Patient-Derived Organoid Biobanks for Translational Research and Precision Medicine: Challenges and Future Perspectives
Abstract
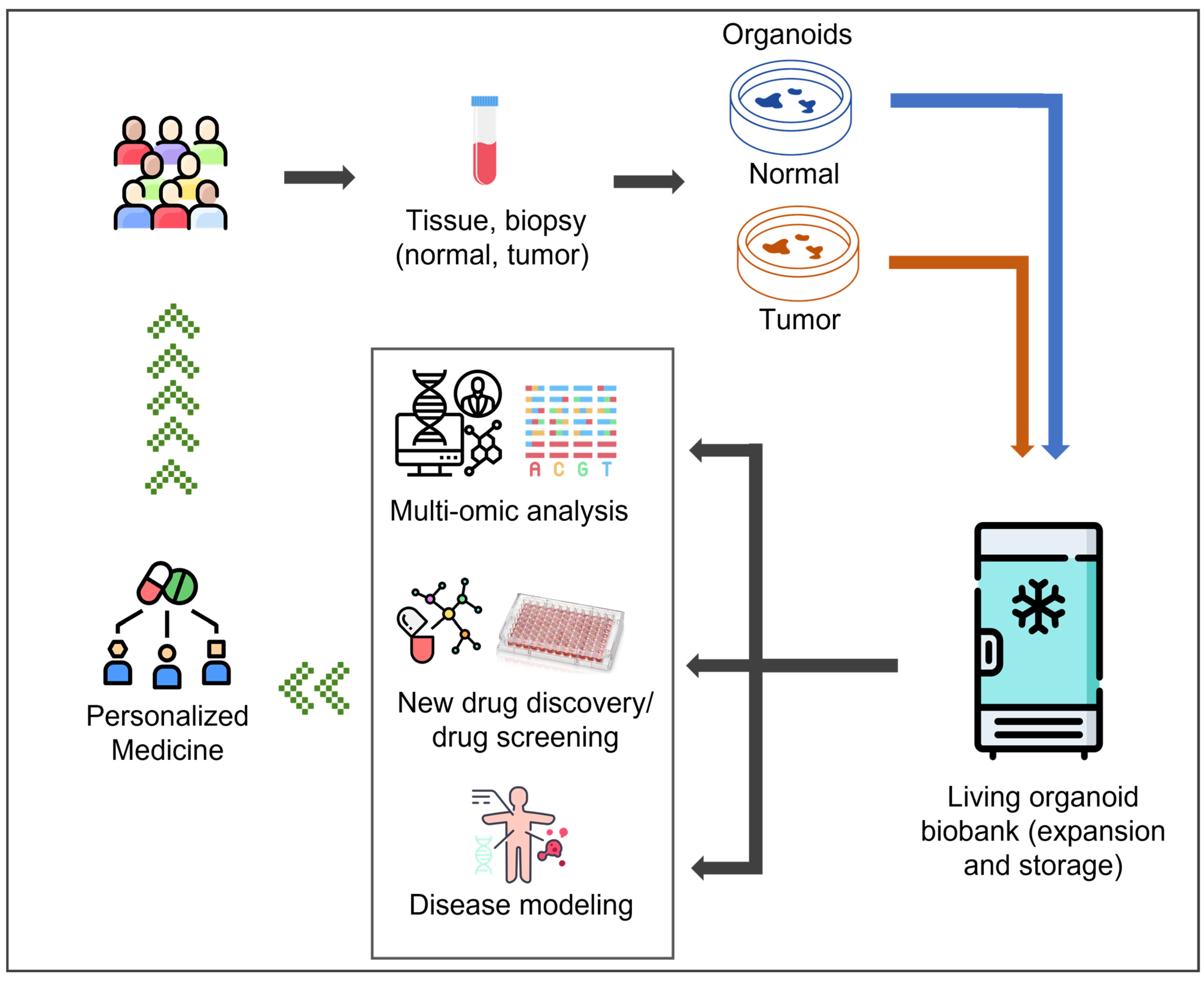
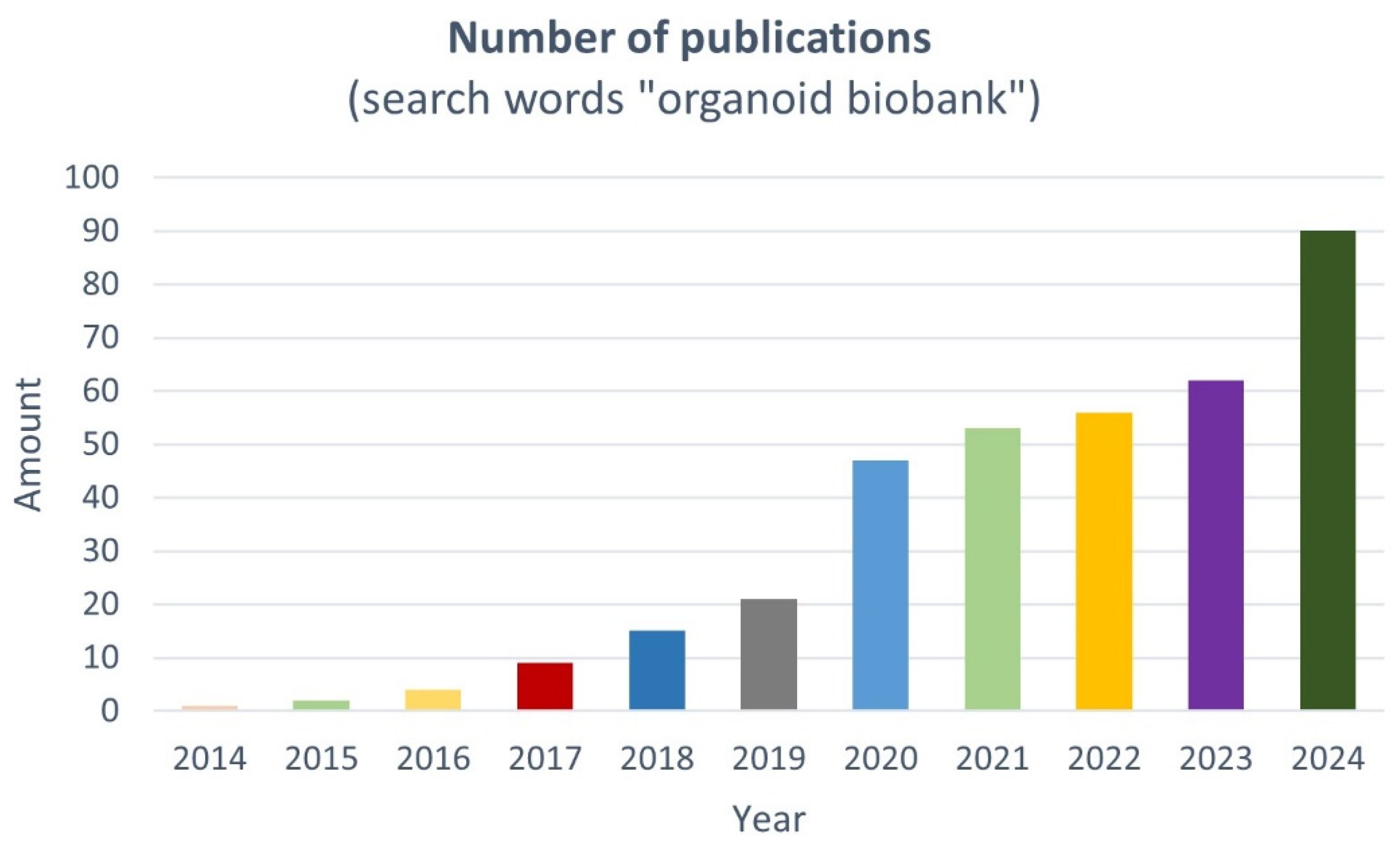
1. Introduction
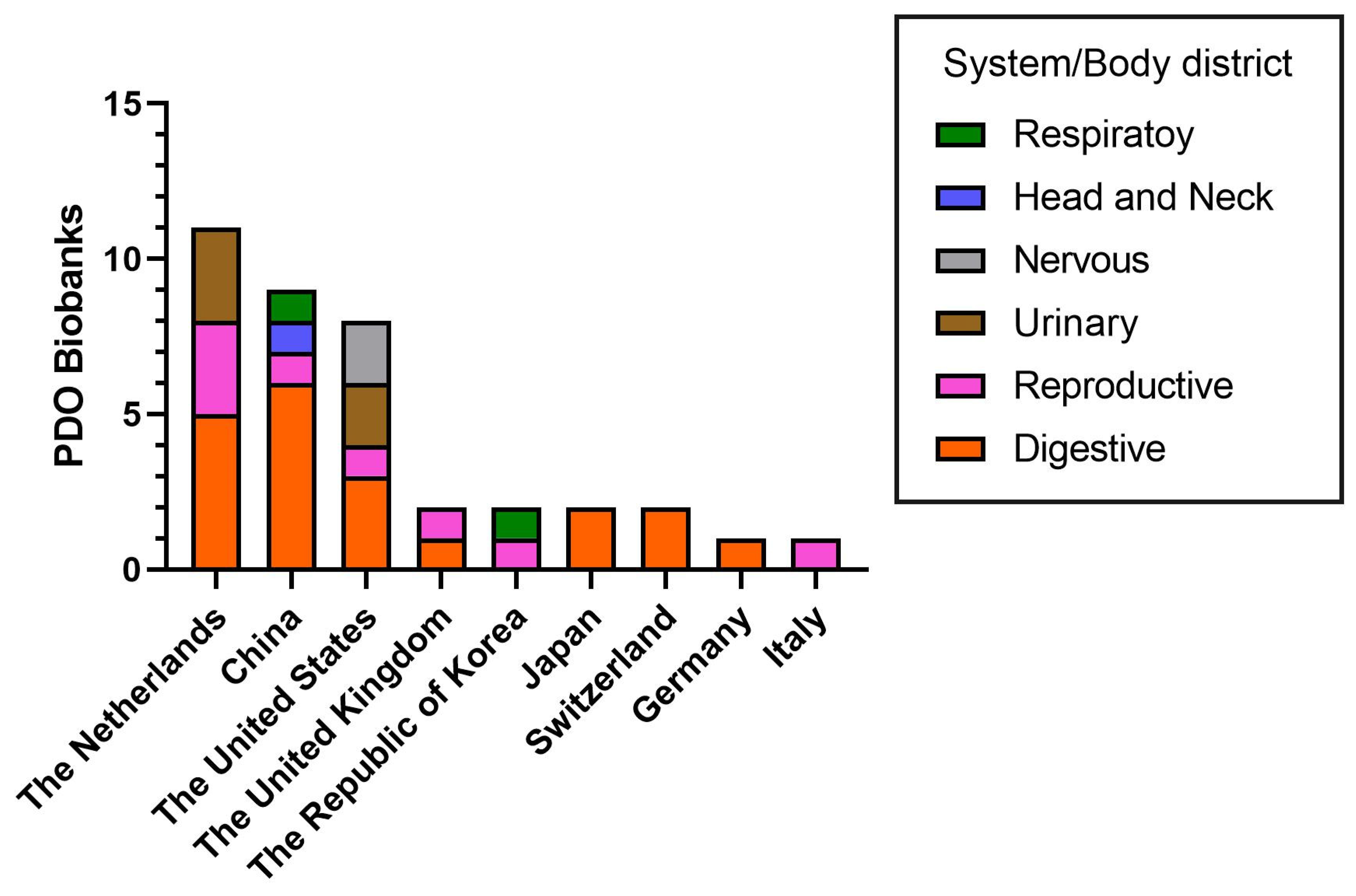
2. Living PDO Biobanks: Classification and Worldwide Distribution
3. Living PDO Biobanks: Translational Research and Personalized Medicine
3.1. Digestive System
3.1.1. Gastrointestinal Tumors
3.1.2. Liver, Pancreatic and Neuroendocrine Tumors
3.2. Reproductive System
3.3. Urinary System
3.4. Nervous System and Head and Neck District
3.5. Respiratory District
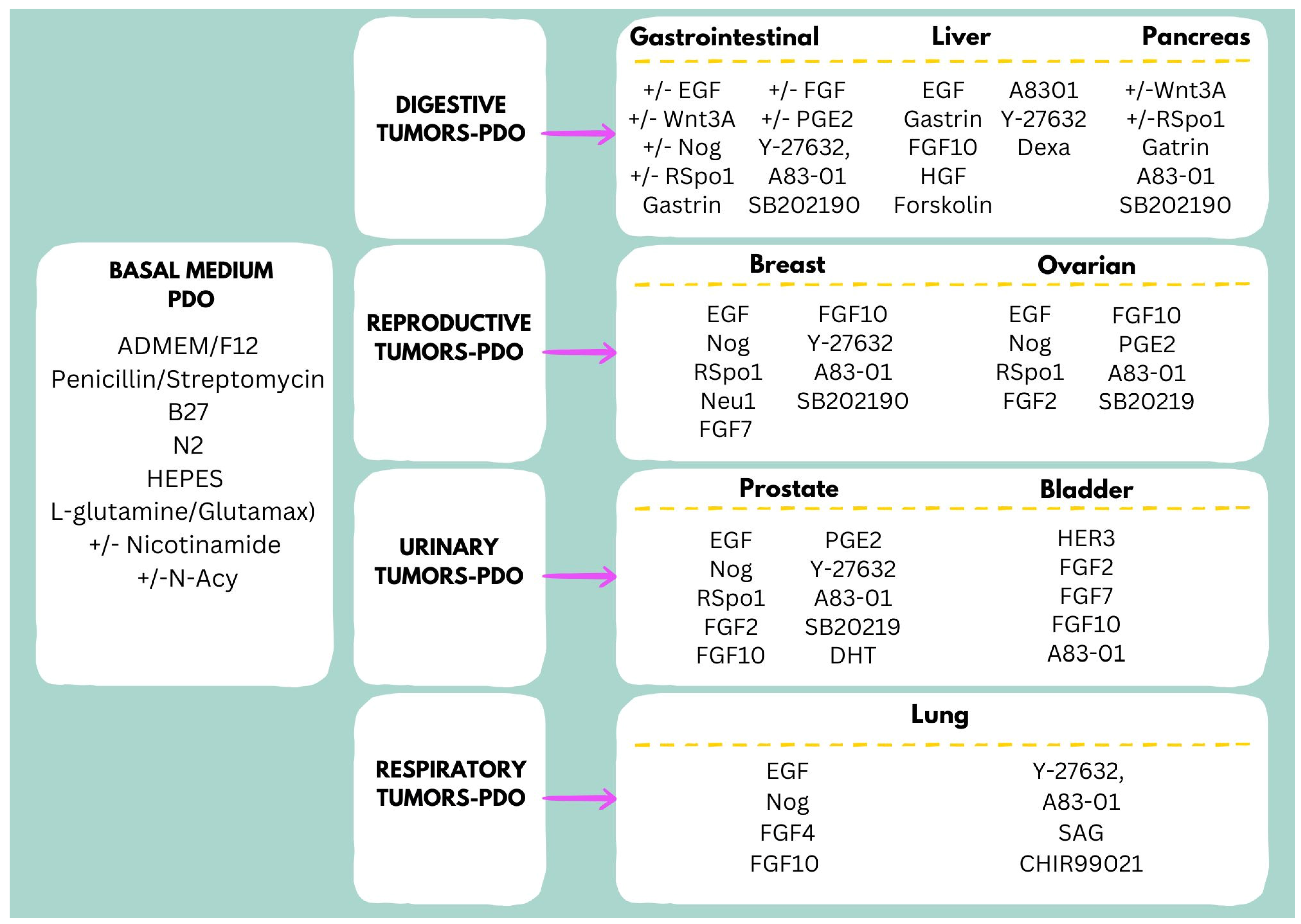
4. Culture Media for Long-Term Expansion of PDOs
5. Challenges and Limitations in the Establishment of PDO Biobanks
5.1. Sample Acquisition and Quality Control of PDO Culture Processes
5.2. Standardization of PDO Culture Processes
5.2.1. Extracellular Matrix (ECM)
5.2.2. Cell Culture Media
5.2.3. Tissue Microenvironment and Vascularization
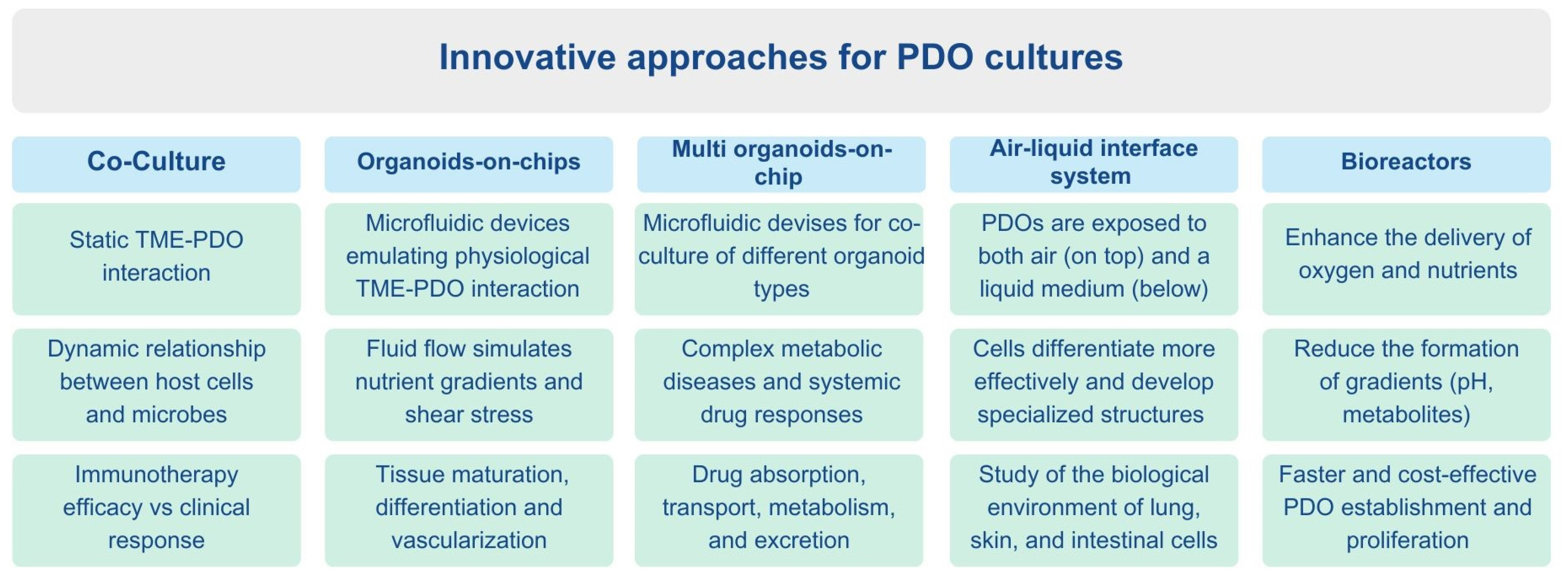
6. Future Perspectives and Solutions
6.1. Co-Culture Systems
6.2. Organoids-on-Chips
6.3. Multi Organoids-on-Chip
6.4. Air–Liquid Interface System
6.5. Bioreactors
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-Term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Loomans, C.J.M.; Williams Giuliani, N.; Balak, J.; Ringnalda, F.; van Gurp, L.; Huch, M.; Boj, S.F.; Sato, T.; Kester, L.; de Sousa Lopes, S.M.C.; et al. Expansion of Adult Human Pancreatic Tissue Yields Organoids Harboring Progenitor Cells with Endocrine Differentiation Potential. Stem Cell Rep. 2018, 10, 712–724. [Google Scholar] [CrossRef]
- Turco, M.Y.; Gardner, L.; Hughes, J.; Cindrova-Davies, T.; Gomez, M.J.; Farrell, L.; Hollinshead, M.; Marsh, S.G.E.; Brosens, J.J.; Critchley, H.O.; et al. Long-Term, Hormone-Responsive Organoid Cultures of Human Endometrium in a Chemically Defined Medium. Nat. Cell Biol. 2017, 19, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.-I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.A.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human Primary Liver Cancer–Derived Organoid Cultures for Disease Modeling and Drug Screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Codrich, M.; Dalla, E.; Mio, C.; Antoniali, G.; Malfatti, M.C.; Marzinotto, S.; Pierobon, M.; Baldelli, E.; Di Loreto, C.; Damante, G.; et al. Integrated Multi-Omics Analyses on Patient-Derived CRC Organoids Highlight Altered Molecular Pathways in Colorectal Cancer Progression Involving PTEN. J. Exp. Clin. Cancer Res. 2021, 40, 198. [Google Scholar] [CrossRef]
