1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is the 6th most common adult cancer worldwide, accounting for 5.3% of all cancers [
1,
2]. Global HNSCC incidence has decreased significantly over the past three decades, primarily attributed to reduced tobacco and alcohol consumption [
1,
2]. Among HNSCC, the incidence of oropharyngeal squamous cell carcinoma (OPSCC) has progressively increased with annual incidence rates rising by 2.7% and 0.5% in males and females in the United States [
3]. The rising incidence of OPSCC can be attributed to human papillomavirus (HPV) infection, which increased in the past few decades [
4]. While HPV-associated OPSCC has become predominant in the United States, it is noteworthy that in many regions outside the US, particularly where anti-tobacco campaigns have been less successful or implemented more recently, tobacco and alcohol-related OPSCC continues to predominate, with significantly lower proportions of HPV-induced cases in countries across Europe, Asia, and South America [
5]. The mechanisms underlying the interactions between HPV and the host environment in the OPSCC development are incompletely elucidated [
4]. The number of studies dedicated to the role of microbial communities in respiratory and gastrointestinal pathophysiology increased over the past decade regarding the development of culture-independent metagenomic techniques [
6]. In OPSCC, research has identified differential microbial signatures between HPV+, HPV− malignant, and healthy tissues, suggesting that dysbiosis-associated alterations influence tumor-promoting inflammation, carcinogenic metabolism, and chemoradiotherapy response patterns [
7].
This systematic review aimed to investigate existing evidence regarding the implications of the microbiome in OPSCC tumorigenesis, progression, clinical phenotypes, and treatment-related endpoints.
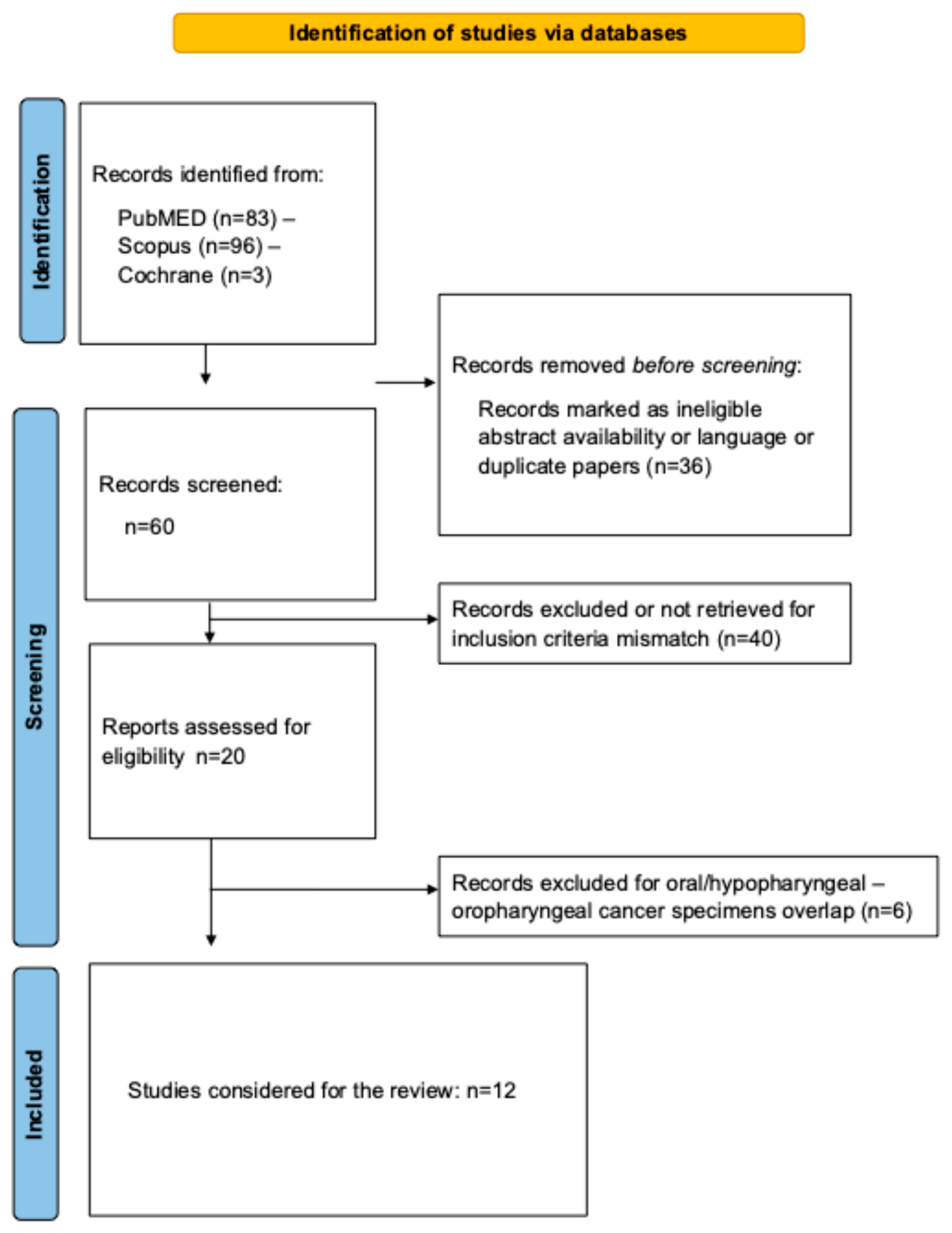
2. Materials and Methods
Two researchers conducted the review using the Preferred Reporting Items for a Systematic Review and Meta-analysis (PRISMA) checklist [
8]. Note that the protocol of review was not registered. Study selection criteria were defined with the PICOTS (Population, Intervention, Comparison, Outcome, Timing, and Setting) framework [
9].
