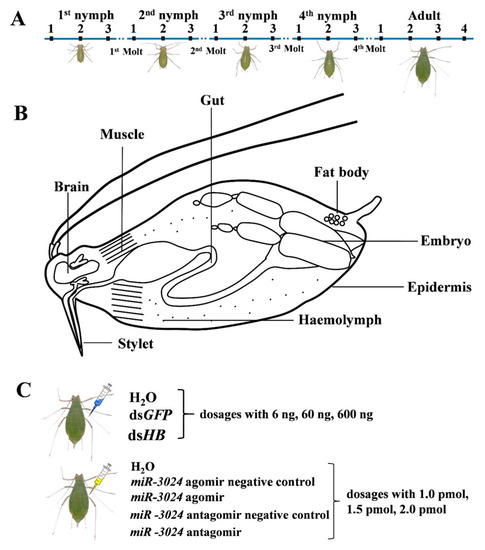
Figure 1.
Experimental design. (A) Sample collection from different developmental stages. N1st-1: new-born nymphs at 4 h; N1st-2: middle of first instar nymphs; N1st-3: first instar nymphs 4 h before the first molt; N2nd-1: second instar nymphs 4 h after the first molt; N2nd-2: middle of second instar nymphs; N2nd-3: second instar nymphs 4 h before the second molt; N3rd-1: third instar nymphs 4 h after the second molt; N3rd-2: middle of third instar nymphs; N3rd-3: third instar nymphs 4 h before the third molt; N4th-1: fourth instar nymphs 4 h after the third molt; N4th-2: middle of fourth instar nymphs; N4th-3: fourth instar nymphs 4 h before the fourth molt; Ad-1: adult 4 h after the fourth molt; Ad-2: aphids at 9 d; Ad-3: aphids at 12 d; Ad-4: aphids at 15 d. (B) Sample collection of different tissues. Eight tissues (gut, epidermis, muscle, brain, fat body, embryo, stylet, and hemolymph) of pea aphids were dissected and collected. (C) Sample collections from different dosages of dsRNA treatments and miRNA agomir/antagomir treatments. For microinjection, each pea aphid was injected with 10 nL dsHB or miR-3024 agomir/antagomir. dsGFP and miR-3024 agomir negative control/antagomir negative control as control. 1 pmol: 15.4 ng (agomir), 8 ng (antagomir); 1.5 pmol: 23.1 ng (agomir), 12 ng (antagomir); 2 pmol: 30.8 ng (agomir), 16 ng (antagomir).
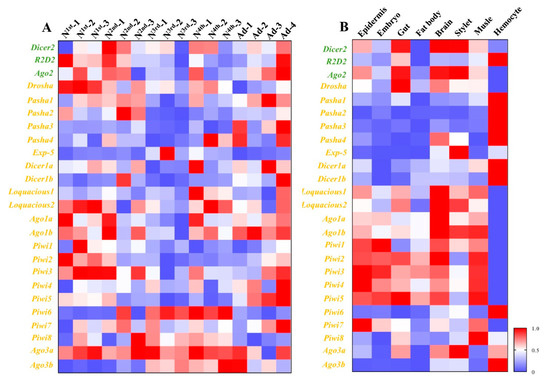
Figure 2.
Spatiotemporal expression profiles of core genes of three RNA interference (RNAi) pathways in different developmental stages and different tissues. (A) Expression profiles of core genes of three RNAi pathways among different developmental stages. (B) Expression profiles of core genes of three RNAi pathways among different tissues. Mean (±SE) expression level was based on four biological replicates. The relative expression was normalized based on two reference genes, A. pisum elongation factor-1 alpha (EF1a) and ribosomal protein S2-like (RPS2). All values are homogenized using the minimum maximum method. The green letters, orange letters, and purple letters respectively represent the genes in siRNA pathway, miRNA pathway, and piRNA pathway.
Epidermis, embryo, gut, fat body, brain, stylet, muscle, and hemolymph were dissected from adults for expression pattern analyses (Figure 2). The greatest expression change of Pasha4 was observed in the hemolymph. Exp-5, Ago2, Ago3b, and Piwi6 were highly expressed in the brain, stylet and hemolymph. Ago1a, Ago1b, Dicer1b, Drosha, Piwi3, Piwi4, Piwi5, and Piwi7 had high expression in seven tissues, except for the hemolymph. In contrast, Dicer1b, Pasha1, Pasha2, Pasha3, and R2D2 had high expression levels in hemolymph but not in the other tissues. The expression levels of Dicer1a and Dicer2 were similar among the sampled tissues (Figure 2).
