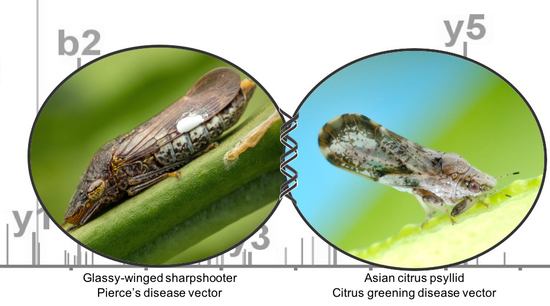
Lessons from One Fastidious Bacterium to Another: What Can We Learn about Liberibacter Species from Xylella fastidiosa
Abstract
1. Introduction and Historical Context
2. Pathogen
2.1. Background and genomic resources
2.2. Virulence
2.3. Biofilm formation
2.4. Biocontrol
2.5. Global Outlook
3. Vector
3.1. Path through vector
3.2. OMICs Resources
3.3. Transmission determinants
3.4. Feeding
3.5. Biocontrol
4. Host
4.1. OMICs as Resources for Breeding
4.2. Transgenic Strategies
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Da Graca, J.; French, J.; Haslem, P.; Skaria, M.; Sétamou, M.; Salas, B. Survey for the Asian citrus psyllid, Diaphorina citri, and citrus huanglongbing (greening disease) in Texas. Subtrop. Plant Sci. 2008, 60, 21–26. [Google Scholar]
- Capoor, S.P. Decline of citrus trees in India. Bull. Natl. Inst. Sci. India 1963, 24, 48–64. [Google Scholar]
- Jagoueix, S.; Bove, J.-M.; Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 1994, 44, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Jagoueix, S.; Bove, J.M.; Garnier, M. Comparison of the 16S/23S ribosomal intergenic regions of “Candidatus Liberobacter asiaticum” and “Candidatus Liberobacter africanum,” the two species associated with citrus huanglongbing (greening) disease. Int. J. Syst. Evol. Microbiol. 1997, 47, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Jagoueix, S.; Bove, J.M.; Garnier, M. PCR detection of the two ‘Candidatus’ Liberobacter species associated with greening disease of citrus. Mol. Cell. Probes 1996, 10, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Coletta-Filho, H.D.; Takita, M.A.; Targon, M.L.P.N.; Machado, M.A. Analysis of 16S rDNA Sequences from Citrus Huanglongbing Bacteria Reveal a Different “Ca. Liberibacter” Strain Associated with Citrus Disease in São Paulo. Plant Dis. 2005, 89, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Da Graça, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef]
- Do Carmo Teixeira, D.; Luc Danet, J.; Eveillard, S.; Cristina Martins, E.; de Jesus Junior, W.C.; Takao Yamamoto, P.; Aparecido Lopes, S.; Beozzo Bassanezi, R.; Juliano Ayres, A.; Saillard, C.; et al. Citrus huanglongbing in Sao Paulo State, Brazil: PCR detection of the ‘Candidatus’ Liberibacter species associated with the disease. Mol. Cell. Probes 2005, 19, 173–179. [Google Scholar] [CrossRef]
- Teixeira, D.C.; Saillard, C.; Couture, C.; Martins, E.C.; Wulff, N.A.; Eveillard-Jagoueix, S.; Yamamoto, P.T.; Ayres, A.J.; Bové, J.M. Distribution and quantification of Candidatus Liberibacter americanus, agent of huanglongbing disease of citrus in São Paulo State, Brasil, in leaves of an affected sweet orange tree as determined by PCR. Mol. Cell. Probes 2008, 22, 139–150. [Google Scholar] [CrossRef]
- Teixeira, D.C.; Saillard, C.; Eveillard, S.; Danet, J.L.; da Costa, P.I.; Ayres, A.J.; Bove, J. ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing (greening disease) in Sao Paulo State, Brazil. Int. J. Syst. Evol. Microbiol. 2005, 55, 1857–1862. [Google Scholar] [CrossRef]
- Graca, J.V. Citrus Greening Disease. Annu. Rev. Phytopathol. 1991, 29, 109–136. [Google Scholar] [CrossRef]
- EPPO; CABI. Liberobacter africanum. In Distribution Maps of Plant Diseases, 1st ed.; CAB International: Oxfordshire, UK, 1998. [Google Scholar]
- Gottwald, T.R. Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.L.; Halbert, S.; Lee, R.; Hoy, M.; Clark, R.; Kesinger, M. The Asian citrus psyllid and citrus greening disease. Citrus Ind. 1998, 79, 1028–1029. [Google Scholar]
- Stelinski, L.L. Ecological aspects of the vector-borne bacterial disease, Citrus Greening (Huanglongbing): Dispersal and host use by Asian Citrus Psyllid, Diaphorina Citri Kuwayama. Insects 2019, 10, 208. [Google Scholar] [CrossRef]
- Gottwald, T.R.; McCollum, T.G. Huanglongbing solutions and the need for anti-conventional thought. J. Citrus Pathol. 2017, 4. [Google Scholar]
- Pierce, N.B. The California Vine Disease: A Preliminary Report of Investigations; US Government Printing Office: Washington, DC, USA, 1892.
- Chang, C.J.; Garnier, M.; Zreik, L.; Rossetti, V.; Bové, J.M. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr. Microbiol. 1993, 27, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Blua, M.; Phillips, P.P.; Redak, R.A. A new sharpshooter threatens both crops and ornamentals. Calif. Agric. 1999, 53, 22–25. [Google Scholar] [CrossRef][Green Version]
- Davis, M.J.; Purcell, A.H.; Thomson, S.V. Pierce’s disease of grapevines: Isolation of the causal bacterium. Science 1978, 199, 75–77. [Google Scholar] [CrossRef]
- Purcell, A. Paradigms: Examples from the bacterium Xylella fastidiosa. Annu. Rev. Phytopathol. 2013, 51, 339–356. [Google Scholar] [CrossRef]
- Purcell, A.H.; Saunders, S.R.; Hendson, M.; Grebus, M.E.; Henry, M.J. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 1999, 89, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Stilwagen, S.; Ivanova, N.; D’Souza, M.; Bernal, A.; Lykidis, A.; Kapatral, V.; Anderson, I.; Larsen, N.; Los, T.; et al. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. USA 2002, 99, 12403–12408. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Silva-Stenico, M.E.; Gomes, J.E.; Lopes, J.R.S.; Tsai, S.M. Detection and diversity assessment of Xylella fastidiosa in field-collected plant and insect samples by using 16S rRNA and gyrB sequences. Appl. Environ. Microbiol. 2003, 69, 4249–4255. [Google Scholar] [CrossRef] [PubMed]
- Bruening, G.E.; Kirkpatrick, B.C.; Esser, T.; Webster, R.K. Managing Newly Established Pests: Cooperative efforts contained spread of Pierce’s disease and found genetic resistance. Calif. Agric. 2014, 68, 134–141. [Google Scholar] [CrossRef]
- Kyrkou, I.; Pusa, T.; Ellegaard-Jensen, L.; Sagot, M.-F.; Hansen, L.H. Pierce’s Disease of Grapevines: A Review of Control Strategies and an Outline of an Epidemiological Model. Front. Microbiol. 2018, 9, 2141–2141. [Google Scholar] [CrossRef] [PubMed]
- Bendix, C.; Lewis, J.D. The enemy within: Phloem-limited pathogens. Mol. Plant Pathol. 2018, 19, 238–254. [Google Scholar] [CrossRef] [PubMed]
- Purcell, A.H.; Hopkins, D.L. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 1996, 34, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Heck, M.; Brault, V. Targeted disruption of aphid transmission: A vision for the management of crop diseases caused by Luteoviridae members. Curr. Opin. Virol. 2018, 33, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Reinach, F.C.; Arruda, P.; Abreu, F.A.; Acencio, M.; Alvarenga, R.; Alves, L.M.; Araya, J.E.; Baia, G.S.; Baptista, C.S.; et al. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 2000, 406, 151–159. [Google Scholar] [CrossRef]
