Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation
Abstract
1. Introduction
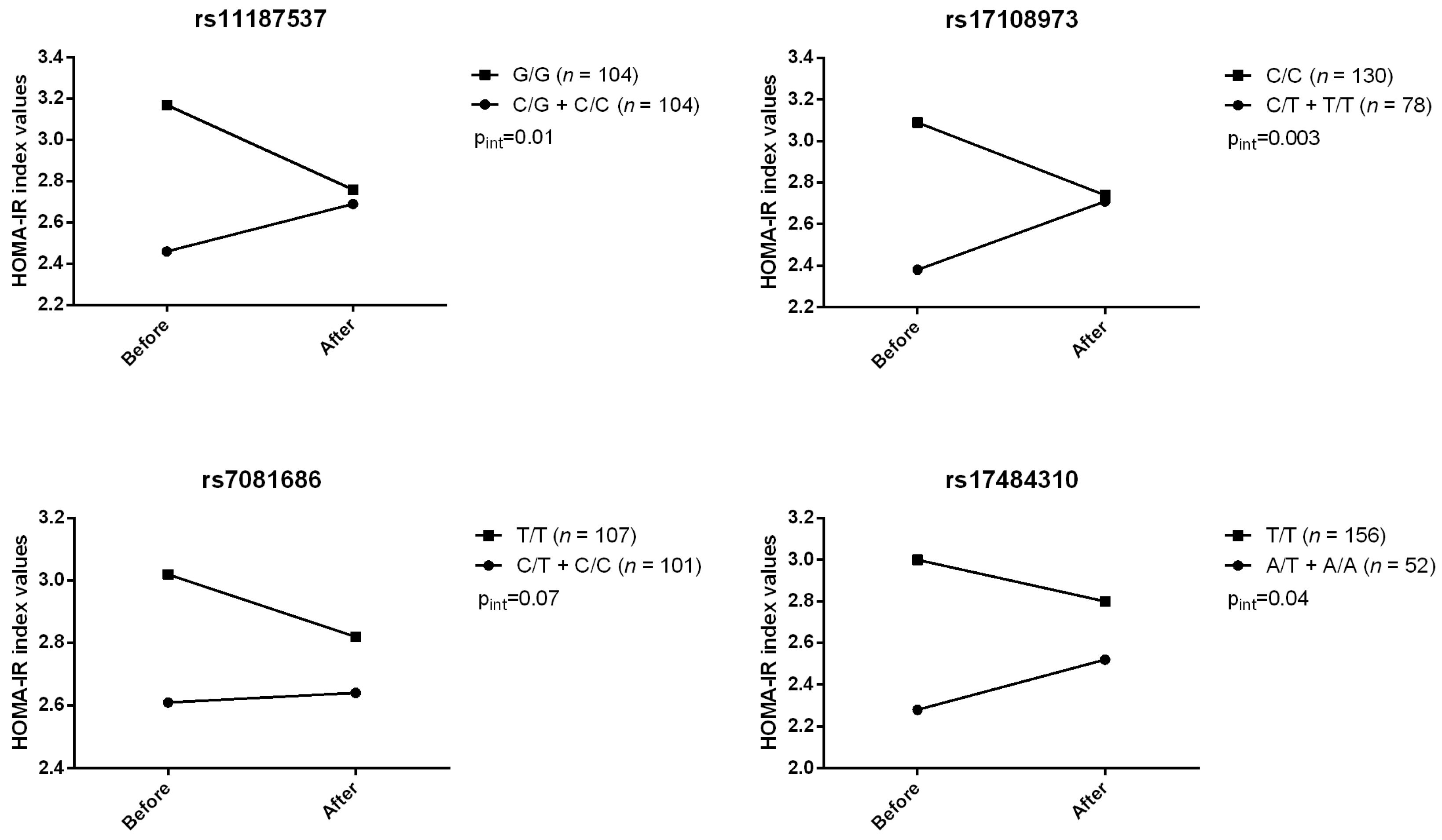
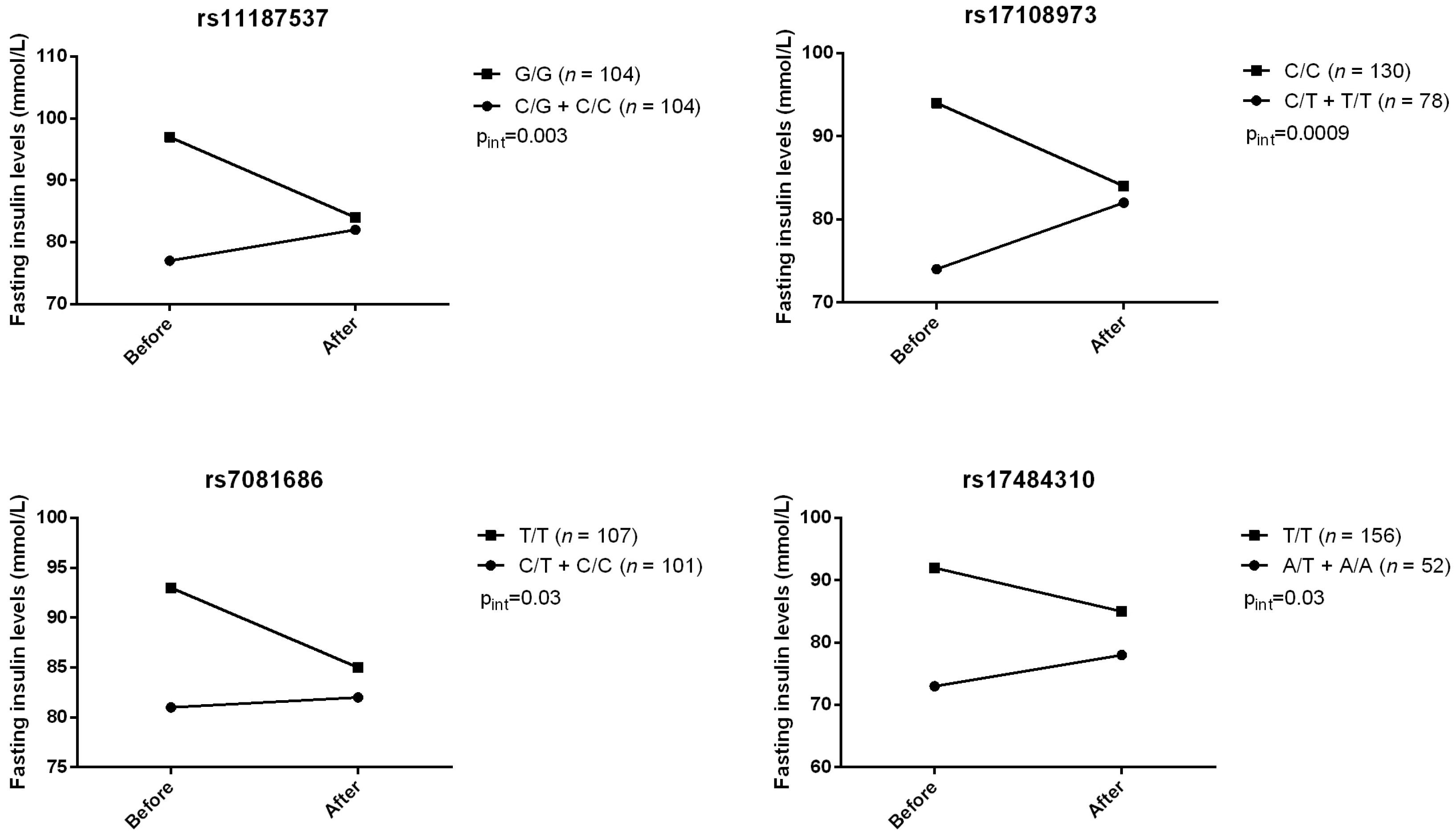
2. Results
3. Discussion
Strengths and Limitations
4. Materials and Methods
4.1. Subjects
4.2. Study Design and Diets
4.3. Anthropometric Measurements
4.4. Biochemical Parameters
4.5. Genotyping of FFAR4 SNPs
4.6. Statistical Analyses
4.7. Transcriptomic and in Silico Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lebovitz, H.E. Insulin resistance: Definition and consequences. Exp. Clin. Endocrinol. Diabetes 2001, 109 (Suppl. 2), S135–S148. [Google Scholar] [CrossRef] [PubMed]
- Henkin, L.; Bergman, R.N.; Bowden, D.W.; Ellsworth, D.L.; Haffner, S.M.; Langefeld, C.D.; Mitchell, B.D.; Norris, J.M.; Rewers, M.; Saad, M.F.; et al. Genetic epidemiology of insulin resistance and visceral adiposity. The iras family study design and methods. Ann. Epidemiol. 2003, 13, 211–217. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States, 2011; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011.