3.2. Expression Profiles of Core Genes of Three RNAi Pathways Upon dsRNA Treatment
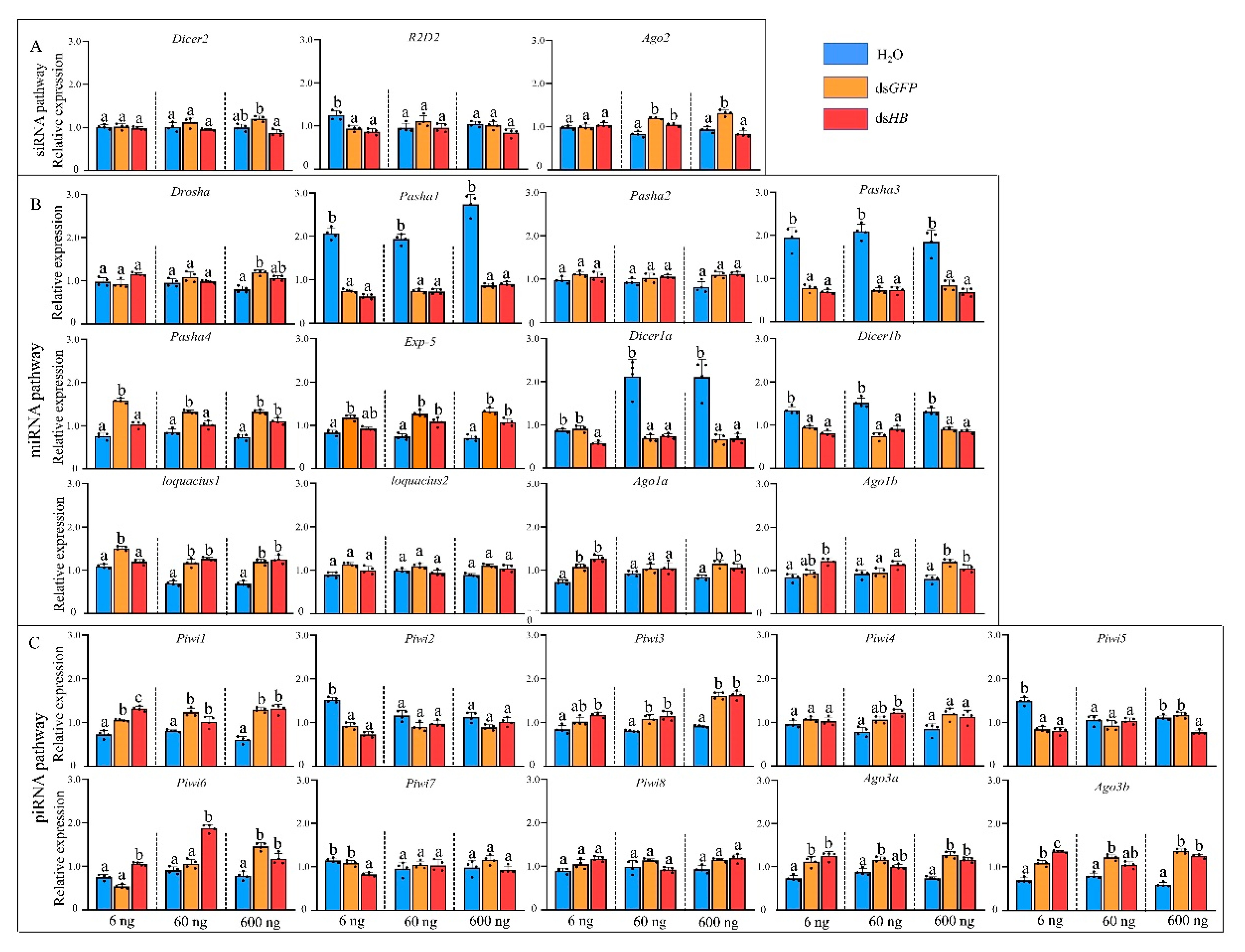
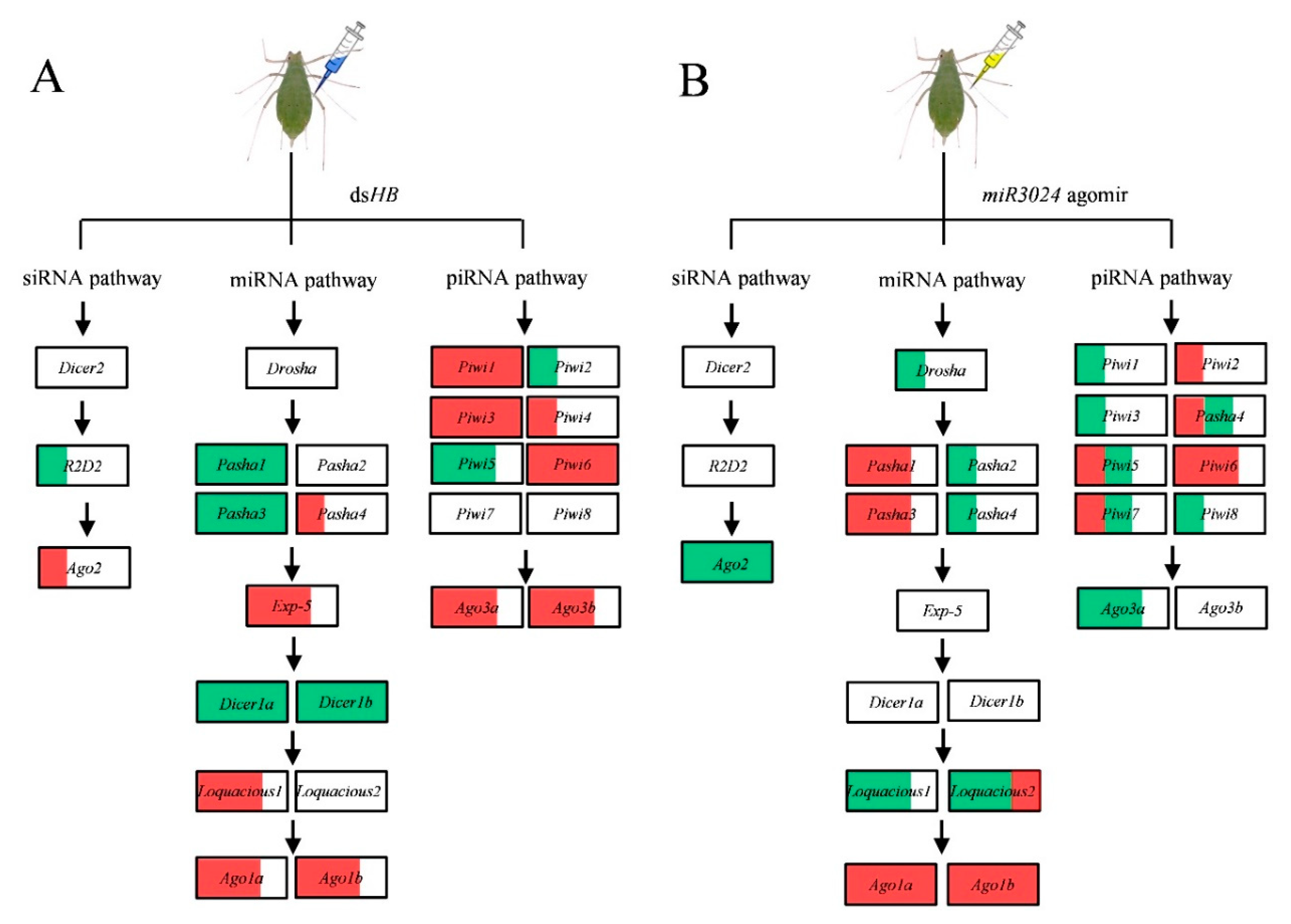
To study the responses of three RNAi pathways (siRNA, miRNA, and piRNA) after dsRNA treatment (Figure 1), the expressions of 25 genes of the core machinery of RNA pathways were examined after injection of different dsRNA doses. After 6 ng dsHB treatment, a gene, R2D2, in the siRNA pathway, was downregulated. Two miRNA genes (Ago1a and Ago1b) and five piRNA pathway genes (Piwi1, Piwi3, Piwi6, Ago3a, and Ago3b) were significantly upregulated. Surprisingly, four miRNA pathway genes (Pasha1, Pasha3, Dicer1a, and Dicer1b) and two piRNA pathway genes (Piwi2, Piwi5, and Piwi7) were significantly downregulated (Figure 3). After 60 ng dsHB treatment, Ago2 was upregulated and the expression levels of miRNA and piRNA pathway genes were altered. Among these, Exp-5, Loquacious1, Piwi1, Piwi3, Piwi4, and Piwi6 were significantly upregulated, while Pasha1, Pasha3, Dicer1a, and