- Van Sluys, M.A.; de Oliveira, M.C.; Monteiro-Vitorello, C.B.; Miyaki, C.Y.; Furlan, L.R.; Camargo, L.E.; da Silva, A.C.; Moon, D.H.; Takita, M.A.; Lemos, E.G.; et al. Comparative analyses of the complete genome sequences of Pierce’s disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 2003, 185, 1018–1026. [Google Scholar] [CrossRef]
- Zhang, S.; Flores-Cruz, Z.; Kumar, D.; Chakrabarty, P.; Hopkins, D.L.; Gabriel, D.W. The Xylella fastidiosa biocontrol strain EB92-1 genome is very similar and syntenic to Pierce’s disease strains. Am. Soc. Microbiol. 2011. [Google Scholar] [CrossRef]
- Vanhove, M.; Retchless, A.C.; Sicard, A.; Rieux, A.; Coletta-Filho, H.D.; De La Fuente, L.; Stenger, D.C.; Almeida, R.P.P. Genomic Diversity and Recombination among Xylella fastidiosa Subspecies. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Poelchau, M.; Childers, C.; Moore, G.; Tsavatapalli, V.; Evans, J.; Lee, C.-Y.; Lin, H.; Lin, J.-W.; Hackett, K. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2014, 43, D714–D719. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Williams, K.P.; et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 2009, 22, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Han, C.S.; Liu, B.; Lou, B.; Bai, X.; Deng, C.; Civerolo, E.L.; Gupta, G. Complete Genome Sequence of a Chinese Strain of “Candidatus Liberibacter asiaticus”. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Miyata, S.-I.; Inoue, H.; Iwanami, T. Unique features of a Japanese ‘Candidatus Liberibacter asiaticus’ strain revealed by whole genome sequencing. PLoS ONE 2014, 9, e106109. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Deng, X.; Chen, J. Whole-Genome Sequence of “Candidatus Liberibacter asiaticus” from Guangdong, China. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Wulff, N.A.; Zhang, S.; Setubal, J.C.; Almeida, N.F.; Martins, E.C.; Harakava, R.; Kumar, D.; Rangel, L.T.; Foissac, X.; Bove, J.M.; et al. The complete genome sequence of ‘Candidatus Liberibacter americanus’, associated with Citrus huanglongbing. Mol. Plant Microbe Interact. 2014, 27, 163–176. [Google Scholar] [CrossRef]
- Lin, H.; Pietersen, G.; Han, C.; Read, D.A.; Lou, B.; Gupta, G.; Civerolo, E.L. Complete genome sequence of “Candidatus Liberibacter africanus,” a bacterium associated with citrus huanglongbing. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Lin, H.; Lou, B.; Glynn, J.M.; Doddapaneni, H.; Civerolo, E.L.; Chen, C.; Duan, Y.; Zhou, L.; Vahling, C.M. The complete genome sequence of ‘Candidatus Liberibacter solanacearum’, the bacterium associated with potato zebra chip disease. PLoS ONE 2011, 6, e19135. [Google Scholar] [CrossRef]
- Leonard, M.T.; Fagen, J.R.; Davis-Richardson, A.G.; Davis, M.J.; Triplett, E.W. Complete genome sequence of Liberibacter crescens BT-1. Stand. Genomic Sci. 2012, 7, 271–283. [Google Scholar] [CrossRef]
- Fagen, J.R.; Leonard, M.T.; McCullough, C.M.; Edirisinghe, J.N.; Henry, C.S.; Davis, M.J.; Triplett, E.W. Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of Liberibacter. PLoS ONE 2014, 9, 11. [Google Scholar] [CrossRef]
- Hartung, J.S.; Shao, J.; Kuykendall, L.D. Comparison of the ‘Ca. Liberibacter asiaticus’ genome adapted for an intracellular lifestyle with other members of the rhizobiales. PLoS ONE 2011, 6, e23289. [Google Scholar] [CrossRef]
- Fagen, J.R.; Leonard, M.T.; Coyle, J.F.; McCullough, C.M.; Davis-Richardson, A.G.; Davis, M.J.; Triplett, E.W. Liberibacter crescens, the first cultured member of the genus Liberibacter. Int. J. Syst. Evol. Microbiol. 2014, 64, 2461–2466. [Google Scholar] [CrossRef]
- Arp, A.P.; Hunter, W.B.; Pelz-Stelinski, K.S. Annotation of the Asian Citrus Psyllid genome reveals a reduced innate immune system. Front. Physiol. 2016, 7, 570. [Google Scholar] [CrossRef]
- Saha, S.; Hosmani, P.S.; Villalobos-Ayala, K.; Miller, S.; Shippy, T.; Rosendale, A.; Cordola, C.; Bell, T.; Mann, H.; DeAvila, G.; et al. Improved annotation of the insect vector of Citrus greening disease: Biocuration by a diverse genomics community. bioRxiv 2017, 2017, bax032. [Google Scholar] [CrossRef]
- Saha, S.; Hosmani, P.S.; Villalobos-Ayala, K.; Miller, S.; Shippy, T.; Rosendale, A.; Cordola, C.; Bell, T.; Mann, H.; DeAvila, G.; et al. Diaphorina citri Official Gene Set v1.0. Ag Data Commons 2017. [Google Scholar] [CrossRef]
- Araujo, K.; Godfrey, K.; Vidalakis, G. Do CTV and Spiroplasma citri Impact CLas Establishment. Citrograph 2019, 10, 50–54. [Google Scholar]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Ciraulo, M.B.; Santos, D.S.; Rodrigues, A.C.; de Oliveira, M.V.; Rodrigues, T.; de Oliveira, R.C.; Nunes, L.R. Transcriptome analysis of the phytobacterium Xylella fastidiosa growing under xylem-based chemical conditions. J. Biomed. Biotechnol. 2010, 2010, 781365. [Google Scholar] [CrossRef]
- Pashalidis, S.; Moreira, L.M.; Zaini, P.A.; Campanharo, J.C.; Alves, L.M.; Ciapina, L.P.; Vencio, R.Z.; Lemos, E.G.; Da Silva, A.M.; Da Silva, A.C. Whole-genome expression profiling of Xylella fastidiosa in response to growth on glucose. Omics J. Integrat. Biol. 2005, 9, 77–90. [Google Scholar] [CrossRef]
- Da Silva Neto, J.F.; Koide, T.; Gomes, S.L.; Marques, M.V. Global gene expression under nitrogen starvation in Xylella fastidiosa: Contribution of the sigma54 regulon. BMC Microbiol. 2010, 10, 231. [Google Scholar] [CrossRef]
- Nandety, R.S.; Kamita, S.G.; Hammock, B.D.; Falk, B.W. Sequencing and De novo assembly of the transcriptome of the glassy-winged sharpshooter (Homalodisca vitripennis). PLoS ONE 2013, 8, e81681. [Google Scholar] [CrossRef]
- Nandety, R.S.; Sharif, A.; Kamita, S.G.; Ramasamy, A.; Falk, B.W. Identification of novel and conserved microRNAs in Homalodisca vitripennis, the glassy-winged sharpshooter by expression profiling. PLoS ONE 2015, 10, e0139771. [Google Scholar] [CrossRef]
- Zaini, P.A.; Nascimento, R.; Gouran, H.; Cantu, D.; Chakraborty, S.; Phu, M.; Goulart, L.R.; Dandekar, A.M. Molecular profiling of pierce’s disease outlines the response circuitry of Vitis vinifera to Xylella fastidiosa infection. Front. Plant Sci. 2018, 9, 771. [Google Scholar] [CrossRef]
- Reese, J.; Christenson, M.K.; Leng, N.; Saha, S.; Cantarel, B.; Lindeberg, M.; Tamborindeguy, C.; Maccarthy, J.; Weaver, D.; Trease, A.J.; et al. Characterization of the Asian Citrus Psyllid Transcriptome. J. Genom. 2014, 2, 54–58. [Google Scholar] [CrossRef]