- Oh, D.Y.; Walenta, E. Omega-3 fatty acids and ffar4. Front. Endocrinol. 2014, 5, 115. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.D.; Wang, W.B.; Xu, Z.G.; Liu, C.H.; He, D.F.; Du, L.P.; Li, M.Y.; Yu, X.; Sun, J.P. FFA4 receptor (GPR120): A hot target for the development of anti-diabetic therapies. Eur. J. Pharmacol. 2015, 763, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, A.; Tsumaya, K.; Awaji, T.; Katsuma, S.; Adachi, T.; Yamada, M.; Sugimoto, Y.; Miyazaki, S.; Tsujimoto, G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 2005, 11, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Curto, E.; Milligan, G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem. Pharmacol. 2016, 114, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Mobraten, K.; Haug, T.M.; Kleiveland, C.R.; Lea, T. Omega-3 and omega-6 PUFAs induce the same GPR120-mediated signalling events, but with different kinetics and intensity in Caco-2 cells. Lipids Health Dis. 2013, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Leung, P.S. Potential roles of GPR120 and its agonists in the management of diabetes. Drug Des. Dev. Ther. 2014, 8, 1013–1027. [Google Scholar]
- Ichimura, A.; Hirasawa, A.; Poulain-Godefroy, O.; Bonnefond, A.; Hara, T.; Yengo, L.; Kimura, I.; Leloire, A.; Liu, N.; Iida, K.; et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 2012, 483, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Talukdar, S.; Bae, E.J.; Imamura, T.; Morinaga, H.; Fan, W.; Li, P.; Lu, W.J.; Watkins, S.M.; Olefsky, J.M. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Beuerlein, K.; Eckart, C.; Weiser, H.; Dickkopf, B.; Muller, H.; Sakurai, H.; Kracht, M. Identification and functional characterization of novel phosphorylation sites in TAK1-binding protein (TAB) 1. PLoS ONE 2011, 6, e29256. [Google Scholar] [CrossRef] [PubMed]
- Danforth, E., Jr. Failure of adipocyte differentiation causes type II diabetes mellitus? Nat. Genet. 2000, 26, 13. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yano, T.; Adachi, T.; Koshimizu, T.A.; Hirasawa, A.; Tsujimoto, G. Cloning and characterization of the rat free fatty acid receptor GPR120: In vivo effect of the natural ligand on GLP-1 secretion and proliferation of pancreatic β cells. Naunyn Schmiedebergs Arch. Pharmacol. 2008, 377, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.P.; Liu, P.; Yu, T.; Hansen, D.R.; Gilbertson, T.A. TRPM5 is critical for linoleic acid-induced CCK secretion from the enteroendocrine cell line, STC-1. Am. J. Physiol. Cell Physiol. 2012, 302, C210–C219. [Google Scholar] [CrossRef] [PubMed]
- Waguri, T.; Goda, T.; Kasezawa, N.; Yamakawa-Kobayashi, K. The combined effects of genetic variations in the GPR120 gene and dietary fat intake on obesity risk. Biomed. Res. 2013, 34, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Cormier, H.; Rudkowska, I.; Thifault, E.; Lemieux, S.; Couture, P.; Vohl, M.C. Polymorphisms in fatty acid desaturase (FADS) gene cluster: Effects on glycemic controls following an omega-3 polyunsaturated fatty acids (PUFA) supplementation. Genes 2013, 4, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, E.; Watterson, K.R.; Stocker, C.J.; Sokol, E.; Jenkins, L.; Simon, K.; Grundmann, M.; Petersen, R.K.; Wargent, E.T.; Hudson, B.D.; et al. Activity of dietary fatty acids on FFA1 and FFA4 and