Dicer1b were significantly downregulated (Figure 3). Upon 600 ng dsHB treatment, the expression of HB, was significantly reduced (Figure S1). Within the same treatment, expressions of siRNA pathway genes were not significantly different. However, five miRNA pathway genes (Pasha4, Exp-5, Loquacious1, Ago1a, Ago1b) and five piRNA pathway genes (Piwi1, Piwi3, Piwi6, Ago3a, Ago3b) were significantly upregulated whereas four miRNA pathway genes (Pasha1, Pasha3, Dicer1a, Dicer1b) and one piRNA pathway gene (Piwi5) were significantly downregulated (Figure 3). Compared to the dsHB, three miRNA pathway genes (Pasha4, Dicer1a, and Loquacious1) and one piRNA pathway gene (Piwi7) were significantly downregulated while one piRNA pathway gene (Piwi6) was upregulated in 6ng. One miRNA pathway gene (Pasha4) was downregulated while one piRNA pathway gene (Piwi6) was upregulated in 60 ng. As for 600 ng, two siRNA pathway genes (Dicer2, Ago2) and one piRNA gene (Piwi5) were downregulated.
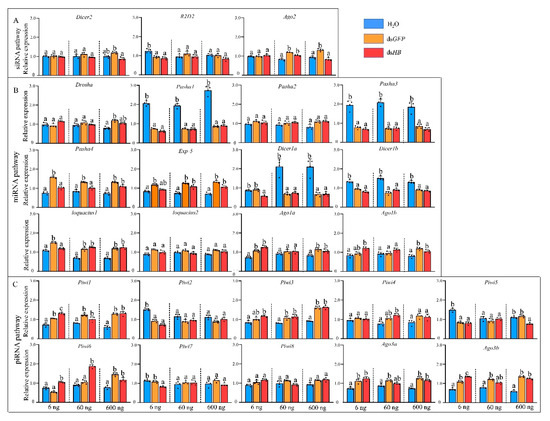
Figure 3.