- Kruse, A.; Fattah-Hosseini, S.; Saha, S.; Johnson, R.; Warwick, E.; Sturgeon, K.; Mueller, L.; MacCoss, M.J.; Shatters, R.G., Jr.; Cilia Heck, M. Combining ‘omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS ONE 2017, 12, e0179531. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, H.; Bin, S.; Chen, L.; Han, Q.; Lin, J. Antennal and abdominal Transcriptomes reveal chemosensory genes in the Asian Citrus Psyllid, Diaphorina citri. PLoS ONE 2016, 11, e0159372. [Google Scholar] [CrossRef]
- Fan, J.; Chen, C.; Yu, Q.; Khalaf, A.; Achor, D.S.; Brlansky, R.H.; Moore, G.A.; Li, Z.G.; Gmitter, F.G., Jr. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ‘Candidatus Liberibacter asiaticus’ infection. Mol. Plant Microbe Interact. 2012, 25, 1396–1407. [Google Scholar] [CrossRef]
- Kim, J.S.; Sagaram, U.S.; Burns, J.K.; Li, J.L.; Wang, N. Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: Microscopy and microarray analyses. Phytopathology 2009, 99, 50–57. [Google Scholar] [CrossRef]
- Smolka, M.B.; Martins-de-Souza, D.; Winck, F.V.; Santoro, C.E.; Castellari, R.R.; Ferrari, F.; Brum, I.J.; Galembeck, E.; Della Coletta Filho, H.; Machado, M.A.; et al. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics 2003, 3, 224–237. [Google Scholar] [CrossRef]
- Silva, M.S.; De Souza, A.A.; Takita, M.A.; Labate, C.A.; Machado, M.A. Analysis of the biofilm proteome of Xylella fastidiosa. Proteome Sci. 2011, 9, 58. [Google Scholar] [CrossRef]
- Yang, L.; Lin, H.; Takahashi, Y.; Chen, F.; Walker, M.A.; Civerolo, E.L. Proteomic analysis of grapevine stem in response to Xylella fastidiosa inoculation. Physiol. Mol. Plant Pathol. 2011, 75, 90–99. [Google Scholar] [CrossRef]
- Basha, S.M.; Mazhar, H.; Vasanthaiah, H.K.N. Proteomics Approach to identify unique Xylem Sap proteins in Pierce’s disease-tolerant Vitis species. Appl. Biochem. Biotechnol. 2010, 160, 932–944. [Google Scholar] [CrossRef]
- Ramsey, J.S.; Johnson, R.S.; Hoki, J.S.; Kruse, A.; Mahoney, J.; Hilf, M.E.; Hunter, W.B.; Hall, D.G.; Schroeder, F.C.; MacCoss, M.J.; et al. Metabolic interplay between the Asian Citrus Psyllid and its profftella symbiont: An Achilles’ Heel of the Citrus Greening insect vector. PLoS ONE 2015, 10, e0140826. [Google Scholar] [CrossRef]
- Kruse, A.; Ramsey, J.S.; Johnson, R.; Hall, D.G.; MacCoss, M.J.; Heck, M. Candidatus Liberibacter asiaticus minimally alters expression of immunity and metabolism proteins in hemolymph of Diaphorina citri, the insect vector of Huanglongbing. J. Prot. Res. 2018, 17, 2995–3011. [Google Scholar] [CrossRef]
- Katz, E.; Fon, M.; Lee, Y.J.; Phinney, B.S.; Sadka, A.; Blumwald, E. The citrus fruit proteome: Insights into citrus fruit metabolism. Planta 2007, 226, 989–1005. [Google Scholar] [CrossRef]
- Nwugo, C.C.; Lin, H.; Duan, Y.; Civerolo, E.L. The effect of ‘Candidatus Liberibacter asiaticus’ infection on the proteomic profiles and nutritional status of pre-symptomatic and symptomatic grapefruit (Citrus paradisi) plants. BMC Plant Biol. 2013, 13, 59. [Google Scholar] [CrossRef]
- Girelli, C.R.; Angile, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR metabolite fingerprinting analysis reveals a disease biomarker and a field treatment response in Xylella fastidiosa subsp. pauca-Infected Olive Trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef]
- Killiny, N.; Hijaz, F.; El-Shesheny, I.; Alfaress, S.; Jones, S.E.; Rogers, M.E. Metabolomic analyses of the haemolymph of the Asian citrus psyllid Diaphorina citri, the vector of huanglongbing. Physiol. Entomol. 2017, 42, 134–145. [Google Scholar] [CrossRef]
- Killiny, N.; Jones, S.E. Metabolic alterations in the nymphal instars of Diaphorina citri induced by Candidatus Liberibacter asiaticus, the putative pathogen of huanglongbing. PLoS ONE 2018, 13, e0191871. [Google Scholar] [CrossRef]
- Killiny, N. Metabolomic comparative analysis of the phloem sap of curry leaf tree (Bergera koenegii), orange jasmine (Murraya paniculata), and Valencia sweet orange (Citrus sinensis) supports their differential responses to Huanglongbing. Plant Signal. Behav. 2016, 11, e1249080. [Google Scholar] [CrossRef]
- Nehela, Y.; Killiny, N. ‘Candidatus Liberibacter asiaticus’ and its vector, Diaphorina citri, augment the tricarboxylic acid cycle of their host via the gamma-Aminobutyric acid shunt and polyamines pathway. Mol. Plant Microbe Interact. 2019, 32, 413–427. [Google Scholar] [CrossRef]
- Slisz, A.M.; Breksa, A.P., 3rd; Mishchuk, D.O.; McCollum, G.; Slupsky, C.M. Metabolomic analysis of citrus infection by ‘Candidatus Liberibacter’ reveals insight into pathogenicity. J. Proteome Res. 2012, 11, 4223–4230. [Google Scholar] [CrossRef]
- Parker, J.K.; Wisotsky, S.R.; Johnson, E.G.; Hijaz, F.M.; Killiny, N.; Hilf, M.E.; De La Fuente, L. Viability of ‘Candidatus Liberibacter asiaticus’ prolonged by addition of citrus juice to culture medium. Phytopathology 2014, 104, 15–26. [Google Scholar] [CrossRef]
- Sechler, A.; Schuenzel, E.L.; Cooke, P.; Donnua, S.; Thaveechai, N.; Postnikova, E.; Stone, A.L.; Schneider, W.L.; Damsteegt, V.D.; Schaad, N.W. Cultivation of ‘Candidatus Liberibacter asiaticus’, ‘Ca. L. africanus’, and ‘Ca. L. americanus’ Associated with Huanglongbing. Phytopathology 2009, 99, 480–486. [Google Scholar] [CrossRef]
- Merfa, M.V.; Perez-Lopez, E.; Naranjo, E.; Jain, M.; Gabriel, D.W.; De La Fuente, L. Progress and obstacles in culturing ‘Candidatus Liberibacter asiaticus’, the bacterium associated with Huanglongbing. Phytopathology 2019, 109, 1092–1101. [Google Scholar] [CrossRef]
- Zhou, L.; Powell, C.A.; Li, W.; Irey, M.; Duan, Y. Prophage-mediated dynamics of ‘Candidatus Liberibacter asiaticus’ populations, the destructive bacterial pathogens of citrus huanglongbing. PLoS ONE 2013, 8, e82248. [Google Scholar] [CrossRef]
- Pitino, M.; Hoffman, M.T.; Zhou, L.; Hall, D.G.; Stocks, I.C.; Duan, Y. The phloem-sap feeding Mealybug (Ferrisia virgata) Carries ‘Candidatus Liberibacter asiaticus’ populations that do not cause disease in host plants. PLoS ONE 2014, 9, e85503. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Stilwagen, S.; Reznik, G.; Feil, H.; Feil, W.S.; Anderson, I.; Bernal, A.; D’Souza, M.; Ivanova, N.; Kapatral, V.; et al. Draft sequencing and comparative genomics of Xylella fastidiosa strains reveal novel biological insights. Genome Res. 2002, 12, 1556–1563. [Google Scholar] [CrossRef]
- Reddy, J.D.; Reddy, S.L.; Hopkins, D.L.; Gabriel, D.W. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol. Plant Microbe Interact. 2007, 20, 403–410. [Google Scholar] [CrossRef]
- Cianciotto, N.P.; White, R.C. Expanding role of Type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 2017, 85, e00014–e00017. [Google Scholar] [CrossRef]