- Ha, D.; Kong, J.; Kim, D.; Lee, K.; Lee, J.; Park, M.; Ahn, H.; Oh, Y.; Kim, S. Development of Bioinformatics and Multi-Omics Analyses in Organoids. BMB Rep. 2023, 56, 43–48. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, F.; Jin, Y.; Ma, Y. Applications of Human Organoids in the Personalized Treatment for Digestive Diseases. Signal Transduct. Target. Ther. 2022, 7, 336. [Google Scholar] [CrossRef]
- Tong, L.; Cui, W.; Zhang, B.; Fonseca, P.; Zhao, Q.; Zhang, P.; Xu, B.; Zhang, Q.; Li, Z.; Seashore-Ludlow, B.; et al. Patient-Derived Organoids in Precision Cancer Medicine. Med 2024, 5, 1351–1377. [Google Scholar] [CrossRef]
- Annaratone, L.; De Palma, G.; Bonizzi, G.; Sapino, A.; Botti, G.; Berrino, E.; Mannelli, C.; Arcella, P.; Di Martino, S.; Steffan, A.; et al. Basic Principles of Biobanking: From Biological Samples to Precision Medicine for Patients. Virchows Arch. 2021, 479, 233–246. [Google Scholar] [CrossRef]
- Huang, W.; Xu, Z.; Li, S.; Zhou, J.; Zhao, B. Living Biobanks of Organoids: Valuable Resource for Translational Research. Biopreserv. Biobank. 2024, 22, 543–549. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.N.; Siu, H.C.; Ho, S.L.; Yue, S.S.K.; Gao, Y.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Wong, J.W.H.; Man, A.H.Y.; et al. Organoid Cultures of Early-Onset Colorectal Cancers Reveal Distinct and Rare Genetic Profiles. Gut 2020, 69, 2165–2179. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26.e6. [Google Scholar] [CrossRef] [PubMed]
- Geevimaan, K.; Guo, J.-Y.; Shen, C.-N.; Jiang, J.-K.; Fann, C.S.J.; Hwang, M.-J.; Shui, J.-W.; Lin, H.-T.; Wang, M.-J.; Shih, H.-C.; et al. Patient-Derived Organoid Serves as a Platform for Personalized Chemotherapy in Advanced Colorectal Cancer Patients. Front. Oncol. 2022, 12, 883437. [Google Scholar] [CrossRef]
- Laoukili, J.; Constantinides, A.; Wassenaar, E.C.E.; Elias, S.G.; Raats, D.A.E.; van Schelven, S.J.; van Wettum, J.; Volckmann, R.; Koster, J.; Huitema, A.D.R.; et al. Peritoneal Metastases from Colorectal Cancer Belong to Consensus Molecular Subtype 4 and Are Sensitised to Oxaliplatin by Inhibiting Reducing Capacity. Br. J. Cancer 2022, 126, 1824–1833. [Google Scholar] [CrossRef]
- Mo, S.; Tang, P.; Luo, W.; Zhang, L.; Li, Y.; Hu, X.; Ma, X.; Chen, Y.; Bao, Y.; He, X.; et al. Patient-Derived Organoids from Colorectal Cancer with Paired Liver Metastasis Reveal Tumor Heterogeneity and Predict Response to Chemotherapy. Adv. Sci. 2022, 9, e2204097. [Google Scholar] [CrossRef]
- Yao, L.; Zao, X.-L.; Pan, X.-F.; Zhang, H.-G.; Wang, F.-J.; Qiao, P.-F. Application of Tumoroids Derived from Advanced Colorectal Cancer Patients to Predict Individual Response to Chemotherapy. J. Chemother. 2023, 35, 104–116. [Google Scholar] [CrossRef]