Studies: Studies published in peer-reviewed English-language journals between January 2000 and January 2025 were considered, including prospective and retrospective cohorts, cross-sectional investigations of cancer registries, and controlled trials exploring microbiome findings in OPSCC. Case reports, conference papers, preprints, and experimental animal studies were excluded.
Participants and inclusion criteria: Eligible studies provided OPSCC-specific patient data. Microbiome assessments derived from oropharyngeal or oral fluid or tumor specimens were considered eligible. There was no selection criteria based on the treatments, microbiome characterization methods, or demographic factors.
Outcomes: The primary outcomes consisted of the diversity and composition of the microbiome (taxonomic classification: phylum, class, order, family, genus, and species) and their associations with OPSCC. Secondary outcomes included study features (design, evidence-based level), demographics (mean/median age, sex ratio), oncological findings (grade, cTNM staging, treatments), and methodological approaches for microbiome evaluations (DNA extraction, amplification, quantification, and sequencing).
Intervention and comparison: There was no criterion for intervention. The data related to the type of treatments (surgery, chemo/radiotherapy) were extracted in studies investigating the prognostic value of microbiome findings.
Timing and Setting: There were no criteria for specific timing in the disease process.
2.1. Search Strategy
A University librarian and the author of the paper independently conducted the systematic literature search using PubMed, Scopus, and Cochrane Library databases. The following keywords were used for the literature research: Oropharynx; Oropharyngeal; Cancer; Squamous Cell Carcinoma; Oncological; Microbiome; Microbiota; Bacteria; and Outcomes. The investigators considered research reporting database abstracts, available full-texts, or titles with the search terms. The reference list of some articles, particularly reviews or meta-analyses, were considered for additional valuable studies. The studies were evaluated for the number of subjects, study design, inclusion and exclusion criteria, quality of trial, evidence-based level [
10], demographics, and outcomes. Cohort studies from the same research team were carefully investigated for potential overlaps. Ethics committee approval was not required.
2.2. Bias Analysis
The bias analysis was conducted using the methodological index for non-randomized studies (MINORS), a validated tool for assessing study quality [
11]. MINORS evaluates key methodological aspects on a scale of 0 (absent), 1 (inadequate/partial), or 2 (adequate). Items include study aim clarity, consecutive patient inclusion, prospective data collection, endpoint appropriateness, adequate follow-up period (for predictive studies), and acceptable lost-to-follow-up rates (<5%). For prospective studies, sample size calculation was also evaluated. The optimal MINORS score is 16 for non-comparative studies and 24 for comparative studies [
11].
4. Discussion
Multi-omic analysis and microbiome dynamics characterization are emerging in medicine and surgery due to the accessibility of metagenomic shotgun sequencing and microbiome functional analyses [
30].
The present systematic review identified specific phyla/bacteria that may be significantly associated with the development/progression of OPSCC. Precisely, the transversal analysis of the literature demonstrated a predominance of Spirochaetes and Bacteroidetes, while Proteobacteria were predominant in control tissues compared to tumor specimens. Among the genera, Leptotrichia, Selenomonas, and Treponema showed higher representation in OPSCC compared to control specimens, whereas Neisseria, Porphyromonas, Rothia, Streptococcus, and Veillonella were more abundant in normal versus OPSCC tissue specimens.
The overrepresentation of
Bacteroidetes in carcinoma tissues corroborates the findings found for laryngeal squamous cell carcinoma (LSCC) [
31,
32], with the detection of
Bacteroidetes genera in approximately 15% of LSCC tissues. In oral squamous cell carcinomas, a recent review reported that
Bacteroidetes was predominantly found in 13 of the 27 studies exploring microbiome features in oral squamous cell carcinoma [
33].