- Chatterjee, S.; Almeida, R.P.; Lindow, S. Living in two worlds: The plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 2008, 46, 243–271. [Google Scholar] [CrossRef]
- Lindow, S.; Newman, K.; Chatterjee, S.; Baccari, C.; Lavarone, A.T.; Ionescu, M. Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of pierce’s disease. Mol. Plant Microbe Interact. 2014, 27, 244–254. [Google Scholar] [CrossRef]
- Ionescu, M.; Baccari, C.; Da Silva, A.M.; Garcia, A.; Yokota, K.; Lindow, S.E. Diffusible signal factor (DSF) synthase RpfF of Xylella fastidiosa is a multifunction protein also required for response to DSF. J. Bacteriol. 2013, 195, 5273–5284. [Google Scholar] [CrossRef]
- Nascimento, R.; Gouran, H.; Chakraborty, S.; Gillespie, H.W.; Almeida-Souza, H.O.; Tu, A.; Rao, B.J.; Feldstein, P.A.; Bruening, G.; Goulart, L.R.; et al. The Type II secreted Lipase/Esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Sci. Rep. 2016, 6, 18598. [Google Scholar] [CrossRef]
- Li, W.; Cong, Q.; Pei, J.; Kinch, L.N.; Grishin, N.V. The ABC transporters in Candidatus Liberibacter asiaticus. Proteins Struct. Funct. Bioinf. 2012, 80, 2614–2628. [Google Scholar] [CrossRef]
- Hao, G.; Boyle, M.; Zhou, L.; Duan, Y. The intracellular citrus Huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ encodes two novel autotransporters. PLoS ONE 2013, 8, e68921. [Google Scholar] [CrossRef]
- Mann, M.; Fattah-Hosseini, S.; Ammar, E.D.; Stange, R.; Warrick, E.; Sturgeon, K.; Shatters, R.; Heck, M. Diaphorina citri nymphs are resistant to morphological changes induced by “Candidatus Liberibacter asiaticus” in midgut epithelial cells. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Shams-Bakhsh, M.; Mann, M.; Fattah-Hosseini, S.; Bagheri, A.; Mehrabadi, M.; Heck, M. Distribution and variation of bacterial endosymbiont and “Candidatus Liberibacter asiaticus” titer in the Huanglongbing insect vector, Diaphorina citri Kuwayama. Microb. Ecol. 2018, 78, 206–222. [Google Scholar] [CrossRef]
- Rapicavoli, J.N.; Blanco-Ulate, B.; Muszynski, A.; Figueroa-Balderas, R.; Morales-Cruz, A.; Azadi, P.; Dobruchowska, J.M.; Castro, C.; Cantu, D.; Roper, M.C. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 2018, 9, 390. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Lai, K.-K.; Davis-Richardson, A.G.; Dias, R.; Triplett, E.W. Identification of the genes required for the Culture of Liberibacter crescens, the closest cultured relative of the Liberibacter plant pathogens. Front. Microbiol. 2016, 7, 547. [Google Scholar] [CrossRef]
- Naranjo, E.; Merfa, M.V.; Ferreira, V.; Jain, M.; Davis, M.J.; Bahar, O.; Gabriel, D.W.; De La Fuente, L. Liberibacter crescens biofilm formation in vitro: Establishment of a model system for pathogenic ‘Candidatus Liberibacter spp’. Sci. Rep. 2019, 9, 5150. [Google Scholar] [CrossRef]
- Monroe, D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007, 5, e307. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.; Purcell, A.H.; Lindow, S.E. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 2004, 101, 1737–1742. [Google Scholar] [CrossRef]
- Guilhabert, M.R.; Kirkpatrick, B.C. Identification of Xylella fastidiosa antivirulence genes: Hemagglutinin adhesins contribute to X. fastidiosaa biofilm maturation and colonization and attenuate virulence. Mol. Plant Microbe Interact. 2005, 18, 856–868. [Google Scholar] [CrossRef]
- Zhang, S.; Chakrabarty, P.K.; Fleites, L.A.; Rayside, P.A.; Hopkins, D.L.; Gabriel, D.W. Three new Pierce’s disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92-1. PLoS ONE 2015, 10, e0133796. [Google Scholar] [CrossRef]
- Killiny, N.; Almeida, R.P.P. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl. Environ. Microbiol. 2009, 75, 521–528. [Google Scholar] [CrossRef]
- Killiny, N.; Almeida, R.P. Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl. Environ. Microbiol. 2014, 80, 420–426. [Google Scholar] [CrossRef]
- Caserta, R.; Takita, M.A.; Targon, M.L.; Rosselli-Murai, L.K.; de Souza, A.P.; Peroni, L.; Stach-Machado, D.R.; Andrade, A.; Labate, C.A.; Kitajima, E.W.; et al. Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation. Appl. Environ. Microbiol. 2010, 76, 4250–4259. [Google Scholar] [CrossRef]
- Chen, H.; Kandel, P.P.; Cruz, L.F.; Cobine, P.A.; De La Fuente, L. The major outer membrane protein MopB is required for twitching movement and affects biofilm formation and virulence in two Xylella fastidiosa strains. Mol. Plant Microbe Interact. 2017, 30, 896–905. [Google Scholar] [CrossRef]
- Meng, Y.; Li, Y.; Galvani, C.D.; Hao, G.; Turner, J.N.; Burr, T.J.; Hoch, H.C. Upstream migration of Xylella fastidiosa via Pilus-Driven twitching motility. J. Bacteriol. 2005, 187, 5560. [Google Scholar] [CrossRef]
- Von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar] [CrossRef]
- Andrade, M.O.; Wang, N. The Tad pilus apparatus of Candidatus Liberibacter asiaticus and its regulation by VisNR. Mol. Plant Microbe Interact. 2019, 32, 1175–1187. [Google Scholar] [CrossRef]
- Garnier, M.; Bové, J.M. Transmission of the organism associated with citrus greening disease from sweet orange to periwinkle by dodder. Phytopathology 1983, 73, 1358–1363. [Google Scholar] [CrossRef]
- Shokrollah, H.; Abdullah, T.L.; Sijam, K.; Abdullah, S.N.A. Ultrastructures of Candidatus Liberibacter asiaticus and its damage in huanglongbing (HLB) infected citrus. Afr. J. Biotechnol. 2010, 9, 5897–5901. [Google Scholar]
- Moll, J.N.; Martin, M.M. Electron microscope evidence that citrus psylla (Trioza etytreae) is a vector of greening disease in South Africa. Phytophylactica 1973, 5, 41–44. [Google Scholar]
- Patel, H.K.; Suarez-Moreno, Z.R.; Degrassi, G.; Subramoni, S.; Gonzalez, J.F.; Venturi, V. Bacterial LuxR solos have evolved to respond to different molecules including signals from plants. Front. Plant Sci. 2013, 4, 447. [Google Scholar] [CrossRef]
- Ammar, E.-D.; Ramos, J.E.; Hall, D.G.; Dawson, W.O.; Shatters, R.G., Jr. Acquisition, replication and inoculation of Candidatus Liberibacter asiaticus following various acquisition periods on Huanglongbing-infected citrus by nymphs and adults of the Asian Citrus Psyllid. PLoS ONE 2016, 11, e0159594. [Google Scholar] [CrossRef]
- Jain, M.; Fleites, L.A.; Gabriel, D.W. A small Wolbachia protein directly represses Phage Lytic cycle genes in “Candidatus Liberibacter asiaticus” within Psyllids. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Hopkins, D.L. Biological Control of Pierce’s Disease in the vineyard with strains of Xylella fastidiosa benign to grapevine. Plant Dis. 2005, 89, 1348–1352. [Google Scholar] [CrossRef]