Expression profiles of core genes of three RNAi pathways upon dsRNA administration by microinjection. (A) siRNA pathway. (B) miRNA pathway. (C) piRNA pathway. Different colors represent different level of changes. Mean (±SE) expression level was based on four biological replicates. The relative expression was normalized based on two reference genes, A. pisum elongation factor-1 alpha (EF1α) and ribosomal protein S2-like (RPS2). Lowercase letters above each bar indicate significant differences (one-way ANOVA followed by Tukey’s honestly significant difference multiple comparison test; p < 0.01). The Significant difference was analyzed in each dosage, independently.
3.3. Expression Profiles of Core Genes of Three RNAi Pathways Upon miRNA Treatment
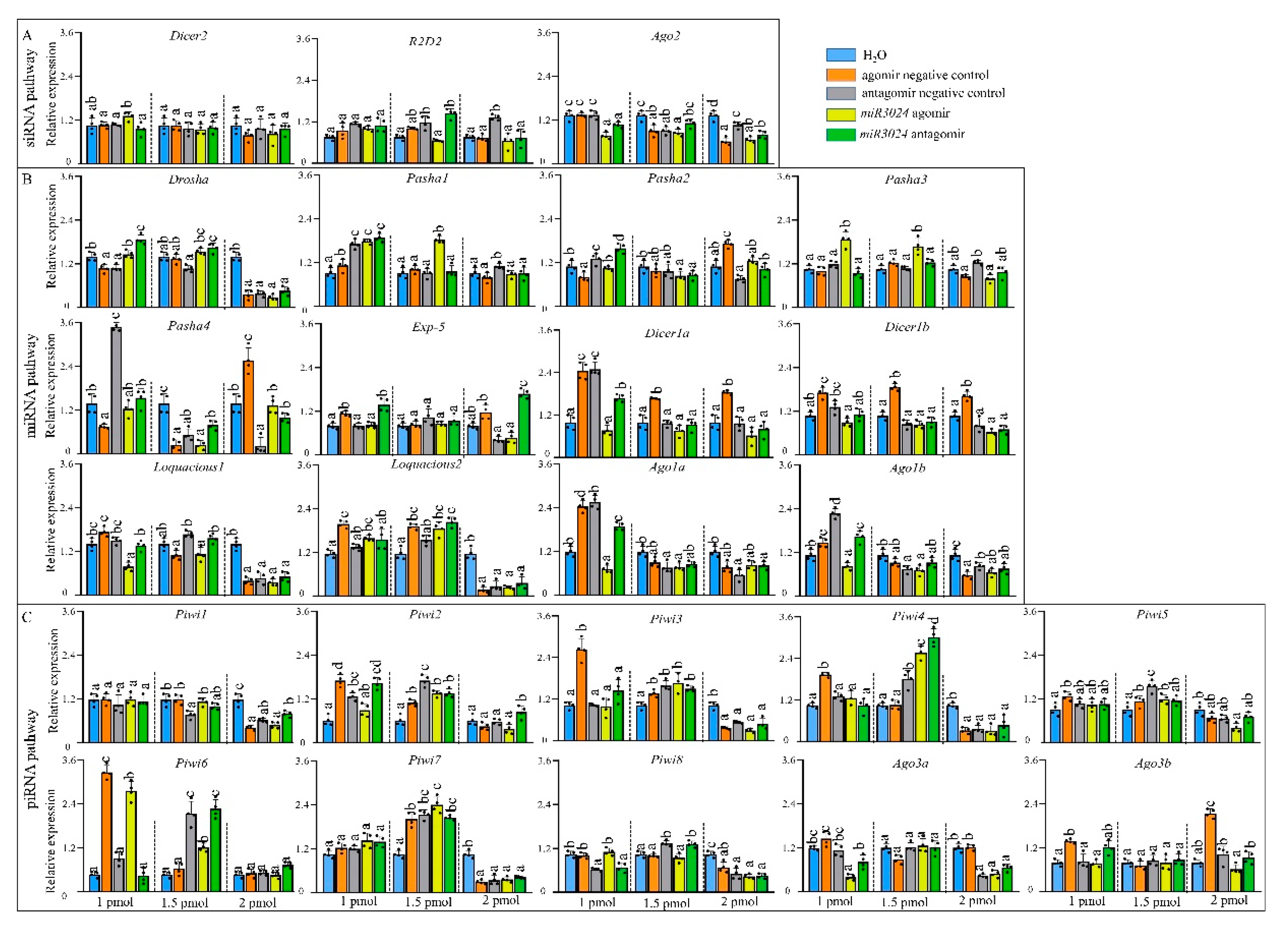
Expression levels of the 25 genes were detected after the treatment with different dosages of miR-3024 agomir/antagomir (Figure 4). The altered expression level of miR-3024 was also confirmed to verify the success of experimental treatments (Figure S2). After 1 pmol agomir treatment, the expression of Ago2 was downregulated while three miRNA pathway genes (Pasha1, Pasha3, Loquacious2) were significantly upregulated and three miRNA pathway genes (Loquacious1, Ago1a, Ago1b) were significantly downregulated. One piRNA pathway genes (Ago3a) were significantly downregulated. Upon 1.5 pmol agomir treatment, the expressions of siRNA pathway genes Ago2 decreased. Three miRNA pathway genes (Pasha1, Pasha3, Loquaciou2) were upregulated and four genes (Pasha2, Pasha4, Ago1a, and Ago1b) were downregulated while five piRNA pathway genes (Piwi2, Piwi4, Piwi5, Piwi6, and Piwi7) were upregulated. In the 2 pmol agomir treatment, the expression of Ago2 in the siRNA pathway was downregulated (Figure 4). Four miRNA pathway genes (Drosha, loquacious1, loquacious2, and Ago1b) were downregulated. In the piRNA pathway, Piwi1, Piwi3, Piwi4, Piwi5, Piwi7, Piwi8, and Ago3a were downregulated. In terms of dosage, the expressions of 9 genes (Ago2, Pasha1, Pasha3, Loquacious1, Loquacious2, Ago1a, Ago1b, Piwi6, and Ago3a) were changed in 1 pmol while 13 genes (Ago2, Pasha1, Pasha2, Pasha3, Pasha4, Loquacious2, Ago1a, Ago1b, Piwi2, Piwi4, Piwi5, Piwi6, and Piwi7) in 1.5 pmol and 12 genes (Ago2, Pasha1, Loquacious, Loquacious2, Ago1a, Ago1b, Piwi1, Piwi3, Piwi4, Piwi5, Piwi7, Piwi8, and Ago3a) in 2 pmol.