Bacteroidetes was similarly involved in the development of gastrointestinal malignancies through multiple mechanistic pathways, including the modulation of WNT/β-catenin signaling, the activation of pro-inflammatory cytokine releases such as IL-8, and the upregulation of MAPK and WNT signaling cascades [
34]. A better understanding of the mechanisms linking the tumor and
Bacteroidetes may lead to the identification of transversal biomarkers across HNSCC. The relative abundance of
Bacteroidetes,
Fusobacteria,
Proteobacteria, and
Actinobacteria was inversely correlated with
Firmicutes in LSCC [
31,
32]. In the present review,
Streptococcus (Firmicutes) was consistently overrepresented in healthy tissues compared to OPSCC specimens, corroborating findings in LSCC and OSCC. Beyond its potential role as a biomarker for HNSCC,
Streptococcus demonstrates prognostic value, as disease-free patients exhibited higher
Streptococcus abundance in HNSCC [
35]. Similarly to other phyla and genera, the mechanistic role of
Streptococcus as protective genera in carcinogenesis remains largely unknown. In the oral cavity, the genus
Streptococcus constitutes approximately 80% of the oral biofilm, where perturbations in oral streptococcal composition can lead to dysbiosis, altering host–pathogen interactions and resulting in oral inflammation [
36]. The mechanistic relationship between decreased
Streptococcus abundance in the upper aerodigestive tract mucosa and the development of chronic inflammation, related DNA damage, and carcinogenesis warrants further investigation. Similar observations can be made for
Rothia genera that was transversally identified as abundant in healthy tissues compared to carcinoma specimens in HNSCC [
31,
33].
Despite the limited available literature, findings from the present review indicate potential distinct microbiome compositional shifts associated with chemoradiation response, HPV status, and tobacco-alcohol intoxications. Oliva et al. observed that the number of species detected in oral samples significantly decreased after chemoradiation [
18], while Bahig et al. reported a potential predictive value of the microbiome on chemoradiation response, with higher baseline oral saliva and tumor tissue α-diversity in complete responders versus partial responders [
23]. The predictive value of microbiome features in chemoradiotherapy response was investigated in studies considering all HNSCC sublocations [
6,
37]. Torozan et al. reported that patients with HNSCC exhibited significantly reduced alpha diversity compared to controls before and after chemoradiotherapy with an increase in the relative abundance of
Staphylococcus aureus and
Escherichia coli during chemoradiotherapy [
37]. In a cohort of 52 HNSCC patients with stool samples, Hes et al. demonstrated that gut microbiome composition had predictive value for oral mucositis development, revealing a significant correlation between severe mucositis and reduced overall survival [
6]. To date, the limited number of studies investigating oral or OPSCC microbiome dynamics during chemoradiotherapy and follow-up precludes definitive conclusions. However, such future investigations are important given the rising incidence of OPSCC and the substantial proportion of patients receiving chemoradiation therapy.
Some conflicting results across studies in the present review may be attributed to heterogeneity in inclusion criteria and confounding factors, which represent the primary limitation of the present review. Given the small number of patients, most studies did not investigate the impact of HPV status, laryngopharyngeal reflux disease, tobacco and alcohol consumption on the microbial composition and diversity, although these factors may influence the microbial composition [
18,
23,
38]. In OSCC, alcohol consumption leads to chronic inflammation, dysbiosis, and an increased acetaldehyde level, leading to a tumor-promoting environment [
28,
39]. In LSCC, tobacco consumption was found to be significantly associated with the global community structure, specifically at lower taxonomic levels [
28].
The potential heterogeneity across the studies in the inclusion criteria is an additional limitation. This limitation particularly concerns antibiotic exposure criteria at enrollment. Some authors did not exclude antibiotic use in the days preceding sampling or during surgery (sample collection time) [
14,
15], while others documented antibiotic consumption in the week before sample collection, which can significantly impact microbiome assessment [
18]. Although most authors used saliva samples for microbiome analyses, some assessed the microbiome using tumor samples [
13,
14,
15,
16,
20], which can limit comparisons across studies. The use of tissue from patients with identified diseases as controls (e.g., obstructive sleep apnea tonsils, tumor-adjacent tissue) represents a limitation due to potential dysbiosis related to the underlying disease. Finally, the full understanding of the role of the microbiome in the development of OSCC may require additional examinations, such as secretome and metaproteomic analyses, which provide the specific activity of species, while identifying the mediators influencing the specific activities. Spatial metagenomic and metaproteomic approaches represent another pathway for improvement, as they can determine the three-dimensional relationships among host tissues (tumor, peritumoral tissue) and microbial species. A summary of the key limitations of the literature and related considerations for future studies are provided in
Table 6.