- Herpers, B.; Eppink, B.; James, M.I.; Cortina, C.; Cañellas-Socias, A.; Boj, S.F.; Hernando-Momblona, X.; Glodzik, D.; Roovers, R.C.; van de Wetering, M.; et al. Functional Patient-Derived Organoid Screenings Identify MCLA-158 as a Therapeutic EGFR × LGR5 Bispecific Antibody with Efficacy in Epithelial Tumors. Nat. Cancer 2022, 3, 418–436. [Google Scholar] [CrossRef]
- Schütte, M.; Risch, T.; Abdavi-Azar, N.; Boehnke, K.; Schumacher, D.; Keil, M.; Yildiriman, R.; Jandrasits, C.; Borodina, T.; Amstislavskiy, V.; et al. Molecular Dissection of Colorectal Cancer in Pre-Clinical Models Identifies Biomarkers Predicting Sensitivity to EGFR Inhibitors. Nat. Commun. 2017, 8, 14262. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-Derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef] [PubMed]
- Nuciforo, S.; Fofana, I.; Matter, M.S.; Blumer, T.; Calabrese, D.; Boldanova, T.; Piscuoglio, S.; Wieland, S.; Ringnalda, F.; Schwank, G.; et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018, 24, 1363–1376. [Google Scholar] [CrossRef]
- Beato, F.; Reverón, D.; Dezsi, K.B.; Ortiz, A.; Johnson, J.O.; Chen, D.-T.; Ali, K.; Yoder, S.J.; Jeong, D.; Malafa, M.; et al. Establishing a Living Biobank of Patient-Derived Organoids of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Lab. Invest. 2021, 101, 204–217. [Google Scholar] [CrossRef]
- Hirt, C.K.; Booij, T.H.; Grob, L.; Simmler, P.; Toussaint, N.C.; Keller, D.; Taube, D.; Ludwig, V.; Goryachkin, A.; Pauli, C.; et al. Drug Screening and Genome Editing in Human Pancreatic Cancer Organoids Identifies Drug-Gene Interactions and Candidates for off-Label Treatment. Cell Genom. 2022, 2, 100095. [Google Scholar] [CrossRef]
- Demyan, L.; Habowski, A.N.; Plenker, D.; King, D.A.; Standring, O.J.; Tsang, C.; St Surin, L.; Rishi, A.; Crawford, J.M.; Boyd, J.; et al. Pancreatic Cancer Patient-Derived Organoids Can Predict Response to Neoadjuvant Chemotherapy. Ann. Surg. 2022, 276, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Trujillo, M.A.; Fujikura, K.; Qiu, M.; Chen, F.; Felsenstein, M.; Zhou, C.; Skaro, M.; Gauthier, C.; Macgregor-Das, A.; et al. Molecular Characterization of Organoids Derived from Pancreatic Intraductal Papillary Mucinous Neoplasms. J. Pathol. 2020, 252, 252–262. [Google Scholar] [CrossRef]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic Cancer Organoids Recapitulate Disease and Allow Personalized Drug Screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef]