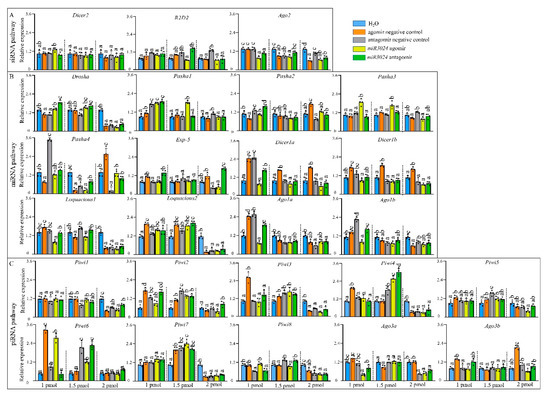
Figure 4.
Expression profiles of core genes of three RNAi pathways after miRNA agomir/antagomir administration by microinjection. (A) siRNA pathway. (B) miRNA pathway. (C) piRNA pathway. Different colors represent different change levels. Mean (±SE) expression level is based on four biological replicates. The relative expression was normalized based on two reference genes, A. pisum elongation factor-1 alpha (EF1a) and ribosomal protein S2-like RPS2. Lowercase letters above each bar indicate significant differences (one-way ANOVA followed by Tukey’s honestly significant difference multiple comparison test; p < 0.01). agoNC: agomir negative control; antNC: antagomir negative control; ago3024: mi-R3024 agomir; ant3024: miR-3024 antagomir. The Significant difference was analyzed in each dosage, independently. 1 pmol: 15.4 ng (agomir), 8 ng (antagomir); 1.5 pmol: 23.1 ng (agomir), 12 ng (antagomir); 2 pmol: 30.8 ng (agomir), 16 ng (antagomir).
4. Discussion
With the expansion of core RNAi genes discovered in pea aphids, it remains unclear how these contribute to the function of RNAi pathways. To explore the function of these genes the transcript levels of a number of 25 core genes in three RNAi pathways (siRNA, miRNA, and piRNA) were screened in three experimental conditions: spatiotemporal samples, artificial synthesized dsRNA treated samples, and artificial synthesized miRNA treated samples.
In the spatiotemporal samples, we determined the transcription level of the 25 genes in developmental stages and tissues. The expression dynamics of these genes were variable. Genes with the presented expression based on the RT-qPCR detection limit, and the variation expression levels in certain developmental stages or tissues provide clues to their involvement in specific physiological processes. Pasha3, Pasha4, Exp-5, Piwi4, Piwi6, and Ago3b had high expression in fourth instar nymphs and adults, which are critical stages in maturation and reproduction, respectively. These genes are closely associated with miRNA and piRNA, and these two pathways are important in the regulation of development and reproduction [42,43]. piRNA pathway is known to function primarily in suppressing transposon expression in the gonads of animals and Ago3 is expressed specifically in germ cells of D. melanogaster [44]. However, the pea aphid used in our study is parthenogenesis. The expression of Ago3 is still unknown in tissues of aphids of sexual reproduction phase.