- Baccari, C.; Antonova, E.; Lindow, S. Biological control of Pierce’s disease of grape by an endophytic bacterium. Phytopathology 2019, 109, 248–256. [Google Scholar] [CrossRef]
- Bextine, B.; Lauzon, C.; Potter, S.; Lampe, D.; Miller, T.A. Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Curr. Microbiol. 2004, 48, 327–331. [Google Scholar] [CrossRef]
- Zhang, S.; Flores-Cruz, Z.; Zhou, L.; Kang, B.H.; Fleites, L.A.; Gooch, M.D.; Wulff, N.A.; Davis, M.J.; Duan, Y.P.; Gabriel, D.W. ‘Ca. Liberibacter asiaticus’ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol. Plant Microbe Interact. 2011, 24, 458–468. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, P.; Pu, X.; Xing, X.; Chen, J.; Deng, X. Analysis of a prophage gene frequency revealed population variation of ‘Candidatus Liberibacter asiaticus’ from two Citrus-Growing provinces in China. Plant Dis. 2010, 95, 431–435. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, F.; Kumagai, L.B.; Polek, M.; Deng, X.; Chen, J. Two ‘Candidatus Liberibacter asiaticus’ strains recently found in California harbor different prophages. Phytopathology 2017, 107, 662–668. [Google Scholar] [CrossRef]
- Feil, H.; Purcell, A.H. Temperature-dependent growth and survival of Xylella fastidiosa in vitro and in potted grapevines. Plant Dis. 2001, 85, 1230–1234. [Google Scholar] [CrossRef]
- Lieth, J.H.; Meyer, M.M.; Yeo, K.H.; Kirkpatrick, B.C. Modeling cold curing of Pierce’s disease in Vitis vinifera ‘Pinot Noir’ and ‘Cabernet Sauvignon’ grapevines in California. Phytopathology 2011, 101, 1492–1500. [Google Scholar] [CrossRef]
- Corlett, R.T.; Westcott, D.A. Will plant movements keep up with climate change? Trends Ecol. Evol. 2013, 28, 482–488. [Google Scholar] [CrossRef]
- Anas, O.; Harrison, U.J.; Brannen, P.M.; Sutton, T.B. The effect of warming winter temperatures on the severity of Pierce’s disease in the Appalachian mountains and Piedmont of the southeastern United States. Plant Health Progr. 2008. [Google Scholar] [CrossRef]
- Lopes, S.A.; Frare, G.F.; Bertolini, E.; Cambra, M.; Fernandes, N.G.; Ayres, A.J.; Marin, D.R.; Bove, J.M. Liberibacters associated with Citrus Huanglongbing in Brazil: ‘Candidatus Liberibacter asiaticus’ is heat tolerant, ‘Ca. L. americanus’ is heat sensitive. Plant Dis. 2009, 93, 257–262. [Google Scholar] [CrossRef]
- Doud, M.M.; Wang, Y.; Hoffman, M.T.; Latza, C.L.; Luo, W.; Armstrong, C.M.; Gottwald, T.R.; Dai, L.; Luo, F.; Duan, Y. Solar thermotherapy reduces the titer of Candidatus Liberibacter asiaticus and enhances canopy growth by altering gene expression profiles in HLB-affected citrus plants. Hortic. Res. 2017, 4, 17054–17054. [Google Scholar] [CrossRef]
- Bove, J.M.; Calavan, E.C.; Capoor, S.P.; Cortez, R.E.; Schwarz, R.E. Influence of temperature on symptoms of California stubborn, South Africa greening, India citrus decline and Philippines leaf mottling diseases. Proc. Int. Organ. Citrus Virol. 1974, 6, 12–15. [Google Scholar]
- Hopkins, D.L.; Purcell, A.H. Xylella fastidiosa: Cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef]
- Ammar, E.D.; Shatters, R.G., Jr.; Lynch, C.; Hall, D.G. Candidatus Liberibacter asiaticus in the salivary glands and alimentary canal of Diaphorina citri (Hemiptera: Psyllidae) vector of citrus Huanglongbing disease. Ann. Entomol. Soc. Am. 2011, 104, 526–533. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering and Medicine. A Review of the Citrus Greening Research and Development Efforts Supported by the Citrus Research and Development Foundation: Fighting a Ravaging Disease; The National Academies Press: Washington, DC, USA, 2018; 25026p. [Google Scholar]
- Burand, J.P.; Hunter, W.B. RNAi: Future in insect management. J. Invert. Pathol. 2013, 112, S68–S74. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double strand RNA delivery system for plant-sap-feeding insects. PLoS ONE 2017, 12, e0171861. [Google Scholar] [CrossRef]
- Killiny, N.; Almeida, R.P.P. Host structural carbohydrate induces vector transmission of a bacterial plant pathogen. Proc. Natl. Acad. Sci. USA 2009, 106, 22416. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Ramsey, J.; Mann, M.; Bennett, L.; Hunter, W.B.; Shams-Bakhsh, M.; Hall, D.G.; Heck, M. Color morphology of Diaphorina citri influences interactions with its bacterial endosymbionts and ‘Candidatus Liberibacter asiaticus’. PLoS ONE 2019, 14, e0216599. [Google Scholar] [CrossRef]
- Ammar, E.D.; Hall, D.G; Hosseinzadeh, S.; Heck, M. The quest for a non-vector psyllid: Natural variation in acquisition and transmission of the huanglongbing pathogen “Candidatus Liberibacter asiaticus” by Asian citrus psyllid isofemale lines. PLoS ONE 2018, 13, e0195804. [Google Scholar] [CrossRef]
- Pelz-Stelinski, K.S.; Killiny, N. Better together: Association with ‘Candidatus Liberibacter Asiaticus’ increases the reproductive fitness of its insect vector, Diaphorina citri (Hemiptera: Liviidae). Ann. Entomol. Soc. Am. 2016, 109, 371–376. [Google Scholar] [CrossRef]
- Wood, M.; McBride, J. Scientists sharpen strategies to sabotage glassy-winged sharpshooter. Agric. Res. 2001, 49, 20–22. [Google Scholar]
- Tubajika, K.; Civerolo, E.; Puterka, G.; Hashim, J.; Luvisi, D. The effects of kaolin, harpin, and imidacloprid on development of Pierce’s disease in grape. Crop. Protect. 2007, 26, 92–99. [Google Scholar] [CrossRef]
- Obradovic, A.; Jones, J.B.; Momol, M.T.; Balogh, B.; Olson, S.M. management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Dis. 2004, 88, 736–740. [Google Scholar] [CrossRef]
- Alarcon, C.; Castro, J.; Munoz, F.; Arce-Johnson, P.; Delgado, J. Protein(s) from the Gram-Positive bacterium Clavibacter michiganensis subsp. michiganensis induces a Hypersensitive response in plants. Phytopathology 1998, 88, 306–310. [Google Scholar] [CrossRef]
- Jackson, B.C.; Blua, M.J.; Bextine, B. Impact of duration versus frequency of probing by Homalodisca vitripennis (Hemiptera: Cicadellidae) on inoculation of Xylella fastidiosa. J. Econ. Entomol. 2008, 101, 1122–1126. [Google Scholar] [CrossRef]
- Daugherty, M.P.; Almeida, R.P.P. Estimating Xylella fastidiosa transmission parameters: Decoupling sharpshooter number and feeding period. Entomol. Exp. Appl. 2009, 132, 84–92. [Google Scholar] [CrossRef]
- Pelz-Stelinski, K.S.; Brlansky, R.H.; Ebert, T.A.; Rogers, M.E. Transmission parameters for Candidatus liberibacter asiaticus by Asian citrus psyllid (Hemiptera: Psyllidae). J. Econ. Entomol. 2010, 103, 1531–1541. [Google Scholar] [CrossRef]
- Alhaddad, H.; Coudron, T.A.; Backus, E.A.; Schreiber, F. Comparative behavioral and protein study of salivary secretions in Homalodisca spp. sharpshooters (Hemiptera: Cicadellidae: Cicadellinae). Ann. Entomol. Soc. Am. 2011, 104, 543–552. [Google Scholar] [CrossRef]