- Vaes, R.D.W.; van Dijk, D.P.J.; Welbers, T.T.J.; Blok, M.J.; Aberle, M.R.; Heij, L.; Boj, S.F.; Olde Damink, S.W.M.; Rensen, S.S. Generation and Initial Characterization of Novel Tumour Organoid Models to Study Human Pancreatic Cancer-Induced Cachexia. J. Cachexia. Sarcopenia Muscle 2020, 11, 1509–1524. [Google Scholar] [CrossRef]
- Kawasaki, K.; Toshimitsu, K.; Matano, M.; Fujita, M.; Fujii, M.; Togasaki, K.; Ebisudani, T.; Shimokawa, M.; Takano, A.; Takahashi, S.; et al. An Organoid Biobank of Neuroendocrine Neoplasms Enables Genotype-Phenotype Mapping. Cell 2020, 183, 1420–1435.e21. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-Term Culture, Genetic Manipulation and Xenotransplantation of Human Normal and Breast Cancer Organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef] [PubMed]
- Mazzucchelli, S.; Piccotti, F.; Allevi, R.; Truffi, M.; Sorrentino, L.; Russo, L.; Agozzino, M.; Signati, L.; Bonizzi, A.; Villani, L.; et al. Establishment and Morphological Characterization of Patient-Derived Organoids from Breast Cancer. Biol. Proced. Online 2019, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Shen, M.; Li, K.; Han, X.; Li, H.; Tan, Z.; Wang, Y.; Peng, Y.; Tang, Z.; Qu, C.; et al. Organoids from Patient Biopsy Samples Can Predict the Response of BC Patients to Neoadjuvant Chemotherapy. Ann. Med. 2022, 54, 2581–2597. [Google Scholar] [CrossRef]
- Kim, J.; Yu, D.; Kwon, Y.; Lee, K.S.; Sim, S.H.; Kong, S.-Y.; Lee, E.S.; Park, I.H.; Park, C. Genomic Characteristics of Triple-Negative Breast Cancer Nominate Molecular Subtypes That Predict Chemotherapy Response. Mol. Cancer Res. 2020, 18, 253–263. [Google Scholar] [CrossRef]
- Bhatia, S.; Kramer, M.; Russo, S.; Naik, P.; Arun, G.; Brophy, K.; Andrews, P.; Fan, C.; Perou, C.M.; Preall, J.; et al. Patient-Derived Triple-Negative Breast Cancer Organoids Provide Robust Model Systems That Recapitulate Tumor Intrinsic Characteristics. Cancer Res. 2022, 82, 1174–1192. [Google Scholar] [CrossRef]
- Lõhmussaar, K.; Oka, R.; Espejo Valle-Inclan, J.; Smits, M.H.H.; Wardak, H.; Korving, J.; Begthel, H.; Proost, N.; van de Ven, M.; Kranenburg, O.W.; et al. Patient-Derived Organoids Model Cervical Tissue Dynamics and Viral Oncogenesis in Cervical Cancer. Cell Stem Cell 2021, 28, 1380–1396.e6. [Google Scholar] [CrossRef]
- Nelson, L.; Tighe, A.; Golder, A.; Littler, S.; Bakker, B.; Moralli, D.; Murtuza Baker, S.; Donaldson, I.J.; Spierings, D.C.J.; Wardenaar, R.; et al. A Living Biobank of Ovarian Cancer Ex Vivo Models Reveals Profound Mitotic Heterogeneity. Nat. Commun. 2020, 11, 822. [Google Scholar] [CrossRef]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; van Ineveld, R.L.; Derakhshan, S.; de Haan, S.; et al. An Organoid Biobank for Childhood Kidney Cancers That Captures Disease and Tissue Heterogeneity. Nat. Commun. 2020, 11, 1310. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef]