RNAi (normally inferred to as the siRNA pathway) is a promising tool for aphid control [32,33,34] but RNAi use in effective pest control remains a challenge. This is due to gene silencing efficacies (both RNA and protein level), pest control efficacy (mortality), field delivery systems and potential resistance development [44]. Therefore, we evaluated the transcriptional level of 25 genes in artificial synthesized dsRNA treated samples. As expected, genes of the siRNA pathway (e.g., Ago2) were upregulated after dsRNA treatment showing that the exogenous application of dsRNA induced the activity of the RNAi pathway [45]. However, two other genes, Dicer2 and R2D2, were not significantly altered after the same dsRNA treatment. Of the three dsRNA doses (6, 60, 600 ng per aphid), only the 600 ng treatment induced significant gene silencing of the target gene (HB). The induced level of Ago2 only had a slight difference. This trend occurred in the dsGFP treated sample but not in the dsHB treated sample. We previously observed that the induction of these three genes occurred 24 h after dsRNA injection (600 ng per aphid) [36], implying that the variation of gene expression level induction is associated with the time of sample collection and the physiological stage of the aphids. We believe the expressions of 25 core genes upon the dsRNA treatment are dose-dependent as well as in sufficient dose presenting a time points-based dynamics. For instance, the response of core machinery was detected in both 24 h [36] and 36 h in the current study. In addition, the RNAi efficiency of the targeted gene was the highest at the 36 h [36]. Under the same treatment, there were significant expression changes in miRNA and piRNA pathway associated genes. For miRNA, reduction of Pasha1, Pasha3, Dicer1a, and Dicer1b were observed in three dsRNA dosages (6, 60, 600 ng), while induction of loquacious 1, Ago1a, and Ago1b, were observed upon dsRNA treatments, reflecting a plasticity of miRNA pathways upon dsRNA in pea aphids. For piRNA, the induction of Piwi1, Piwi3, Piwi6, Ago3a, and Ago3b occurred after dsRNA treatments (Figure 5). The genes in siRNA pathway would respond to the dsRNA [28]. But dsHB target its mRNA while dsGFP act as a non-target negative control, this may differ the response of the siRNA pathway core genes between treatments of dsGFP and dsHB. For example, Ago2 upregulated upon 600 ng of dsGFP while it did not significantly alter upon dsHB. A crosstalk existed among miRNA pathway, siRNA pathway and piRNA pathway [28,29]. Thus, the difference of dsGFP and dsHB treatments could also affect the response of genes in miRNA pathway (e.g., Pasha4) and piRNA pathway (e.g., Piwi6). siRNAs and miRNAs are Dicer-dependent groups and piRNAs are the Dicer-independent Group. Competition with the Argonaute protein could be associated with above-observed phenomenon [46]. In addition, genes such as Loquacious could function as interactive genes to connect the miRNA pathway and the siRNA pathway [35]. Thus, the pea aphid genes altered after dsRNA treatments, which are normally recognized as genes in the miRNA/piRNA pathway, should be studied to determine if they contribute to the siRNA pathway activity. This could enhance our understanding of the siRNA pathway mechanism in aphids and its associated factors. These data will be needed to design an efficient RNAi gene silencing approach for aphid pest control. The study in Drosophila showed that Ago2 and Dicer2 downregulated in 2 ug of dsGFP treatment [28] But in pea aphids Ago2 were upregulated in dsRNA treatments of 60 ng and 600 ng rather than 6 ng. It seems that the change of genes in RNAi pathways may be relative to dose of dsRNA. Dose-dependent upregulation of Dicer2 and Ago2 expression was also found in response to injection with dsRNA in M. sexta [29]. Exogenous dsRNA induced the gene expressions of the piRNA pathway were also reported [30,31] which was also observed in A. pisum in this study. Taken together, it can indicate crosstalk among miRNA pathway, siRNA pathway, and piRNA pathway.
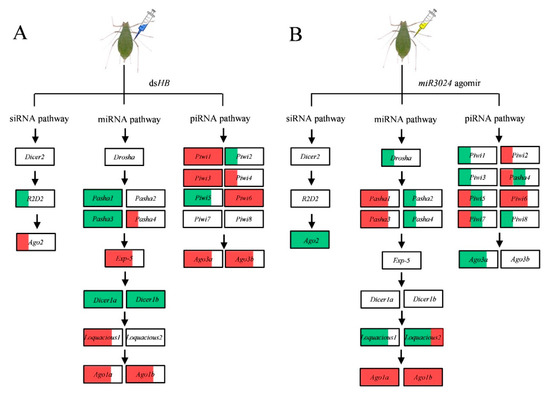
Figure 5.
Schematic of expression profiles of core genes of three RNAi pathways upon dsRNA and miRNA agomir administration by microinjection. (A) dsHB treatment. (B) miR-3024 agomir treatment. The white, red, and green of the box represents the expression of gene are no significance, upregulation, and downregulation, respectively. 1/3, 2/3, and full red/green area of the box represent the ratio of genes upregulation/downregulation in three dose treatments of dsHB (6 ng, 60 ng, 600 ng) or miR-3024 agomir/antagomir (agomir: 15.4 ng, 23.1 ng, 30.8 ng; antagomir: 8 ng, 12 ng, 16 ng).