characterisation of pinolenic acid as a dual FFA1/FFA4 agonist with potential effect against metabolic diseases. Br. J. Nutr. 2015, 113, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Taneera, J.; Lang, S.; Sharma, A.; Fadista, J.; Zhou, Y.; Ahlqvist, E.; Jonsson, A.; Lyssenko, V.; Vikman, P.; Hansson, O.; et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012, 16, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, T.; Kurata, R.; Yoshida, K.; Murayama, M.A.; Cui, X.; Hasegawa, A. Free fatty acids-sensing G protein-coupled receptors in drug targeting and therapeutics. Curr. Med. Chem. 2013, 20, 3855–3871. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Hatae, N.; Yano, T.; Ruike, Y.; Kimura, M.; Hirasawa, A.; Tsujimoto, G. Free fatty acids inhibit serum deprivation-induced apoptosis through GPR120 in a murine enteroendocrine cell line STC-1. J. Biol. Chem. 2005, 280, 19507–19515. [Google Scholar] [CrossRef] [PubMed]
- Shaul, Y.D.; Seger, R. The MEK/ERK cascade: From signaling specificity to diverse functions. Biochim. Biophys. Acta 2007, 1773, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Seger, R. The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors 2006, 24, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, A.; Lamri, A.; Leloire, A.; Vaillant, E.; Roussel, R.; Levy-Marchal, C.; Weill, J.; Galan, P.; Hercberg, S.; Ragot, S.; et al. Contribution of the low-frequency, loss-of-function p.R270H mutation in FFAR4 (GPR120) to increased fasting plasma glucose levels. J. Med. Genet. 2015, 52, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Marzuillo, P.; Grandone, A.; Perrone, L.; Miraglia Del Giudice, E. Understanding the pathophysiological mechanisms in the pediatric non-alcoholic fatty liver disease: The role of genetics. World J. Hepatol. 2015, 7, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Im, D.S. Functions of omega-3 fatty acids and FFA4 (GPR120) in macrophages. Eur. J. Pharmacol. 2016, 785, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Ussar, S.; Tschop, M.H. Breaking fat. Cell 2014, 159, 238–240. [Google Scholar] [CrossRef] [PubMed]
- Stone, V.M.; Dhayal, S.; Brocklehurst, K.J.; Lenaghan, C.; Sorhede Winzell, M.; Hammar, M.; Xu, X.; Smith, D.M.; Morgan, N.G. GPR120 (FFAR4) is preferentially expressed in pancreatic delta cells and regulates somatostatin secretion from murine islets of langerhans. Diabetologia 2014, 57, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Paerregaard, S.I.; Agerholm, M.; Serup, A.K.; Ma, T.; Kiens, B.; Madsen, L.; Kristiansen, K.; Jensen, B.A. FFAR4 (GPR120) signaling is not required for anti-inflammatory and insulin-sensitizing effects of omega-3 fatty acids. Mediat. Inflamm. 2016, 2016, 1536047. [Google Scholar] [CrossRef] [PubMed]
- Pinel, A.; Pitois, E.; Rigaudiere, J.P.; Jouve, C.; De Saint-Vincent, S.; Laillet, B.; Montaurier, C.; Huertas, A.; Morio, B.; Capel, F. EPA prevents fat mass expansion and metabolic disturbances in mice fed with a Western diet. J. Lipid Res. 2016, 57, 1382–1397. [Google Scholar] [CrossRef] [PubMed]
- McEwen, B.; Morel-Kopp, M.C.; Tofler, G.; Ward, C. Effect of omega-3 fish oil on cardiovascular risk in diabetes. Diabetes Educ. 2010, 36, 565–584. [Google Scholar] [CrossRef] [PubMed]
- Health Canada. Eating Well with Canada’s Food Guide; Health Canada: Ottawa, ON, Canada, 2007.