- Cicero, J.M.; Fisher, T.W.; Brown, J.K. Localization of ‘Candidatus Liberibacter solanacearum’ and evidence for surface appendages in the Potato Psyllid vector. Phytopathology 2016, 106, 142–154. [Google Scholar] [CrossRef]
- Ammar, E.-D.; Richardson, M.L.; Abdo, Z.; Hall, D.G.; Shatters, R.G., Jr. Differences in stylet sheath occurrence and the fibrous ring (Sclerenchyma) between xCitroncirus plants relatively resistant or susceptible to adults of the Asian Citrus Psyllid Diaphorina citri (Hemiptera: Liviidae). PLoS ONE 2014, 9, e110919. [Google Scholar] [CrossRef]
- Morgan, J.K.; Luzio, G.A.; Ammar el, D.; Hunter, W.B.; Hall, D.G.; Shatters, R.G., Jr. Formation of Stylet Sheaths in aere (in air) from eight species of phytophagous hemipterans from six families (Suborders: Auchenorrhyncha and Sternorrhyncha). PLoS ONE 2013, 8, e62444. [Google Scholar] [CrossRef]
- Cicero, J.M.; Stansly, P.A.; Brown, J.K. Functional anatomy of the oral region of the Potato Psyllid (Hemiptera: Psylloidea: Triozidae). Ann. Entomol. Soc. Am. 2015, 108, 743–761. [Google Scholar] [CrossRef]
- Yu, X.; Killiny, N. The secreted salivary proteome of Asian citrus psyllid Diaphorina citri. Physiol. Entomol. 2018, 43, 324–333. [Google Scholar] [CrossRef]
- Will, T.; Vilcinskas, A. The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 2015, 57, 34–40. [Google Scholar] [CrossRef]
- California Department of Food and Agriculture (CDFA). Pierce’s Disease Control Program—Report to the Legislature for Calendar Year 2017; CDFA: Sacramento, CA, USA, 2017. [Google Scholar]
- Étienne, J.; Quilici, S.; Marival, D.; Franck, A. Biological control of Diaphorina citri (Hemiptera: Psyllidae) in Guadeloupe by imported Tamarixia radiata (hymenoptera: Eulophidae). Fruits 2001, 56, 307–315. [Google Scholar] [CrossRef]
- Qureshi, J.A.; Rogers, M.E.; Hall, D.G.; Stansly, P.A. Incidence of invasive Diaphorina citri (Hemiptera: Psyllidae) and its introduced parasitoid Tamarixia radiata (Hymenoptera: Eulophidae) in Florida citrus. J. Econ. Entomol. 2009, 102, 247–256. [Google Scholar] [CrossRef]
- McFarland, C.D.; Hoy, M.A. Survival of Diaphorina citri (Homoptera: Psyllidae), and its two parasitoids, Tamarixia radiata (Hymenoptera: Eulophidae) and Diaphorencyrtus aligarhensis (Hymenoptera: Encyrtidae), under different relative humidities and temperature regimes. Fla. Entomol. 2001, 84, 227–233. [Google Scholar] [CrossRef]
- Nigg, J.C.; Nouri, S.; Falk, B.W. Complete genome sequence of a putative densovirus of the Asian Citrus Psyllid, Diaphorina citri. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Nouri, S.; Salem, N.; Falk, B.W. complete genome sequence of Diaphorina citri-associated C virus, a novel putative RNA virus of the Asian Citrus Psyllid, Diaphorina citri. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Avery, P.B.; Hunter, W.B.; Hall, D.G.; Jackson, M.A.; Powell, C.A.; Rogers, M.E. Diaphorina citri (Hemiptera: Psyllidae) infection and dissemination of the entomopathogenic fungus Isaria fumosorosea (Hypocreales: Cordycipitaceae) under laboratory conditions. Fla. Entomol. 2009, 92, 608–619. [Google Scholar] [CrossRef]
- Hall, D.G.; Hentz, M.G.; Meyer, J.M.; Kriss, A.B.; Gottwald, T.R.; Boucias, D.G. Observations on the entomopathogenic fungus Hirsutella citriformis attacking adult Diaphorina citri (Hemiptera: Psyllidae) in a managed citrus grove. BioControl 2012, 57, 663–675. [Google Scholar] [CrossRef]
- Chow, A.; Dunlap, C.A.; Jackson, M.A.; Flores, D.; Patt, J.M.; Setamou, M. Oviposition behavior and survival of Tamarixia radiata (Hymenoptera: Eulophidae), an ectoparasitoid of the Asian Citrus Psyllid, Diaphorina citri (Hemiptera: Liviidae), on hosts exposed to an entomopathogenic fungus, Isaria fumosorosea (Hypocreales: Cordycipitaceae), under laboratory conditions. J. Econ. Entomol. 2016, 109, 1995–2005. [Google Scholar] [CrossRef]
- Tyler, H.L.; Roesch, L.F.; Gowda, S.; Dawson, W.O.; Triplett, E.W. Confirmation of the sequence of Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant Microbe Interact. 2009, 22, 1624–1634. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Dandekar, A.M.; Gouran, H.; Ibanez, A.M.; Uratsu, S.L.; Aguero, C.B.; McFarland, S.; Borhani, Y.; Feldstein, P.A.; Bruening, G.; Nascimento, R.; et al. An engineered innate immune defense protects grapevines from Pierce disease. Proc. Natl. Acad. Sci. USA 2012, 109, 3721–3725. [Google Scholar] [CrossRef]
- Rapicavoli, J.; Ingel, B.; Blanco-Ulate, B.; Cantu, D.; Roper, C. Xylella fastidiosa: An examination of a re-emerging plant pathogen. Mol. Plant Pathol. 2018, 19, 786–800. [Google Scholar] [CrossRef]
- Kirkpatrick, B.C.; Lincoln, J.E.; Roper, C.; Esser, T. Evaluation of Pierce’s disease resistance in transgenic Vitis vinifera grapevines expressing either grape thaumatin-like protein or Xylella fastidiosa hemagglutinin protein. In Proceedings of the Pierce’s Disease Research Symposium, Sacramento, CA, USA; 2012; pp. 130–136. [Google Scholar]
- Gilchrist, D.; Lincoln, J.E.; Esser, T. Systemic control of Pierce’s disease by altered expression of anti-apoptotic genes or their RNA-based regulatory elements. In Proceedings of the Pierce’s Disease Research Symposium, Sacramento, CA, USA; 2008; pp. 252–255. [Google Scholar]
- Dandekar, A.M.; Gilchrist, D.; Miller, T.; Esser, T. Chimeric antimicrobial protein and polygalacturonase-inhibiting protein transgenic grapevine field trial. In Proceedings of the Pierce’s Disease Research Progress Reports, Sacramento, CA USA; 2015; pp. 94–103. [Google Scholar]
- Miller, T.; Daugherty, M.; Mauk, P.; Esser, T. Field trial for resistance to Pierce’s disease. In Proceedings of the Pierce’s Disease Research Progress Reports, Sacramento, CA, USA; 2012; pp. 175–177. [Google Scholar]
- Aguero, C.B.; Uratsu, S.L.; Greve, C.; Powell, A.L.; Labavitch, J.M.; Meredith, C.P.; Dandekar, A.M. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005, 6, 43–51. [Google Scholar] [CrossRef]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic Citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against Huanglongbing (HLB.; Citrus Greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef]
- Mirkov, T.E.; Gonzalez-Ramos, J. Pathogen Resistant Citrus Compositions, Organisms, Systems, and Methods. Google Patents: U.S. Patent Application No. 14/139,791.2013, 2013. [Google Scholar]
