A Transcriptomics Approach Reveals Putative Interaction of Candidatus Liberibacter Solanacearum with the Endoplasmic Reticulum of Its Psyllid Vector
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of Psyllid Colonies and RNA Extraction
2.2. Library Construction and Illumina Sequencing and de novo Assembly of the Reference
2.3. Denovo Transcriptome Assembly
2.4. Mapping of Reads and Sample Specific Gene Quantification
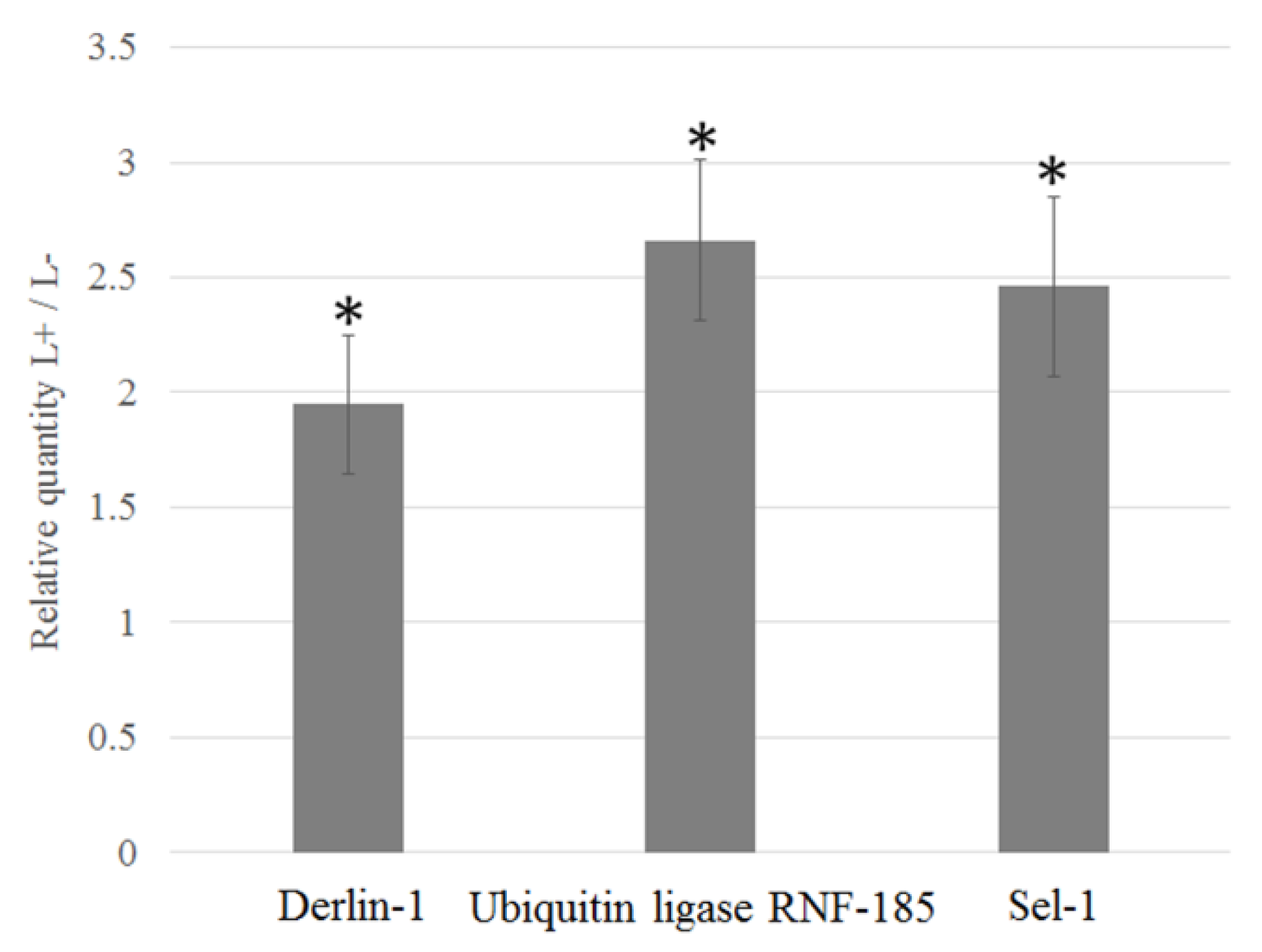
2.5. Relative Quantification of Selected DEGs by qRT-PCR
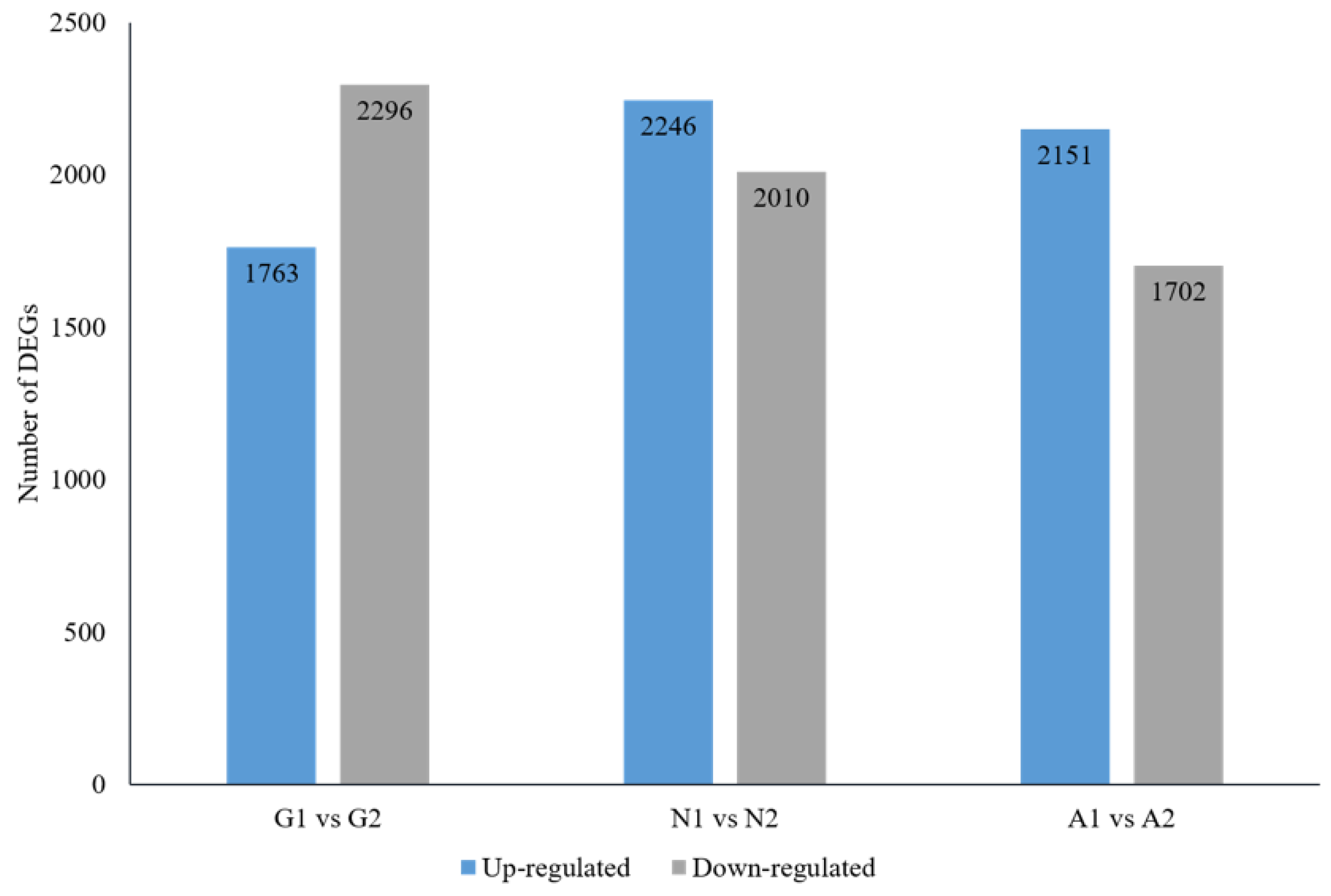
3. Results
3.1. Sequencing of the Reference Transcriptome, De Novo Assembly, and Functional Annotation of Contigs
3.2. RNAseq of Samples
3.3. Screening Differentially Expressed Genes (DEGs) and Annotation
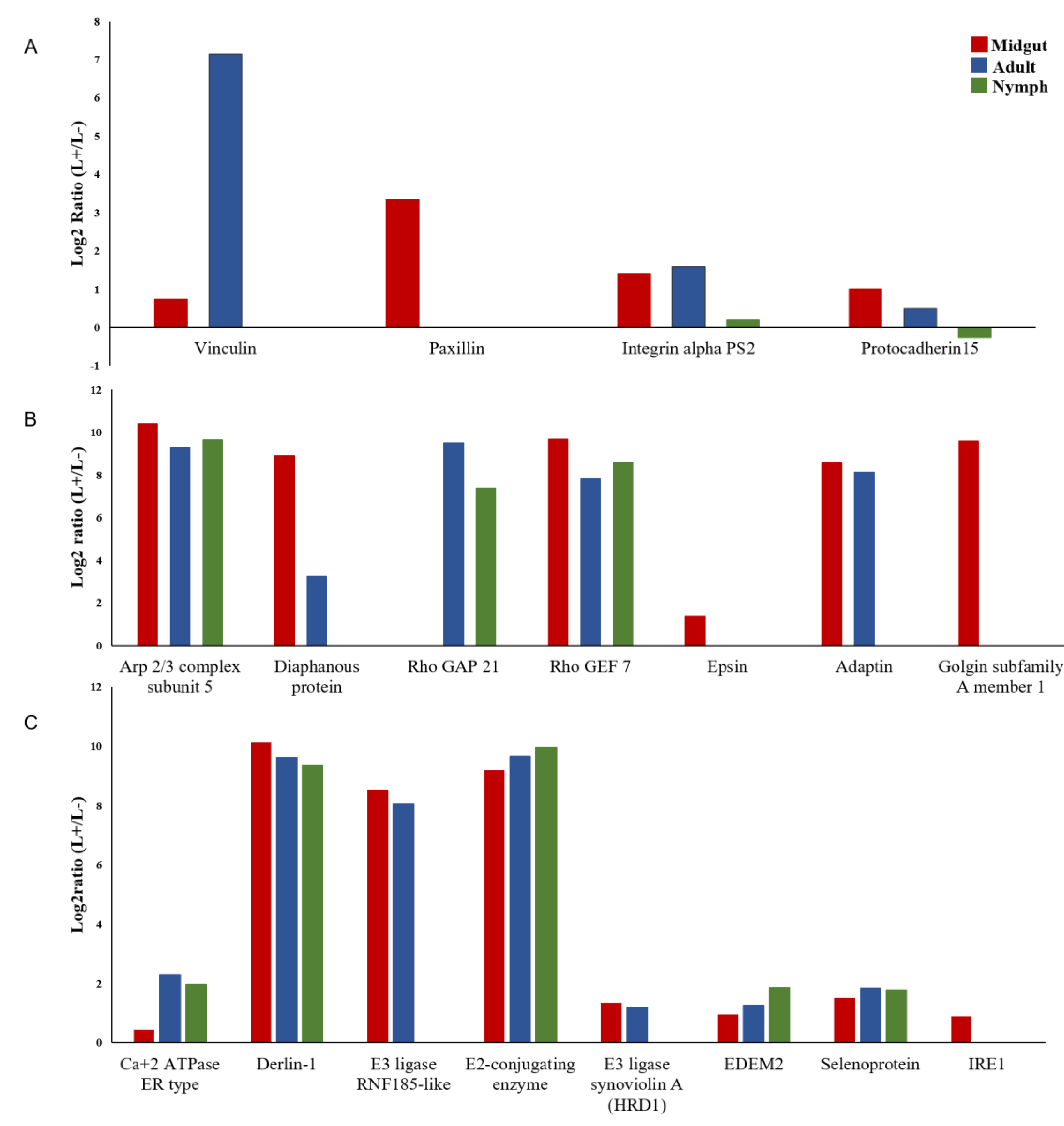
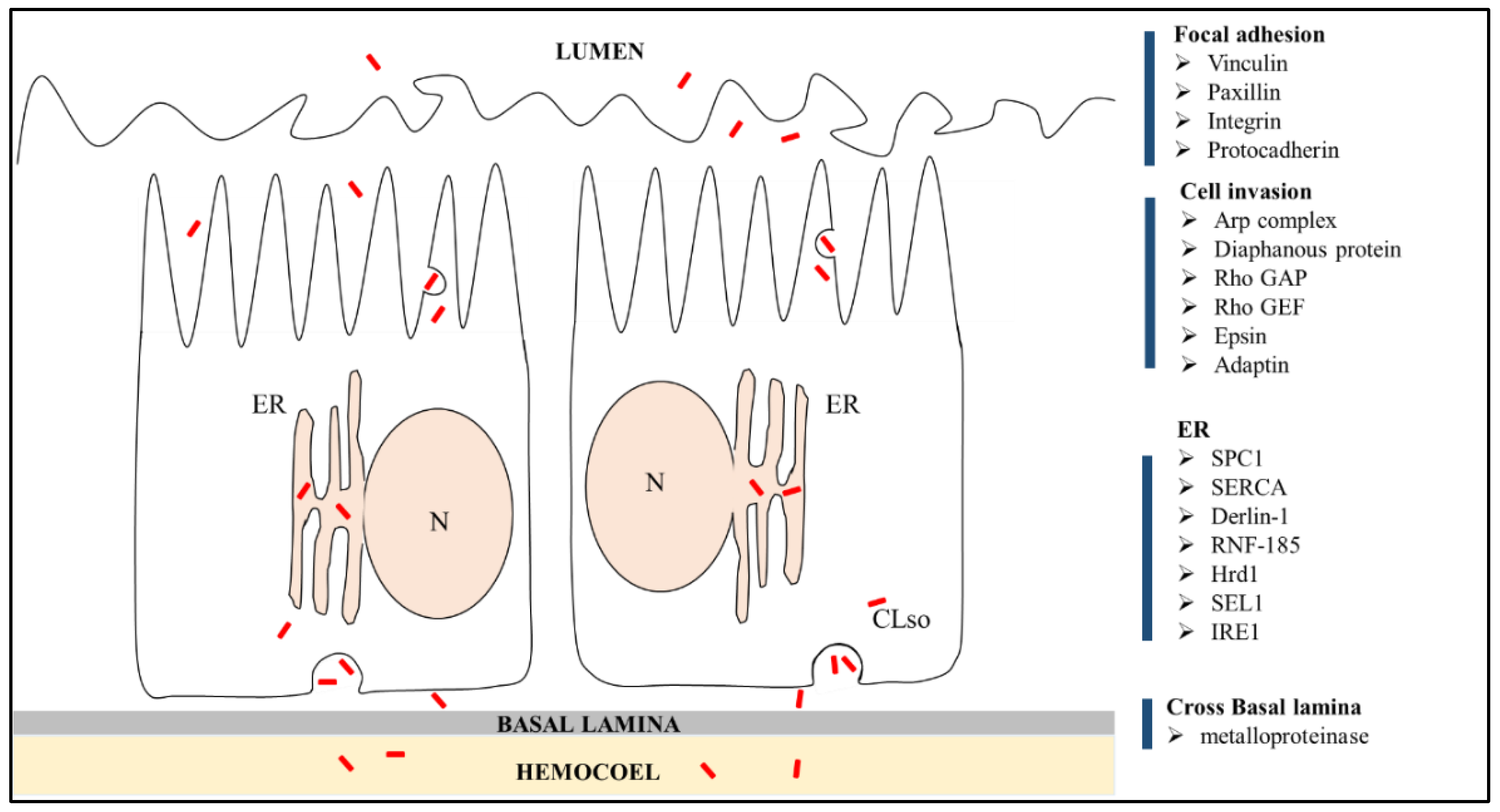
3.4. DEGs of B. Trigonica Putatively Associated with CLso Establishment and Pathogenicity
3.5. Upregulation of D. Citri Genes Related to ER Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Soliman, T.; Mourits, M.C.M.; Van Der Werf, W.; Lansink, A.G.J.M.O.; Werf, W. Economic justification for quarantine status—The case study of ‘Candidatus Liberibacter solanacearum’ in the European Union. Plant Pathol. 2013, 62, 1106–1113. [Google Scholar] [CrossRef]
- Capoor, S.P.; Rao, D.G.; Viswanath, S.M. Diaphorina citri Kuway, a vector of the greening disease of citrus in India. Indian J. Agric. Sci. 1967, 37, 572–576. [Google Scholar]
- Hansen, A.K.; Trumble, J.T.; Stouthamer, R.; Paine, T.D. A New Huanglongbing Species, “Candidatus Liberibacter psyllaurous,” Found to Infect Tomato and Potato, Is Vectored by the Psyllid Bactericera cockerelli (Sulc). Appl. Environ. Microbiol. 2008, 74, 5862–5865. [Google Scholar] [CrossRef] [PubMed]
- Munyaneza, J.E.; Sengoda, V.G.; Stegmark, R.; Arvidsson, A.K.; Anderbrant, O.; Yuvaraj, J.K.; Rämert, B.; Nissinen, A. First Report of “Candidatus Liberibacter solanacearum” Associated with Psyllid-Affected Carrots in Sweden. Plant Dis. 2012, 96, 453. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.R.; Sengoda, V.G.; Alfaro-Fernandez, A.O.; Font, M.I.; Crosslin, J.M.; Munyaneza, J.E. A new haplotype of “Candidatus Liberibacter solanacearum” identified in the Mediterranean region. Eur. J. Plant Pathol. 2013, 135, 633–639. [Google Scholar] [CrossRef]