- Goulet, J.; Nadeau, G.; Lapointe, A.; Lamarche, B.; Lemieux, S. Validity and reproducibility of an interviewer-administered food frequency questionnaire for healthy french-canadian men and women. Nutr. J. 2004, 3, 13. [Google Scholar] [CrossRef] [PubMed]
- Callaway, C.W.; Chumlea, W.C.; Bouchard, C.; Himes, J.H.; Lohman, T.G.; Martin, A.D.; Mitchell, C.D.; Mueller, W.H.; Roche, A.F.; Seefeldt, V.D. The Airlie (va) Consensus Conference; Human Kinetics Publishers: Champaign, IR, USA, 1988; pp. 39–80. [Google Scholar]
- McNamara, J.R.; Schaefer, E.J. Automated enzymatic standardized lipid analyses for plasma and lipoprotein fractions. Clin. Chim. Acta 1987, 166, 1–8. [Google Scholar] [CrossRef]
- Burstein, M.; Samaille, J. On a rapid determination of the cholesterol bound to the serum α- and β-lipoproteins. Clin. Chim. Acta 1960, 5, 609. (In French) [Google Scholar] [CrossRef]
- Albers, J.J.; Warnick, G.R.; Wiebe, D.; King, P.; Steiner, P.; Smith, L.; Breckenridge, C.; Chow, A.; Kuba, K.; Weidman, S.; et al. Multi-laboratory comparison of three heparin-Mn2+ precipitation procedures for estimating cholesterol in high-density lipoprotein. Clin. Chem. 1978, 24, 853–856. [Google Scholar] [PubMed]
- Pirro, M.; Bergeron, J.; Dagenais, G.R.; Bernard, P.M.; Cantin, B.; Despres, J.P.; Lamarche, B. Age and duration of follow-up as modulators of the risk for ischemic heart disease associated with high plasma C-reactive protein levels in men. Arch. Int. Med. 2001, 161, 2474–2480. [Google Scholar] [CrossRef]
- Desbuquois, B.; Aurbach, G.D. Use of polyethylene glycol to separate free and antibody-bound peptide hormones in radioimmunoassays. J. Clin. Endocrinol. Metab. 1971, 33, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Richterich, R.; Dauwalder, H. Determination of plasma glucose by hexokinase-glucose-6-phosphate dehydrogenase method. Schweiz. Med. Wochenschr. 1971, 101, 615–618. [Google Scholar] [PubMed]
- International HapMap Consortium. The international hapmap project. Nature 2003, 426, 789–796. [Google Scholar]
- Livak, K.J. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 1999, 14, 143–149. [Google Scholar] [CrossRef]