- Weber, C. A Biobank for Bladder Cancer. Nat. Cell Biol. 2018, 20, 634. [Google Scholar] [CrossRef]
- Mullenders, J.; de Jongh, E.; Brousali, A.; Roosen, M.; Blom, J.P.A.; Begthel, H.; Korving, J.; Jonges, T.; Kranenburg, O.; Meijer, R.; et al. Mouse and Human Urothelial Cancer Organoids: A Tool for Bladder Cancer Research. Proc. Natl. Acad. Sci. 2019, 116, 4567–4574. [Google Scholar] [CrossRef]
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.; et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4332–4345. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-Tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Abdullah, K.G.; Bird, C.E.; Buehler, J.D.; Gattie, L.C.; Savani, M.R.; Sternisha, A.C.; Xiao, Y.; Levitt, M.M.; Hicks, W.H.; Li, W.; et al. Establishment of Patient-Derived Organoid Models of Lower-Grade Glioma. Neuro. Oncol. 2022, 24, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Xia, T.-L.; Tang, H.-C.; Liu, X.; Han, R.; Zou, X.; Zhao, Y.-T.; Chen, M.-Y.; Li, G. Establishment of a Patient-Derived Organoid Model and Living Biobank for Nasopharyngeal Carcinoma. Ann. Transl. Med. 2022, 10, 526. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Kim, S.-M.; Lim, S.; Lee, J.Y.; Choi, S.-J.; Yang, S.-D.; Yun, M.R.; Kim, C.G.; Gu, S.R.; Park, C.; et al. Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 4397–4409. [Google Scholar] [CrossRef]
- Li, Y.F.; Gao, Y.; Liang, B.W.; Cao, X.Q.; Sun, Z.J.; Yu, J.H.; Liu, Z.D.; Han, Y. Patient-Derived Organoids of Non-Small Cells Lung Cancer and Their Application for Drug Screening. Neoplasma 2020, 67, 430–437. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in Cancer Research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.-D.T.; Hsiao, J.-H.; Tseng, L.-M.; Hou, M.-F.; Li, C.-J. Breast Cancer Organoids Derived from Patients: A Platform for Tailored Drug Screening. Biochem. Pharmacol. 2023, 217, 115803. [Google Scholar] [CrossRef]
- Wang, H.-M.; Zhang, C.-Y.; Peng, K.-C.; Chen, Z.-X.; Su, J.-W.; Li, Y.-F.; Li, W.-F.; Gao, Q.-Y.; Zhang, S.-L.; Chen, Y.-Q.; et al. Using Patient-Derived Organoids to Predict Locally Advanced or Metastatic Lung Cancer Tumor Response: A Real-World Study. Cell Rep. Med. 2023, 4, 100911. [Google Scholar] [CrossRef]
- Yang, R.; Yu, Y. Patient-Derived Organoids in Translational Oncology and Drug Screening. Cancer Lett. 2023, 562, 216180. [Google Scholar] [CrossRef]
- Hubrecht Organoid Biobank Foundation 2025. Available online: https://www.hubrechtorganoidbiobank.org/ (accessed on 15 April 2023).