- Teresani, G.R.; Bertolini, E.; Alfaro-Fernández, A.; Martínez, C.; Tanaka, F.A.O.; Kitajima, E.; Roselló, M.; Sanjuan, S.; Ferrandiz, J.C.; Lopez, M.M.; et al. Association of ’Candidatus Liberibacter solanacearum’ with a vegetative disorder of celery in Spain and development of a real-time PCR method for its detection. Phytopathology 2014, 104, 804–811. [Google Scholar] [CrossRef]
- Tahzima, R.; Maes, M.; Achbani, E.H.; Swisher, K.D.; Munyaneza, J.E.; De Jonghe, K. First Report of ‘Candidatus Liberibacter solanacearum’ on Carrot in Africa. Plant Dis. 2014, 98, 1426. [Google Scholar] [CrossRef]
- Holeva, M.C.; Glynos, P.E.; Karafla, C.D. First report of ‘Candidatus Liberibacter solanacearum’ on carrot in Greece. Plant Dis. 2017, 101, 1819. [Google Scholar] [CrossRef]
- Mawassi, M.; Dror, O.; Bar-Joseph, M.; Piasezky, A.; Sjölund, J.M.; Levitzky, N.; Shoshana, N.; Meslenin, L.; Haviv, S.; Porat, C.; et al. ‘Candidatus Liberibacter solanacearum’ Is Tightly Associated with Carrot Yellows Symptoms in Israel and Transmitted by the Prevalent Psyllid Vector Bactericera trigonica. Phytopathology 2018, 108, 1056–1066. [Google Scholar] [CrossRef]
- Qureshi, J.A.; Kostyk, B.C.; Stansly, P.A. Insecticidal Suppression of Asian Citrus Psyllid Diaphorina citri (Hemiptera: Liviidae) Vector of Huanglongbing Pathogens. PLoS ONE 2014, 9, e112331. [Google Scholar] [CrossRef] [PubMed]
- Page-Weir, N.E.M.; Jamieson, L.E.; Chhagan, A.; Connolly, P.G.; Curtis, C. Efficacy of insecticides against the tomato/potato psyllid (Bactericera cockerelli). New Zeal. Plant Prot. 2011, 64, 276–281. [Google Scholar]
- Tansey, J.A.; Vanaclocha, P.; Monzo, C.; Jones, M.; Stansly, P.A. Costs and benefits of insecticide and foliar nutrient applications to huanglongbing-infected citrus trees. Pest Manag. Sci. 2017, 73, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Guenthner, J.; Goolsby, J.; Greenway, G. Use and Cost of Insecticides to Control Potato Psyllids and Zebra Chip on Potatoes. Southwest. Entomol. 2012, 37, 263–270. [Google Scholar] [CrossRef]
- Tiwari, S.; Mann, R.S.; Rogers, E.M.; Stelinski, L.L. Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 2011, 67, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Ammar, E.D.; Ramos, J.E.; Hall, D.G.; Dawson, W.O.; Shatters, R.G., Jr. Acquisition, replication and inoculation of Candidatus Liberibacter asiaticus following various acquisition periods on huanglongbing-infected citrus by nymphs and adults of the Asian citrus psyllid. PLoS ONE 2016, 11, e0159594. [Google Scholar] [CrossRef] [PubMed]