- Wallace, T.M.; Levy, J.C.; Matthews, D.R. Use and abuse of homa modeling. Diabetes Care 2004, 27, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Rudkowska, I.; Paradis, A.M.; Thifault, E.; Julien, P.; Barbier, O.; Couture, P.; Lemieux, S.; Vohl, M.C. Differences in metabolomic and transcriptomic profiles between responders and non-responders to an n-3 polyunsaturated fatty acids (PUFAs) supplementation. Genes Nutr. 2013, 8, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Handsaker, R.E.; Pulit, S.L.; Nizzari, M.M.; O’Donnell, C.J.; de Bakker, P.I. SNAP: A web-based tool for identification and annotation of proxy SBPs using HapMap. Bioinformatics 2008, 24, 2938–2939. [Google Scholar] [CrossRef] [PubMed]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef] [PubMed]
- Manke, T.; Roider, H.G.; Vingron, M. Statistical modeling of transcription factor binding affinities predicts regulatory interactions. PLoS Comput. Biol. 2008, 4, e1000039. [Google Scholar] [CrossRef] [PubMed]
- Roider, H.G.; Kanhere, A.; Manke, T.; Vingron, M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics 2007, 23, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Thomas-Chollier, M.; Hufton, A.; Heinig, M.; O’Keeffe, S.; Masri, N.E.; Roider, H.G.; Manke, T.; Vingron, M. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 2011, 6, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
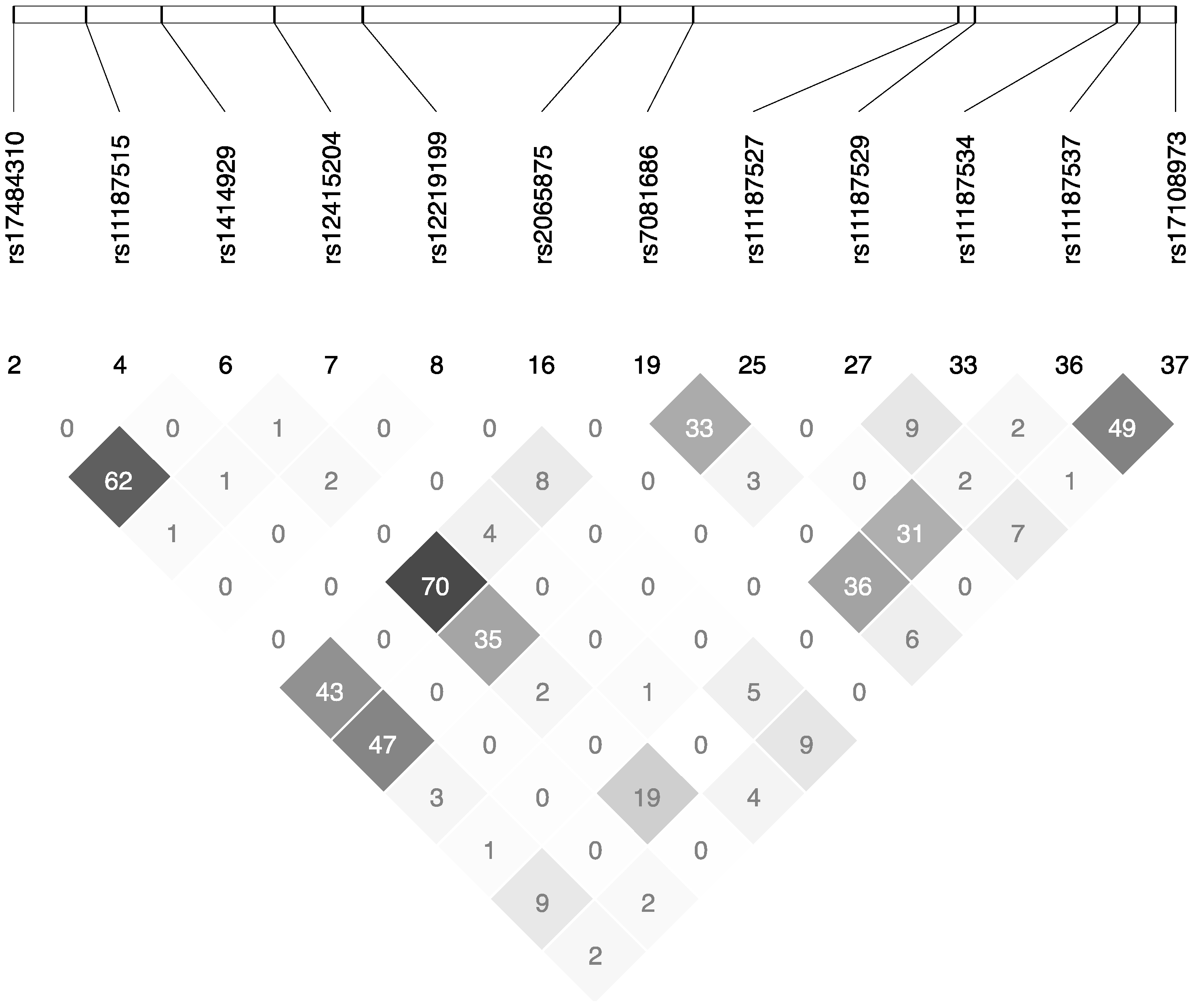
| dbSNP No. 1 | Sequence 2 | Position | Alleles (Major/Minor) | AA | CC | CA | CG | CT | GA | GG | GT | AT | TT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n 3 (%) | |||||||||||||
| rs1414929 | GCA[A/T]GGA | Intron | A/T | 127 (60.5) | 69 (32.9) | 14 (6.7) | |||||||
| rs12219199 | ATG[C/T]GCC | Intron | C/T | 176 (83.8) | 31 (14.8) | 3 (1.4) | |||||||
| rs2065875 | TGC[C/T]CAC | Intron | C/T | 194 (92.4) | 16 (7.6) | ||||||||
| rs11187527 | TTT[G/T]TAA | Intron | G/T | 175 (83.3) | 33 (15.7) | 2 (1.0) | |||||||
| rs11187537 | AGT[C/G]CAG | Intron | C/G | 28 (13.4) | 76 (36.4) | 105 (50.2) | |||||||
| rs17108973 | AAC[C/T]TAT | UTR-3 | C/T | 131 (62.7) | 62 (29.7) | 16 (7.7) | |||||||
| rs7081686 | TGA[C/T]GCT | Intron | C/T | 19 (9.1) | 84 (40.0) | 107 (51.0) | |||||||
| rs17484310 | AAA[A/T]CCG | UTR-5 | A/T | 6 (2.9) | 47 (22.4) | 157 (74.8) | |||||||