- Shi, Q.; Pitino, M.; Zhang, S.; Krystel, J.; Cano, L.M.; Shatters, R.G., Jr.; Hall, D.G.; Stover, E. Temporal and spatial detection of Candidatus Liberibacter asiaticus putative effector transcripts during interaction with Huanglongbing-susceptible, -tolerant, and -resistant citrus hosts. BMC Plant Biol. 2019, 19, 122. [Google Scholar] [CrossRef]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G., Jr. Construction of high-density genetic maps and detection of QTLs associated with Huanglongbing tolerance in Citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef]
- Dawson, W.O.; Bar-Joseph, M.; Garnsey, S.M.; Moreno, P. Citrus tristeza virus: Making an ally from an enemy. Annu. Rev. Phytopathol. 2015, 53, 137–155. [Google Scholar] [CrossRef]
- Domínguez, A.; Guerri, J.; Cambra, M.; Navarro, L.; Moreno, P.; Peña, L. Efficient production of transgenic citrus plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep. 2000, 19, 427–433. [Google Scholar] [CrossRef]
- Pandey, S.S.; Wang, N. Targeted early detection of Citrus Huanglongbing causal agent ‘Candidatus Liberibacter asiaticus’ before symptom expression. Phytopathology 2019, 109, 952–959. [Google Scholar] [CrossRef]
- Riaz, S.; Tenscher, A.C.; Graziani, R.; Krivanek, A.F.; Ramming, D.W.; Walker, M.A. Using marker-assisted selection to breed Pierce’s Disease-resistant grapes. Am. J. Enol. Vitic. 2009, 60, 199–207. [Google Scholar]
- Chowell, G.; Mizumoto, K.; Banda, J.M.; Poccia, S.; Perrings, C. Assessing the potential impact of vector-borne disease transmission following heavy rainfall events: A mathematical framework. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180272. [Google Scholar] [CrossRef]
- Perrings, C. Options for managing the infectious animal and plant disease risks of international trade. Food Secur. 2016, 8, 27–35. [Google Scholar] [CrossRef]
- McRoberts, N.; Thomas, C.; Grafton-Cardwell, B. Minimizing the Dispersal of Asian Citrus Psyllid in California: The Key Role for Regulating Transport Corridors; Policy Briefing Paper; University of California, Davis: Riverside, CA, USA, 2016. [Google Scholar]
- De Macedo Lemos, E.G.; Alves, L.M.C.; Campanharo, J.C. Genomics-based design of defined growth media for the plant pathogen Xylella fastidiosa. FEMS Microbiol. Lett. 2003, 219, 39–45. [Google Scholar] [CrossRef]
- Ammar, E.-D.; Shatters, R.G.; Hall, D.G. Localization of Candidatus Liberibacter asiaticus, associated with Citrus Huanglongbing Disease, in its Psyllid vector using fluorescence in situ hybridization. J. Phytopathol. 2011, 159, 726–734. [Google Scholar] [CrossRef]
| Xylella fastidiosa | Candidatus Liberibacter asiaticus |
|---|---|
| Pierce’s disease was first thought to be caused by a virus. | HLB was first thought to be caused by a virus. |
| Xylem-limited | Phloem-limited |
| Gammaprotobacteria (includes other Xanthomonads) | Alphaprotobacteria (includes Rickettsia, Agrobacterium, Rhizobium, Wolbachia) |
| Transmitted by hemipteran insect | Transmitted by hemipteran insect |
| Lacks a type III secretion system | Lacks a type III secretion system |
| Genome may contain four predicted prophages | Genome may contain prophage |
| Forms biofilms in insect, plant, and in vitro | Forms biofilms in insect, not observed in plant |
| Culturable | Non-culturable |
| Propagative, foregut-borne transmission | Propagative, circulative transmission |
| Generalist pathogen, in which addition of a small number of genes or plasmids can alter host specificity | High level of host and vector specificity |
| OMIC Resource | Bacteria | Insect Vector | Plant |
|---|---|---|---|
| Genome | X. fastidiosa genome, CVC strain 9a5c [30] X. fastidiosa genome, Pierce’s disease strain Temecula1 and comparative genomics analyses [31] X. fastidiosa biocontrol strain EB92 genome and comparative genomics analyses [32] Comparative genome analysis of 72 X. fastidiosa genomes, with 36 newly sequenced genomes presented [33] | GWSS genome [34] | Draft genome of Vitis vinifera [35] |
| C. Las genome [36,37,38,39] C. Lam genome [40] C. Laf genome [41] C. Lso genome [42] L. crescens genome [43] Comparative genomics among Liberibacter species and relatives [36,43,44,45,46] | D. citri genome [47,48,49] Metagenomics analysis of infected citrus phloem [50] | Draft genome of Citrus sinensis [51] | |
| Transcriptome | X. fastidiosa transcriptome, CVC strain 9a5c [52,53] and J1a12 [54] | GWSS de novo transcriptome and mRNA profile [55,56] | Infected grapevine transcriptome [57] |
| Transcriptome not available due to culture challenges | D. citri whole-body, antenna, abdominal, gut transcriptome [58,59,60] | Comparative transcriptome of infected rough lemon and sweet orange [61] Microarray comparison of healthy and infected sweet orange [62] | |
| Proteome | X. fastidiosa proteome, CVC strain 9a5c [63] X. fastidiosa biofilm proteome, CVC strain 9a5c [64] | Not available | Infected grapevine proteome [57] Comparison of proteomes in infected and healthy grapevine [65] Proteomic comparison of tolerant and susceptible grapevine [66] |
| Proteome not available due to culture challenges | D. citri whole-body, gut, and hemolymph proteome [59,67,68] | Citrus fruit proteome [69] Proteomic analysis of infected pre-symptomatic and symptomatic grapefruit (Citrus paradisi) [70] | |
| Metabolome | Metabolome not available | Not available | Infected grapevine metabolome [57] Metabolomics response of olive trees to X. fastidiosa subsp. pauca [71] |
| Metabolome not available due to culture challenges | D. citri hemolymph metabolome [72] Metabolic comparison of infected and healthy nymphs [73] | Metabolic comparison of phloem sap from Murraya paniculata, Citrus sinensis, and Bergera koenegii [74] Metabolic analysis of citrus leaves infected or uninfected with C. Las, fed on by healthy D. citri [75] Metabolic comparison of juice from healthy and infected Citrus sinensis [76] |
| Gene Name | Function | Relevant Bacterium | Importance |
|---|---|---|---|
| tolC | Outer membrane component of type I secretion system | X. fastidiosa | Knockout causes avirulence and hypersensitivity to phytoalexins |
| rpf gene cluster | Diffusible signal factor (XfDSF) synthesis and recognition | X. fastidiosa | Expression in grapevine reduces X. fastidiosa spread |
| lesA | Lipase/esterase | X. fastidiosa | Key pathogenicity factor for X. fastidiosa |
| pilA2 & pilC | Type 4 pili proteins | X. fastidiosa | Involved in biofilm formation |
| xadA1 & xadA2 | Afimbrial adhesins | X. fastidiosa | Involved in biofilm formation |
| mopB | X. fastidiosa outer membrane protein | X. fastidiosa | Deletion affects biofilm formation and virulence, eliminates twitching motility |