- Cicero, J.M.; Fisher, T.W.; Qureshi, J.A.; Stansly, P.A.; Brown, J.K. Colonization and intrusive invasion of potato psyllid by ‘Candidatus Liberibacter solanacearum. Phytopathology 2016, 107, 36–49. [Google Scholar] [CrossRef]
- Ghanim, M.; Fattah-Hosseini, S.; Levy, A.; Cilia, M. Morphological abnormalities and cell death in the Asian citrus psyllid (Diaphorina citri) midgut associated with Candidatus Liberibacter asiaticus. Sci. Rep. 2016, 6, 33418. [Google Scholar] [CrossRef]
- Ghanim, M.; Achor, D.; Ghosh, S.; Kontsedalov, S.; Lebedev, G.; Levy, A. ‘Candidatus Liberibacter asiaticus’ Accumulates inside Endoplasmic Reticulum Associated Vacuoles in the Gut Cells of Diaphorina citri. Sci. Rep. 2017, 7, 16945. [Google Scholar] [CrossRef]
- Perilla-Henao, L.M.; Casteel, C.L. Vector-Borne Bacterial Plant Pathogens: Interactions with Hemipteran Insects and Plants. Front. Plant Sci. 2016, 7, 1457. [Google Scholar] [CrossRef]
- Roy, C.R.; Salcedo, S.P.; Gorvel, J.P.E. Pathogen–endoplasmic-reticulum interactions: In through the out door. Nat. Rev. Immunol. 2006, 6, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Tsai, B. How Viruses Use the Endoplasmic Reticulum for Entry, Replication, and Assembly. Cold Spring Harb. Perspect. Biol. 2013, 5, a013250. [Google Scholar] [CrossRef] [PubMed]
- Celli, J.; Tsolis, R.M. Bacteria, the endoplasmic reticulum and the unfolded protein response: Friends or foes? Nat. Rev. Microbiol. 2015, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Morito, D.; Nagata, K. Pathogenic Hijacking of ER-Associated Degradation: Is ERAD Flexible? Mol. Cell 2015, 59, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.R. Exploitation of the endoplasmic reticulum by bacterial pathogens. Trends Microbiol. 2002, 10, 418–424. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; López, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 99, 138–148. [Google Scholar]