| rs11187529 | GAG[C/T]GAT | Intron | C/T | 194 (92.4) | 16 (7.6) | ||||||||
| rs11187515 | GAA[C/T]CAG | Intron | C/T | 165 (78.6) | 40 (19.1) | 5 (2.4) | |||||||
| rs12415204 | GGG[A/C]GAA | Intron | A/C | 14 (6.7) | 130 (61.2) | 66 (31.4) | |||||||
| rs11187534 | CCC[A/G]AGC | Intron | A/G | 2 (1.0) | 42 (20.0) | 166 (79.1) | |||||||
| Characteristics | Pre-Suppl. | Post-Suppl. | p-Value |
|---|---|---|---|
| Age, years | 30.9 ± 8.7 | - | |
| Body mass index, kg/m2 b,c | 27.8 ± 3.7 | 27.9 ± 3.8 | 0.005 |
| Waist circumference, cm c | 93.3 ± 10.5 | 93.4 ± 10.7 | 0.53 |
| Total cholesterol, mmol/L b,d | 4.75 ± 0.9 | 4.72 ± 0.94 | 0.42 |
| HDL-cholesterol, mmol/L b,d | 1.44 ± 0.36 | 1.47 ± 0.40 | 0.004 |
| LDL-cholesterol, mmol/L d | 2.76 ± 0.81 | 2.78 ± 0.85 | 0.47 |
| TG, mmol/L b,d | 1.21 ± 0.63 | 1.02 ± 0.52 | <0.0001 |
| Apolipoprotein B, g/L d | 0.84 ± 0.24 | 0.87 ± 0.24 | 0.003 |
| Fasting insulin, ρmol/mL b,d | 87.2 ± 75.7 | 83.6 ± 40.8 | 0.81 |
| Fasting glucose, mmol/L d | 4.95 ± 0.46 | 5.05 ± 0.49 | 0.0004 |
| CRP, mg/L b,d | 2.59 ± 3.92 | 2.66 ± 4.32 | 0.93 |
| TNF-α, ρg/mL b,d | 1.68 ± 1.42 | 1.68 ± 1.26 | 0.96 |
| IL-6, ρg/mL b,d | 1.38 ± 1.13 | 1.34 ± 0.99 | 0.63 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallée Marcotte, B.; Cormier, H.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.-C. Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation. J. Pers. Med. 2017, 7, 15. https://doi.org/10.3390/jpm7040015
Vallée Marcotte B, Cormier H, Rudkowska I, Lemieux S, Couture P, Vohl M-C. Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation. Journal of Personalized Medicine. 2017; 7(4):15. https://doi.org/10.3390/jpm7040015
Chicago/Turabian StyleVallée Marcotte, Bastien, Hubert Cormier, Iwona Rudkowska, Simone Lemieux, Patrick Couture, and Marie-Claude Vohl. 2017. "Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation" Journal of Personalized Medicine 7, no. 4: 15. https://doi.org/10.3390/jpm7040015
APA StyleVallée Marcotte, B., Cormier, H., Rudkowska, I., Lemieux, S., Couture, P., & Vohl, M.-C. (2017). Polymorphisms in FFAR4 (GPR120) Gene Modulate Insulin Levels and Sensitivity after Fish Oil Supplementation. Journal of Personalized Medicine, 7(4), 15. https://doi.org/10.3390/jpm7040015