- Lung Cancer Organoid Biobank. Available online: https://www.sigmaaldrich.com/technical-documents/technical-article/cell-culture-and-cell-culture-analysis/mammalian-cell-culture/lung-organoid-pdo-biobank (accessed on 15 April 2023).
- Organoids. Available online: https://www.atcc.org/cell-products/cell-models/organoids#t=productTab&numberOfResults=24 (accessed on 15 April 2023).
- Cellesce. Available online: http://www.cellesce.com/ (accessed on 15 April 2023).
- DefiniGEN. Available online: https://www.definigen.com/ (accessed on 15 April 2023).
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Author Correction: Establishment of Patient-Derived Cancer Organoids for Drug-Screening Applications. Nat. Protoc. 2021, 16, 5739. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, D.; Andrieux, G.; Boehnke, K.; Keil, M.; Silvestri, A.; Silvestrov, M.; Keilholz, U.; Haybaeck, J.; Erdmann, G.; Sachse, C.; et al. Heterogeneous Pathway Activation and Drug Response Modelled in Colorectal-Tumor-Derived 3D Cultures. PLOS Genet. 2019, 15, e1008076. [Google Scholar] [CrossRef]
- Aboulkheyr Es, H.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar] [CrossRef]
- Qu, J.; Kalyani, F.S.; Liu, L.; Cheng, T.; Chen, L. Tumor Organoids: Synergistic Applications, Current Challenges, and Future Prospects in Cancer Therapy. Cancer Commun. 2021, 41, 1331–1353. [Google Scholar] [CrossRef]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential Cancer Mutations in Cultured Human Intestinal Stem Cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, S.; Zhu, L.; Huang, M.; Xie, Y.; Song, X.; Chen, Z.; Lau, H.C.-H.; Sung, J.J.-Y.; Xu, L.; et al. Personalized Drug Screening Using Patient-Derived Organoid and Its Clinical Relevance in Gastric Cancer. Cell Rep. Med. 2024, 5, 101627. [Google Scholar] [CrossRef]
- He, G.-W.; Lin, L.; DeMartino, J.; Zheng, X.; Staliarova, N.; Dayton, T.; Begthel, H.; van de Wetering, W.J.; Bodewes, E.; van Zon, J.; et al. Optimized Human Intestinal Organoid Model Reveals Interleukin-22-Dependency of Paneth Cell Formation. Cell Stem Cell 2022, 29, 1333–1345.e6. [Google Scholar] [CrossRef] [PubMed]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e6. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.J.; Decker, B.; Roberts, E.A.; Horowitz, N.S.; Muto, M.G.; Worley, M.J.J.; Feltmate, C.M.; Nucci, M.R.; Swisher, E.M.; Nguyen, H.; et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov. 2018, 8, 1404–1421. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, C.; Zhang, Y.; Shi, C. Application of Organoid Models in Prostate Cancer Research. Front. Oncol. 2021, 11, 736431. [Google Scholar] [CrossRef]
- Whyard, T.; Liu, J.; Darras, F.S.; Waltzer, W.C.; Romanov, V. Organoid Model of Urothelial Cancer: Establishment and Applications for Bladder Cancer Research. Biotechniques 2020, 69, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non-Small Cell Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Jensen, K.B.; Little, M.H. Organoids Are Not Organs: Sources of Variation and Misinformation in Organoid Biology. Stem Cell Rep. 2023, 18, 1255–1270. [Google Scholar] [CrossRef]
- Walsh, A.J.; Cook, R.S.; Sanders, M.E.; Arteaga, C.L.; Skala, M.C. Drug Response in Organoids Generated from Frozen Primary Tumor Tissues. Sci. Rep. 2016, 6, 18889. [Google Scholar] [CrossRef]
- He, A.; Powell, S.; Kyle, M.; Rose, M.; Masmila, E.; Estrada, V.; Sicklick, J.K.; Molinolo, A.; Kaushal, S. Cryopreservation of Viable Human Tissues: Renewable Resource for Viable Tissue, Cell Lines, and Organoid Development. Biopreserv. Biobank. 2020, 18, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Restivo, G.; Tastanova, A.; Balázs, Z.; Panebianco, F.; Diepenbruck, M.; Ercan, C.; Preca, B.-T.; Hafner, J.; Weber, W.P.; Kurzeder, C.; et al. Live Slow-Frozen Human Tumor Tissues Viable for 2D, 3D, Ex Vivo Cultures and Single-Cell RNAseq. Commun. Biol. 2022, 5, 1144. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhou, J.-B.; Chu, X.-P.; Feng, Y.-Y.; Zeng, Q.-B.; Lei, J.-H.; Wong, K.-P.; Chan, T.-I.; Lam, C.-W.; Zhu, W.-L.; et al. Establishing a Cryopreserved Biobank of Living Tumor Tissues for Drug Sensitivity Testing. Bioact. Mater. 2025, 46, 582–596. [Google Scholar] [CrossRef]
- Jiang, K.-L.; Wang, X.-X.; Liu, X.-J.; Guo, L.-K.; Chen, Y.-Q.; Jia, Q.-L.; Yang, K.-M.; Ling, J.-H. Success Rate of Current Human-Derived Gastric Cancer Organoids Establishment and Influencing Factors: A Systematic Review and Meta-Analysis. World J. Gastrointest. Oncol. 2024, 16, 1626–1646. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A Complex Protein Mixture Required for Optimal Growth of Cell Culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Passaniti, A.; Kleinman, H.K.; Martin, G.R. Matrigel: History/Background, Uses, and Future Applications. J. Cell Commun. Signal. 2022, 16, 621–626. [Google Scholar] [CrossRef]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordóñez-Morán, P.; Clevers, H.; Lutolf, M.P. Designer Matrices for Intestinal Stem Cell and Organoid Culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef]
- Cruz-Acuña, R.; Quirós, M.; Farkas, A.E.; Dedhia, P.H.; Huang, S.; Siuda, D.; García-Hernández, V.; Miller, A.J.; Spence, J.R.; Nusrat, A.; et al. Synthetic Hydrogels for Human Intestinal Organoid Generation and Colonic Wound Repair. Nat. Cell Biol. 2017, 19, 1326–1335. [Google Scholar] [CrossRef]