- Ibáñez, F.; Tamborindeguy, C. Selection of reference genes for expression analysis in the potato psyllid, Bactericera cockerelli. Insect Mol. Biol. 2016, 25, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ogawa, M.; Mimuro, H.; Sasakawa, C. Reinforcement of epithelial cell adhesion to basement membrane by a bacterial pathogen as a new infectious stratagem. Virulence 2010, 1, 52–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fisher, T.W.; Vyas, M.; He, R.; Nelson, W.; Cicero, J.M.; Willer, M.; Kim, R.; Kramer, R.; May, G.A.; Crow, J.A.; et al. Comparison of Potato and Asian Citrus Psyllid Adult and Nymph Transcriptomes Identified Vector Transcripts with Potential Involvement in Circulative, Propagative Liberibacter Transmission. Pathogens 2014, 3, 875–907. [Google Scholar] [CrossRef] [PubMed]
- Vyas, M.; Fisher, T.W.; He, R.; Nelson, W.; Yin, G.; Cicero, J.M.; Willer, M.; Kim, R.; Kramer, R.; May, G.A.; et al. Asian Citrus Psyllid Expression Profiles Suggest Candidatus Liberibacter Asiaticus-Mediated Alteration of Adult Nutrition and Metabolism, and of Nymphal Development and Immunity. PLoS ONE 2015, 10, e0130328. [Google Scholar] [CrossRef] [PubMed]
- Means, J.C.; Passarelli, A.L. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl. Acad. Sci. USA 2010, 107, 9825–9830. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Balaraman, V.; Kantor, A.M.; Lin, J.; Grant, D.G.; Held, N.L.; Franz, A.W.E. Chikungunya virus dissemination from the midgut of Aedes aegypti is associated with temporal basal lamina degradation during bloodmeal digestion. PLoS Negl. Trop. Dis. 2017, 11, e0005976. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Matsuda, M.; Watashi, K.; Aizaki, H.; Matsuura, Y.; Wakita, T.; Suzuki, T. Signal Peptidase Complex Subunit 1 Participates in the Assembly of Hepatitis C Virus through an Interaction with E2 and NS. PLoS Pathog. 2013, 9, e1003589. [Google Scholar]
- Ashby, M.C.; Tepikin, A.V. ER calcium and the functions of intracellular organelles. Semin. Cell Dev. Biol. 2001, 12, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Benali-Furet, N.L.; Chami, M.; Houel, L.; De Giorgi, F.; Vernejoul, F.; Lagorce, D.; Buscail, L.; Bartenschlager, R.; Ichas, F.; Rizzuto, R.; et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 2005, 24, 4921–4933. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Castillo, K.; Armisén, R.; Stutzin, A.; Soto, C.; Hetz, C. Prion Protein Misfolding Affects Calcium Homeostasis and Sensitizes Cells to Endoplasmic Reticulum Stress. PLoS ONE 2010, 5, e15658. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and Genomic Analyses Reveal an Essential Coordination between the Unfolded Protein Response and ER-Associated Degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Ye, Y.; Shibata, Y.; Yun, C.; Ron, D.; Rapoport, T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004, 429, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, Z.H.; Xu, D.; Lin, G.; Li, D.R.; Wan, T.; Guo, S.L. Expression of Derlin-1 and its effect on expression of autophagy marker genes under endoplasmic reticulum stress in lung cancer cells. Cancer Cell Int. 2014, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Gilbert, J.M.; Ploegh, H.L.; Benjamin, T.L. Murine Polyomavirus Requires the Endoplasmic Reticulum Protein Derlin-2 To Initiate Infection. J. Virol. 2006, 80, 8739–8744. [Google Scholar] [CrossRef] [PubMed]
- Schelhaas, M.; Malmström, J.; Pelkmans, L.; Haugstetter, J.; Ellgaard, L.; Grünewald, K.; Helenius, A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 2007, 131, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M.; Doolman, R.; Dai, M.; Avner, R.; Hassink, G.; Van Voorden, S.; Thanedar, S.; Roitelman, J.; Chau, V.; Wiertz, E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 3525–3534. [Google Scholar] [CrossRef]
- Sun, S.; Shi, G.; Han, X.; Francisco, A.B.; Ji, Y.; Mendonça, N.; Liu, X.; Locasale, J.W.; Simpson, K.W.; Duhamel, G.E.; et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc. Natl. Acad. Sci. USA 2014, 111, E582–E591. [Google Scholar] [CrossRef]
- Mast, S.W.; Diekman, K.; Karaveg, K.; Davis, A.; Sifers, R.N.; Moremen, K.W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 2004, 15, 421–436. [Google Scholar] [CrossRef]
- El Khouri, E.; Le Pavec, G.; Toledano, M.B.; Delaunay-Moisan, A. RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR). J. Biol. Chem. 2013, 288, 31177–31191. [Google Scholar] [CrossRef]
- Byun, H.; Gou, Y.; Zook, A.; Lozano, M.M.; Dudley, J.P. ERAD and how viruses exploit it. Front. Microbiol. 2014, 5, 330. [Google Scholar] [CrossRef]
- Wiertz, E.J.H.J.; Tortorella, D.; Bogyo, M.; Yu, J.; Mothes, W.; Jones, T.R.; Rapoport, T.A.; Ploegh, H.L. Sec6l-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 1996, 384, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Magadán, J.G.; Pérez-Victoria, F.J.; Sougrat, R.; Ye, Y.; Strebel, K.; Bonifacino, J.S. Multilayered Mechanism of CD4 Downregulation by HIV-1 Vpu Involving Distinct ER Retention and ERAD Targeting Steps. PLoS Pathog. 2010, 6, e1000869. [Google Scholar] [CrossRef] [PubMed]