| luxI | Encodes enzymes that produce acyl-homoserine lactone (AHL) molecules | C. Las | C. Las lacks a luxI gene |
| luxR | AHL-responsive regulatory gene | C. Las | C. Las possesses a luxR gene |
| hxfA | Hemagglutinin-like | X. fastidiosa | Deletion results in hypervirulence; plants expressing the gene had decreased disease development |
| lasAI & lasAII | Type V autotransporters | C. Las | Found in C. Las genome; may target plant mitochondria |
| Challenge | Pierce’s Disease | HLB | Proposed Strategy for HLB Field |
|---|---|---|---|
| Pathogen culturability | Pathogen can be cultured | Culture is currently not possible | Leverage ‘omic data from D. citri to replicate nutritional environment from insect for C. Las growth; in lieu of culture, test candidate gene functions using L. crescens, delivery of RNA and other inhibitory molecules [130,131] |
| Presence of insect vector | Management via monitoring of nursery stocks; scouting for GWSS; biological control of GWSS using parasitic wasps [150]; eradication has been achieved in specific areas of California [150] | Management via monitoring of nursery stocks; scouting for D. citri; early detection of infected trees; biological control of D. citri using parasitic wasps [151,152,153] and entomopathogenic fungi [156,157] | Continued aggressive scouting for D. citri; test D. citri nymphs via PCR for detection of early C. Las infection; target plant sampling to sites of D. citri feeding by monitoring stylet sheath deposition [174]; evaluate use of D. citri-infecting viruses and fungi for wide-scale use; apply control strategies in holistic manner with consideration for potential interactions between biological control agents [158] |
| Bacterial biofilm formation | Forms biofilm in both insect and plant [98]; adhesion proteins play a role in biofilm formation [103]; outer membrane protein MopB is important for biofilm formation, systemic colonization of xylem [104,105] | Bacterium has a luxR but not a luxI gene; reported to form biolfilm in insect but not plant [108,109,110]; interaction with other bacterial species may facilitate biofilm formation [58,112,113] | Evaluate importance of outer membrane proteins for biofilm formation; investigate interactions between C. Las and D. citri endosymbionts; L. crescens as a model to study biofilm formation [96] |
| Transmission by insect vector | Paratransgenesis shows promise to reduce bacterial titer in insect foregut [116]; hemagglutinin and adhesion proteins are involved in transmission [132]; lectins and N-acetylglucosamine reduce transmission [101] White kaolin increases GWSS mortality [136,137] | Color morphology impacts vectoring capacity [133]; vector competency varies naturally among D. citri populations | Evaluation of Wolbachia repressor protein to control C. Las [113]; evaluate induction of C. Las prophage; further research into the molecular basis for vector manipulation; delivery of molecules that inhibit feeding structures of D. citri; mine secreted salivary proteome for target proteins with potential roles in insect feeding [148] |
| Infection of host plant | Harpin reduces disease incidence [137]; several conventionally bred and transgenic plants show increased resistance [161,175]; asymptomatic strain of X. fastidiosa as biological control | Transgenic citrus shows increased tolerance to C. Las [168]; studies of citrus varieties’ tolerance provides resources for breeding and engineering [170,171]; viral-based vector systems allow therapeutics to be delivered into trees [172,173] | Evaluation of L. crescens as biological control agent; induction of C. Las phages [117,118,119]; use of delivery systems to deliver RNA and other therapeutics into trees based on OMIC and other functional studies; traditional breeding based on tolerance information; generation of transgenic lines expected to have increased tolerance |
| Climate change | Changing climate resulted in expanded range of GWSS; severity of Pierce’s disease is negatively associated with severity of winter [120,121]; warming climate is expected to expand distribution of pathogen and insect vector | C. Las is very heat tolerant relative to other pathogenic Liberibacter species [124]; rising temperature may expand the range of C. Las and D. citri, and could allow C. Las to outcompete other Liberibacters associated with HLB disease | Monitor geographical range of D. citri and apply predictive models to anticipate spread of the vector and pathogen over time; adapt existing mathematical models to predict effects of extreme weather events in a strategy analogous to that used for human epidemiology [176] |
| Non-biological factors | Regulation and certification of nursery stock and bulk grape material can prevent spread the GWSS and/or X. fastidiosa; changes in trade agreements among nations may force countries to look to new markets for these products, bringing with them different strains and isolates [177] | Movement of plant material can contribute to spread of D. citri and/or C. Las; disease spread has been observed to follow truck routes in California [178]; changes in trade agreements among nations may force countries to look to new markets for these products, bringing with them different strains and isolates [177] | Continued quarantine and regulation of citrus material to prevent spread of D. citri and C. Las; regulation of routes of transport; engagement with legislators to reduce inadvertent disease spread via new trade relationships; development of an international framework for enhanced collaboration among afflicted countries, including sharing information about pathogen detection and disease management strategies |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruse, A.; Fleites, L.A.; Heck, M. Lessons from One Fastidious Bacterium to Another: What Can We Learn about Liberibacter Species from Xylella fastidiosa. Insects 2019, 10, 300. https://doi.org/10.3390/insects10090300
Kruse A, Fleites LA, Heck M. Lessons from One Fastidious Bacterium to Another: What Can We Learn about Liberibacter Species from Xylella fastidiosa. Insects. 2019; 10(9):300. https://doi.org/10.3390/insects10090300
Chicago/Turabian StyleKruse, Angela, Laura A. Fleites, and Michelle Heck. 2019. "Lessons from One Fastidious Bacterium to Another: What Can We Learn about Liberibacter Species from Xylella fastidiosa" Insects 10, no. 9: 300. https://doi.org/10.3390/insects10090300
APA StyleKruse, A., Fleites, L. A., & Heck, M. (2019). Lessons from One Fastidious Bacterium to Another: What Can We Learn about Liberibacter Species from Xylella fastidiosa. Insects, 10(9), 300. https://doi.org/10.3390/insects10090300