- Dorgau, B.; Felemban, M.; Hilgen, G.; Kiening, M.; Zerti, D.; Hunt, N.C.; Doherty, M.; Whitfield, P.; Hallam, D.; White, K.; et al. Decellularised Extracellular Matrix-Derived Peptides from Neural Retina and Retinal Pigment Epithelium Enhance the Expression of Synaptic Markers and Light Responsiveness of Human Pluripotent Stem Cell Derived Retinal Organoids. Biomaterials 2019, 199, 63–75. [Google Scholar] [CrossRef]
- Kozlowski, M.T.; Crook, C.J.; Ku, H.T. Towards Organoid Culture without Matrigel. Commun. Biol. 2021, 4, 1387. [Google Scholar] [CrossRef]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.-H.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.-G.; et al. Tissue Extracellular Matrix Hydrogels as Alternatives to Matrigel for Culturing Gastrointestinal Organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Carpentier, N.; Ye, S.; Delemarre, M.D.; Van der Meeren, L.; Skirtach, A.G.; van der Laan, L.J.W.; Schneeberger, K.; Spee, B.; Dubruel, P.; Van Vlierberghe, S. Gelatin-Based Hybrid Hydrogels as Matrices for Organoid Culture. Biomacromolecules 2024, 25, 590–604. [Google Scholar] [CrossRef]
- Cherne, M.D.; Sidar, B.; Sebrell, T.A.; Sanchez, H.S.; Heaton, K.; Kassama, F.J.; Roe, M.M.; Gentry, A.B.; Chang, C.B.; Walk, S.T.; et al. A Synthetic Hydrogel, VitroGel(®) ORGANOID-3, Improves Immune Cell-Epithelial Interactions in a Tissue Chip Co-Culture Model of Human Gastric Organoids and Dendritic Cells. Front. Pharmacol. 2021, 12, 707891. [Google Scholar] [CrossRef] [PubMed]
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; De Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular Matrix Hydrogel Derived from Decellularized Tissues Enables Endodermal Organoid Culture. Nat. Commun. 2019, 10, 5658. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hu, H.; Kung, H.; Zou, R.; Dai, Y.; Hu, Y.; Wang, T.; Lv, T.; Yu, J.; Li, F. Organoids: The Current Status and Biomedical Applications. MedComm 2023, 4, e274. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Prim. 2022, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Yokota, J.; Yamashita, T.; Inui, T.; Nomoto, R.; Kishimoto, W.; Nakase, H.; Mizuguchi, H. Comparison of Culture Media for Human Intestinal Organoids from the Viewpoint of Pharmacokinetic Studies. Biochem. Biophys. Res. Commun. 2021, 566, 115–122. [Google Scholar] [CrossRef]
- Wilson, S.S.; Mayo, M.; Melim, T.; Knight, H.; Patnaude, L.; Wu, X.; Phillips, L.; Westmoreland, S.; Dunstan, R.; Fiebiger, E.; et al. Optimized Culture Conditions for Improved Growth and Functional Differentiation of Mouse and Human Colon Organoids. Front. Immunol. 2020, 11, 547102. [Google Scholar] [CrossRef]
- Calafato, G.; Alquati, C.; Bernardi, A.; Di Paola, F.J.; Ricciardiello, L. Comparative Analysis of Commercial and Home-Made Media on RSPO1/S6R Axis in Organoids with Different Wnt Backgrounds: A Methodological Guide for the Selection of Intestinal Patient-Derived Organoids Culture Media. Int. J. Mol. Sci. 2024, 25, 1526. [Google Scholar] [CrossRef] [PubMed]
- Hogenson, T.L.; Xie, H.; Phillips, W.J.; Toruner, M.D.; Li, J.J.; Horn, I.P.; Kennedy, D.J.; Almada, L.L.; Marks, D.L.; Carr, R.M.; et al. Culture Media Composition Influences Patient-Derived Organoid Ability to Predict Therapeutic Responses in Gastrointestinal Cancers. JCI Insight 2022, 7, 158060. [Google Scholar] [CrossRef]
- Shin, W.; Wu, A.; Min, S.; Shin, Y.C.; Fleming, R.Y.D.; Eckhardt, S.G.; Kim, H.J. Spatiotemporal Gradient and Instability of Wnt Induce Heterogeneous Growth and Differentiation of Human Intestinal Organoids. iScience 2020, 23, 101372. [Google Scholar] [CrossRef]
- Nwokoye, P.N.; Abilez, O.J. Bioengineering Methods for Vascularizing Organoids. Cell Rep. Methods 2024, 4, 100779. [Google Scholar] [CrossRef] [PubMed]
- Polak, R.; Zhang, E.T.; Kuo, C.J. Cancer Organoids 2.0: Modelling the Complexity of the Tumour Immune Microenvironment. Nat. Rev. Cancer 2024, 24, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Lemme-Dumit, J.M.; Latanich, R.; Pasetti, M.F.; Zachos, N.C. Co-Culture System of Human Enteroids/Colonoids with Innate Immune Cells. Curr. Protoc. Immunol. 2020, 131, e113. [Google Scholar] [CrossRef]
- Luo, X.; Fong, E.L.S.; Zhu, C.; Lin, Q.X.X.; Xiong, M.; Li, A.; Li, T.; Benoukraf, T.; Yu, H.; Liu, S. Hydrogel-Based Colorectal Cancer Organoid Co-Culture Models. Acta Biomater. 2021, 132, 461–472. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, E.; Shang, Z. 3D Co-Culture of Cancer-Associated Fibroblast with Oral Cancer Organoids. J. Dent. Res. 2021, 100, 201–208. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; O’Connor, R. Studying Cryptosporidium Infection in 3D Tissue-Derived Human Organoid Culture Systems by Microinjection. J. Vis. Exp. 2019, e59610. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I.; et al. Intestinal Organoid Cocultures with Microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef] [PubMed]
- Holthaus, D.; Delgado-Betancourt, E.; Aebischer, T.; Seeber, F.; Klotz, C. Harmonization of Protocols for Multi-Species Organoid Platforms to Study the Intestinal Biology of Toxoplasma Gondii and Other Protozoan Infections. Front. Cell. Infect. Microbiol. 2020, 10, 610368. [Google Scholar] [CrossRef]
- Votanopoulos, K.I.; Forsythe, S.; Sivakumar, H.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967. [Google Scholar] [CrossRef]