We used the same experimental approach, with artificially synthesized miRNA agomir and antagomir (e.g., miR-3024), to detect the responses of these 25 genes. The gene expression responses to dsRNA treatment were complex. It seems that the treatment of artificially synthesized miRNA induced dosage dependent gene expression. At the low dose (1 pmol), gene expressions such as Pasha1, Pasha3, Ago1a, Ago1b, and Piwi6 were induced. With the 1.5 pmol dose, expression of genes such as Piwi4 and Piwi7 were induced. In the 2 pmol dose, genes such as Drosha, Loquacious1, Loquacious2, Piwi4, Piwi7, and Ago3a were down regulated. These data indicate that miRNA and piRNA-associated genes can interact with these expanded genes in a way similar to the siRNA pathway. This suggests a complex network of RNAi core genes in the three sub-pathways. siRNAs injected into the embryos of zebrafish competed for cofactors that were required in the miRNA pathway [46]. Similarly, in mammalian cells, transfection of siRNAs can alter the levels of many miRNAs by competition for Ago2 [47]. For piRNA, the endo-siRNA pathway may suppress the expression of piRNAs in the ovary somatic sheet cell line of Drosophila [15]. In summary, the expression profiles of 25 genes of the RNAi core machinery (including genes from miRNA pathway, siRNA pathway and piRNA pathway) were screened in three experimental conditions, spatiotemporal samples, artificial synthesized dsRNA treated samples, and artificial synthesized miRNA treated samples. The expanded genes such as Pasha, Dicer1, loquacious, Ago1, Piwi, and Ago3, showed significant response to dsRNA and/or miRNA, implying a complex network of RNAi core machinery in aphids. Further studies should investigate the contribution of these expanded genes (loquacious, Ago1, Piwi, and Ago3) to the activity of the siRNA pathway. This information will help determine the potential of RNAi for aphid pest control. In addition, the siRNA pathway is reported to be involved in defense of plant viruses in aphids [48] and these genes could be responsible for immunity that may be able to against the viruses before they are transmitted to plants. This possibility will also require further studies.
5. Conclusions
The expressions of 25 genes of RNAi in three RNAi pathways (siRNA, miRNA, and piRNA) were detected in spatiotemporal samples, artificially synthesized dsRNA and miRNA-treated samples. The results from different development stages and different tissues indicated that the miRNA pathway and piRNA pathways are important in the regulation of insect development and reproduction. A complex network of RNAi cores genes in the three sub-pathways was found based on data from the artificially synthesized dsRNA and miRNA treated samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/2/70/s1, Table S1: Primers used for dsRNA synthesis and RT-qPCR, Table S2: Accession number of genes used in this study, Figure S1: The relative mRNA expression profiles of Hunchback upon dsHB administration, Figure S2: Relative expression profiles of miR-3024 upon miR-3024 agomir/antagomir administration.
Author Contributions
L.Y., J.N., and J.-J.W. conceived and designed the experiments. L.Y., Y.-Y.P., and Y.T. contributed materials and performed the experiments; L.Y. and J.N. analyzed the data; L.Y., J.N., and J.-J.W. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Fundamental Research Funds for the Central Universities (No. XDJK2019B043).
Conflicts of Interest
The authors declare no conflict of interest. The funding agency had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Islam, M.N.; Choudhury, N.R.; Haq, Q.R. The Fascinating world of RNA interference. Int. J. Biol. Sci. 2009, 5, 97–117. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.J.; Shen, G.M.; Christiaens, O.; Smagghe, G.; He, L.; Wang, J.J. Beyond insects: Current status and achievements of RNA interference in mite pests and future perspectives. Pest Manag. Sci. 2018, 74, 2680–2687. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Hannon, G.J. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef]
- Hoehener, C.; Hug, I.; Nowacki, M. Dicer-like enzymes with sequence cleavage preferences. Cell 2018, 173, 234–247. [Google Scholar] [CrossRef]
- Emily, B.; Caudy, A.A.; Hammond, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.H.; Kim, H.E.; Smith, D.P.; Wang, X.D. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013, 43, 24–38. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Han, J. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Palli, S.R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 2020, 65, 14.1–14.19. [Google Scholar] [CrossRef] [PubMed]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Burglin, T.R.; Gatto, J.; Ruvkun, G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991, 5, 1813–1824. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Robine, N.; Martin, R.; Chung, W.J.; Niki, Y.; Berezikov, E.; Lai, E.C. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009, 19, 1776–1785. [Google Scholar] [CrossRef]
- Biryukova, I.; Ye, T. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genom. 2015, 16, 278. [Google Scholar] [CrossRef]
- Jaubert-Possamai, S.; Rispe, C.; Tanguy, S.; Gordon, K.; Walsh, T.; Edwards, O.; Tagu, D. Expansion of the miRNA pathway in the hemipteran insect Acyrthosiphon pisum. Mol. Biol. Evol. 2010, 27, 979–987. [Google Scholar] [CrossRef]