- van den Boomen, D.J.H.; Timms, R.T.; Grice, G.L.; Stagg, H.R.; Skødt, K.; Dougan, G.; Nathan, J.A.; Lehner, P.J. TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc. Natl. Acad. Sci. USA 2014, 111, 11425–11430. [Google Scholar] [CrossRef] [PubMed]
- Dupzyk, A.; Tsai, B. How Polyomaviruses Exploit the ERAD Machinery to Cause Infection. Viruses 2016, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, R.L.; Mackenzie, J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2011, 85, 2723–2732. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Suzuki, R.; Watanabe, N.; Masaki, T.; Tomonaga, M.; Muhammad, A.; Kato, T.; Matsuura, Y.; Watanabe, H.; Wakita, T.; et al. Role of the Endoplasmic Reticulum-associated Degradation (ERAD) Pathway in Degradation of Hepatitis C Virus Envelope Proteins and Production of Virus Particles. J. Biol. Chem. 2011, 286, 37264–37273. [Google Scholar] [CrossRef] [PubMed]
- Pillich, H.; Loose, M.; Zimmer, K.-P.; Chakraborty, T. Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 2012, 14, 949–964. [Google Scholar] [CrossRef]


| Target | Sequence (5’→3’) | Product Size (bp) | Reference | Organism |
|---|---|---|---|---|
| Derlin 1 | F- GGATGGTGTGGCCAGTAA R- CGCACACAGTTCAAAGCA ATG | 174 | This study | B. trigonica |
| F- GGTGGTGGTGTCATGGAACT R- CTCAGCTGGTCTTCTTGGCA | D. citri | |||
| E3 Ligase RNF-185 | F- GCAAATACCACCGAACAGAGA R- GACAGGGCCAACAGAATAAGT | 147 | This study | B. trigonica |
| Sel1 | F- ATTGCTCATCTCTTGGCTTGA R- GCAAACTTCTAACCGAGCCT | 167 | This study | B. trigonica |
| IRE1 | F- CTTCACTAGTGTGGCGGAGG R- CCAGCAGCATGAGAGGTGAA | D. citri | ||
| Elongation factor 1 | F- AACATCGTCGTCATTGGACA R- CGCTTGTCAATACCTCCACA | 88 | [31] | B. trigonica |
| GAPDH | F- GACACTCACTCCTCCATCTTT R- GTATCCGTACTCGTTGTCATACC | D. citri |
| Parameter | Total Number | Total Length (bp) | Mean Length (bp) | N50 | N70 | N90 | GC% |
|---|---|---|---|---|---|---|---|
| Assembled transcripts | 152,247 | 77,708,661 | 510 | 716 | 372 | 218 | 40.54 |
| Assembled contigs | 57,736 | 43,076,597 | 746 | 1251 | 634 | 290 | 41.08 |
| Predicted CDS-BLAST | 23,501 | 17,708,597 | 753 | 1131 | 696 | 327 | 46.68% |
| Predicted CDS-ESTScan | 3143 | 1,063,341 | 338 | 339 | 264 | 216 | 41.61% |
| NR annotated | 23,567 (40.82%) | ||||||
| NT annotated | 14,804 (25.64%) | ||||||
| SwissProt annotated | 16,071 (27.84%) | ||||||
| KEGG annotated | 16,333 (28.29%) | ||||||
| COG annotated | 7034 (12.18%) | ||||||
| InterPro annotated | 12,869 (22.29%) | ||||||
| GO ontology | 2093 (3.63%) |
| Sample | Raw Data Size (bp) | Raw Read Number | Clean Data Size (bp) | Clean Reads Number | Clean Data (%) | Q30 (%) | Total Mapped Reads to Reference (%) |
|---|---|---|---|---|---|---|---|
| G1 | 639,180,108 | 13,044,492 | 632,119,355 | 12,900,395 | 98.89 | 96.9 | 75.96 |
| G2 | 639,167,172 | 13,044,228 | 633,795,057 | 12,934,593 | 99.15 | 97.0 | 78.76 |
| N1 | 639,165,261 | 13,044,189 | 633,472,294 | 12,928,006 | 99.10 | 96.8 | 71.32 |
| N2 | 639,191,770 | 13,044,730 | 633,688,629 | 12,932,421 | 99.13 | 97.0 | 74.61 |
| A1 | 639,193,240 | 13,044,760 | 631,462,069 | 12,886,981 | 98.79 | 97.1 | 80.12 |
| A2 | 639,161,439 | 13,044,111 | 633,148,796 | 12,921,404 | 99.05 | 97.1 | 79.91 |
| Function | Contig ID | Description | Log2ratio (L+/L-) | ||
|---|---|---|---|---|---|
| Midgut | Adult | Nymph | |||
| Focal adhesion | CL3226.Contig3 | Vinculin | 0.74 * | 7.15 | - |
| CL3426.Contig1 | Paxillin | 3.34 | - | - | |
| Unigene6272 | Integrin alpha PS2 | 1.41 | 1.58 | 0.21 * | |
| CL6794.Contig1 | Protocadherin 15-like | 1.01 | 0.50* | −0.26 * | |
| Cell invasion | CL3523.Contig2 | Arp 2/3 complex subunit 2 | - | 7.57 | - |
| CL1981.Contig3 | Arp 2/3 complex subunit 5 | 10.40 | 9.29 | 9.66 | |
| CL112.Contig | Formin binding protein 4 | 8.89 | 3.23 | - | |
| CL5016.Contig2/3 | Rho GTPase-activating protein 21 | - | 9.50 | 7.39 | |
| Unigene9133 | Actin-binding Rho-activating protein-like | - | 2.02 | - | |
| CL3896.Contig2 | Rho guanine nucleotide exchange factor 7 | 9.68 | 7.80 | 8.58 | |
| CL7101.Contig2 | Epsin | 1.36 | - | - | |
| CL1283.Contig1 | Clathrin adaptor protein/Adaptin | 8.55 | 8.12 | - | |
| Unigene18634 | Filamin | - | 9.96 | - | |
| CL2699.Contig1 | Dynamin | - | 1.32 | - | |
| Unigene33000 | phosphatidylinositol-binding clathrin assembly protein LAP | 0.75 | 0.86 | - | |
| Basal lamina egress | CL5995.Contig1 | Disintegrin and metalloproteinase with thrombospondin motifs 18 | 6.59 | 7.83 | 6.77 |
| Vesicular trafficking | CL121.Contig2 | Golgin subfamily A member 1 | 9.61 | - | - |
| Autophagy | CL3340.Contig3 | Cathepsin B | 2.51 | 1.39 | |
| CL6200.Contig3 | Cathepsin L-like | 0.34 * | - | ||
| Unigene18908 | Microtubule-associated protein 4 | - | 2.80 | ||