- Lu, H.L.; Tanguy, S.; Rispe, C.; Gauthier, J.P.; Walsh, T.; Gordon, K.; Edwards, O.; Tagu, D.; Chang, C.C.; Jaubert-Possamai, S. Expansion of genes encoding piRNA-associated argonaute proteins in the pea aphid: Diversification of expression profiles in different plastic morphs. PLoS ONE 2011, 6, e28051. [Google Scholar] [CrossRef]
- Okamura, K.; Robine, N.; Liu, Y.; Liu, Q.; Lai, E.C. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol. Cell Biol. 2011, 31, 884–896. [Google Scholar] [CrossRef]
- Campbell, C.L.; Black, W.C.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom. 2008, 9, 425. [Google Scholar] [CrossRef]
- Lewis, S.H.; Salmela, H.; Obbard, D.J. Duplication and diversification of dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol. Evol. 2016, 8, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep. 2016, 6, 22587. [Google Scholar] [CrossRef] [PubMed]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef] [PubMed]
- Mongelli, V.; Saleh, M.C. Bugs are not to be silenced: Small RNA pathways and antiviral responses in insects. Annu. Rev. Virol. 2016, 3, 573–589. [Google Scholar] [CrossRef]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Ralph, S.M.; Mayer, C.; Liu, S.L.; Zhou, X.; et al. Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol. Evol. 2016, 8, 3784–3793. [Google Scholar] [CrossRef]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef]
- Bernhardt, S.A.; Simmons, M.P.; Olson, K.E.; Beaty, B.J.; Blair, C.D.; Black, W.C. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PLoS ONE 2012, 7, e44198. [Google Scholar] [CrossRef]
- Saleh, M.C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O’Farrell, P.H.; Andino, R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 2006, 8, 793–807. [Google Scholar] [CrossRef]
- Garbutt, J.S.; Reynolds, S.E. Induction of RNA interference genes by double-stranded RNA; implications for susceptibility to RNA interference. Insect Biochem. Mol. 2012, 42, 621–628. [Google Scholar] [CrossRef]
- Hoa, N.T.; Keene, K.M.; Olson, K.E.; Zheng, L. Characterization of RNA interference in an Anophelesgambiae cell line. Insect Biochem. Mol. 2003, 33, 949–957. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.J.; Yang, W.J.; Tian, Y.; Fan, J.Y.; Ye, C.; Shang, F.; Ding, B.Y.; Zhang, J.; An, X.; Yang, L.; et al. Topical dsRNA delivery induces gene silencing and mortality in the pea aphid. Pest Manag. Sci. 2019, 75, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.D.; Liu, Z.C.; Huang, S.L.; Chen, Z.Q.; Sun, Y.W.; Duan, P.F.; Ma, Y.Z.; Xia, L.Q. RNAi-mediated plant protection against aphids. Pest Manag. Sci. 2016, 72, 1090–1098. [Google Scholar] [CrossRef]
- Yan, S.; Qian, J.; Cai, C.; Ma, Z.; Li, J.; Yin, M.; Ren, B.; Shen, J. Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J. Pest Sci. 2019, 449–459. [Google Scholar] [CrossRef]
- Mao, J.; Liu, C.; Zeng, F. Hunchback is required for abdominal identity suppression and germband growth in the parthenogenetic embryogenesis of the pea aphid, Acyrthosiphon pisum. Arch. Insect Biochem. 2003, 84, 209–221. [Google Scholar] [CrossRef]
- Ye, C.; Jiang, Y.D.; An, X.; Yang, L.; Shang, F.; Niu, J.; Wang, J.J. Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 2019, 9, 3694. [Google Scholar] [CrossRef]
- Feng, H.L.; Wang, L.Y.; Wuchty, S.; Alex, C.C. microRNA regulation in an ancient obligate endosymbiosis. Mol. Ecol. 2018, 27, 1777–1793. [Google Scholar] [CrossRef]
- Chen, N.; Fan, Y.L.; Bai, Y.; Li, X.D.; Zhang, Z.F.; Liu, T.X. Cytochrome P450 gene, CYP4G51, modulates hydrocarbon production in the pea aphid, Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2016, 76, 84–94. [Google Scholar] [CrossRef]
- Li, Y.; Yang, C.; Pan, H.; Liu, Y.; Zhou, X. Selection of reference genes for expression analysis using quantitative real-Time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS ONE 2014, 9, e110454. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nouzova, M.; Etebari, K.; Noriega, F.G.; Asgari, S. A comparative analysis of corpora allata-corpora cardiaca microRNA repertoires revealed significant changes during mosquito metamorphosis. Insect Biochem. Mol. Biol. 2018, 96, 10–18. [Google Scholar] [CrossRef]
- Park, J.K.; Liu, X.; Strauss, T.J.; McKearin, D.M.; Liu, Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 2007, 17, 533–538. [Google Scholar] [CrossRef]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef] [PubMed]
- Katoch, R.; Sethi, A.; Thakur, N.; Murdock, L.L. RNAi for insect control: Current perspective and future challenges. Appl. Biochem. Biotech. 2013, 171, 847–873. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.J.; Smagghe, G.; Coninck, D.I.; Nieuwerburgh, F.; Deforce, D.; Meeus, I. In vivo study of Dicer-2-mediated immune response of the small interfering RNA pathway upon systemic infections of virulent and avirulent viruses in Bombus terrestris. Insect Biochem. Mol. Biol. 2016, 70, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.F.; Fjose, A.; Larsen, N.; Helvik, J.V.; Øyvind, D. Treatment with small interfering RNA affects the microRNA pathway and causes unspecific defects in zebrafish embryos. FEBS J. 2008, 275, 2177–2184. [Google Scholar] [CrossRef]
- Liang, X.H.; Hart, C.; Crooke, S. Transfection of siRNAs can alter miRNA levels and trigger non-specific protein degradation in mammalian cells. BBA Gene Regul. Mech. 2013, 1829, 455–468. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, T.F.; Guo, M.M.; Chang, T.; Yang, L.; Tan, Y.; Ye, C.; Niu, J.J.; Wang, J.J. Characterization of a new bunyavirus and its derived small RNAs in the brown citrus aphid, Aphis citricidus. Virus Genes 2019, 55, 557–561. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).