| CL179.Contig2 | Syntaxin-17 | 1.65 | 9.37 | ||
| Protein targeting to ER | CL124.Contig2 | Signal peptidase complex subunit 1 | 1.83 | 1.32 | 2.55 |
| ER Ca+2 homeostasis | CL1301.Contig1 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | 0.44 * | 2.3 | 1.98 |
| ERAD | CL3833.Contig2 | Derlin-1 | 10.11 | 9.61 | 9.36 |
| CL7212.Contig6 | E3 ubiquitin-protein ligase RNF185-like | 8.56 | 8.07 | - | |
| CL1088.Contig1 | ubiquitin-conjugating enzyme E2 J1 | 9.18 | 9.64 | 9.96 | |
| CL3991.Contig1 | E3 ubiquitin-protein ligase synoviolin A-like (HRD1) | 1.34 | 1.19* | - | |
| Unigene8036 | ER degradation-enhancing alpha-mannosidase-like protein 2 (EDEM2) | 0.95 * | 1.27* | 1.88 | |
| CL5205.Contig1 | Selenoprotein (Sel1) | 1.52 | 1.85 | 1.78 | |
| UPR | CL1004.Contig2 | Inositol requiring enzyme 1 (IRE1) | 0.90 * | - | - |
| Apoptosis | CL5765.Contig1 | Inhibitor of apoptosis 1-like | −1.56 | −4.47 | −3.17 |
| Chaperone | Unigene15805 | Heat shock protein 70A1 | −2.84 | −2.01 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, S.; Jassar, O.; Kontsedalov, S.; Lebedev, G.; Wang, C.; Turner, D.; Levy, A.; Ghanim, M. A Transcriptomics Approach Reveals Putative Interaction of Candidatus Liberibacter Solanacearum with the Endoplasmic Reticulum of Its Psyllid Vector. Insects 2019, 10, 279. https://doi.org/10.3390/insects10090279
Ghosh S, Jassar O, Kontsedalov S, Lebedev G, Wang C, Turner D, Levy A, Ghanim M. A Transcriptomics Approach Reveals Putative Interaction of Candidatus Liberibacter Solanacearum with the Endoplasmic Reticulum of Its Psyllid Vector. Insects. 2019; 10(9):279. https://doi.org/10.3390/insects10090279
Chicago/Turabian StyleGhosh, Saptarshi, Ola Jassar, Svetlana Kontsedalov, Galina Lebedev, Chunxia Wang, Donielle Turner, Amit Levy, and Murad Ghanim. 2019. "A Transcriptomics Approach Reveals Putative Interaction of Candidatus Liberibacter Solanacearum with the Endoplasmic Reticulum of Its Psyllid Vector" Insects 10, no. 9: 279. https://doi.org/10.3390/insects10090279
APA StyleGhosh, S., Jassar, O., Kontsedalov, S., Lebedev, G., Wang, C., Turner, D., Levy, A., & Ghanim, M. (2019). A Transcriptomics Approach Reveals Putative Interaction of Candidatus Liberibacter Solanacearum with the Endoplasmic Reticulum of Its Psyllid Vector. Insects, 10(9), 279. https://doi.org/10.3390/insects10090279