- Zhou, G.; Lieshout, R.; van Tienderen, G.S.; de Ruiter, V.; van Royen, M.E.; Boor, P.P.C.; Magré, L.; Desai, J.; Köten, K.; Kan, Y.Y.; et al. Modelling Immune Cytotoxicity for Cholangiocarcinoma with Tumour-Derived Organoids and Effector T Cells. Br. J. Cancer 2022, 127, 649–660. [Google Scholar] [CrossRef]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; van der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging Organoid-Immune Co-Culture Models for Cancer Research: From Oncoimmunology to Personalized Immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef] [PubMed]
- Papp, D.; Korcsmaros, T.; Hautefort, I. Revolutionizing Immune Research with Organoid-Based Co-Culture and Chip Systems. Clin. Exp. Immunol. 2024, 218, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, E.; Beenen, A.C.; Kachkin, D.; Regensburger, M.; Zundler, S.; Blumenthal, D.B.; Lutzny-Geier, G.; Winner, B.; Prots, I. Novel Co-Culture Model of T Cells and Midbrain Organoids for Investigating Neurodegeneration in Parkinson’s Disease. npj Park. Dis. 2025, 11, 36. [Google Scholar] [CrossRef]
- Takebe, T.; Zhang, B.; Radisic, M. Synergistic Engineering: Organoids Meet Organs-on-a-Chip. Cell Stem Cell 2017, 21, 297–300. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-Chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, L.; Yu, H.; Yin, F.; Wang, Y.; Liu, H.; Jiang, L.; Qin, J. In Situ Generation of Human Brain Organoids on a Micropillar Array. Lab Chip 2017, 17, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Zhu, Y.; Qin, J. Human Brain Organoid-on-a-Chip to Model Prenatal Nicotine Exposure. Lab Chip 2018, 18, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A Guide to the Organ-on-a-Chip. Nat. Rev. Methods Prim. 2022, 2, 33. [Google Scholar] [CrossRef]
- Man, Y.; Liu, Y.; Chen, Q.; Zhang, Z.; Li, M.; Xu, L.; Tan, Y.; Liu, Z. Organoids-On-a-Chip for Personalized Precision Medicine. Adv. Healthc. Mater. 2024, 13, 2401843. [Google Scholar] [CrossRef]
- Mandrycky, C.J.; Howard, C.C.; Rayner, S.G.; Shin, Y.J.; Zheng, Y. Organ-on-a-Chip Systems for Vascular Biology. J. Mol. Cell. Cardiol. 2021, 159, 1–13. [Google Scholar] [CrossRef]
- Zhang, S.; Wan, Z.; Kamm, R.D. Vascularized Organoids on a Chip: Strategies for Engineering Organoids with Functional Vasculature. Lab Chip 2021, 21, 473–488. [Google Scholar] [CrossRef]
- Bonner, M.G.; Gudapati, H.; Mou, X.; Musah, S. Microfluidic Systems for Modeling Human Development. Development 2022, 149, 199463. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Y.-S.; Chien, S. Shear Stress-Initiated Signaling and Its Regulation of Endothelial Function. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2191–2198. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y. V Effects of Shear Stress on Endothelial Cells: Go with the Flow. Acta Physiol. 2017, 219, 382–408. [Google Scholar] [CrossRef]
- Homan, K.A.; Gupta, N.; Kroll, K.T.; Kolesky, D.B.; Skylar-Scott, M.; Miyoshi, T.; Mau, D.; Valerius, M.T.; Ferrante, T.; Bonventre, J.V.; et al. Flow-Enhanced Vascularization and Maturation of Kidney Organoids in Vitro. Nat. Methods 2019, 16, 255–262. [Google Scholar] [CrossRef]
- Barata, D.; van Blitterswijk, C.; Habibovic, P. High-Throughput Screening Approaches and Combinatorial Development of Biomaterials Using Microfluidics. Acta Biomater. 2016, 34, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Hong, S.; Rhee, W.J. Microfluidic Three-Dimensional Cell Culture of Stem Cells for High-Throughput Analysis. World J. Stem Cells 2019, 11, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Mousavi Shaegh, S.A.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-Integrated Organs-on-Chips Platform for Automated and Continual in Situ Monitoring of Organoid Behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Schuster, B.; Junkin, M.; Kashaf, S.S.; Romero-Calvo, I.; Kirby, K.; Matthews, J.; Weber, C.R.; Rzhetsky, A.; White, K.P.; Tay, S. Automated Microfluidic Platform for Dynamic and Combinatorial Drug Screening of Tumor Organoids. Nat. Commun. 2020, 11, 5271. [Google Scholar] [CrossRef]
- Yin, F.; Zhang, X.; Wang, L.; Wang, Y.; Zhu, Y.; Li, Z.; Tao, T.; Chen, W.; Yu, H.; Qin, J. HiPSC-Derived Multi-Organoids-on-Chip System for Safety Assessment of Antidepressant Drugs. Lab Chip 2021, 21, 571–581. [Google Scholar] [CrossRef]
- Tao, T.; Deng, P.; Wang, Y.; Zhang, X.; Guo, Y.; Chen, W.; Qin, J. Microengineered Multi-Organoid System from HiPSCs to Recapitulate Human Liver-Islet Axis in Normal and Type 2 Diabetes. Adv. Sci. 2022, 9, e2103495. [Google Scholar] [CrossRef]
- Achberger, K.; Probst, C.; Haderspeck, J.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Merging Organoid and Organ-on-a-Chip Technology to Generate Complex Multi-Layer Tissue Models in a Human Retina-on-a-Chip Platform. Elife 2019, 8, e46188. [Google Scholar] [CrossRef]
- Achberger, K.; Cipriano, M.; Düchs, M.J.; Schön, C.; Michelfelder, S.; Stierstorfer, B.; Lamla, T.; Kauschke, S.G.; Chuchuy, J.; Roosz, J.; et al. Human Stem Cell-Based Retina on Chip as New Translational Model for Validation of AAV Retinal Gene Therapy Vectors. Stem Cell Rep. 2021, 16, 2242–2256. [Google Scholar] [CrossRef] [PubMed]
- Ghio, A.J.; Dailey, L.A.; Soukup, J.M.; Stonehuerner, J.; Richards, J.H.; Devlin, R.B. Growth of Human Bronchial Epithelial Cells at an Air-Liquid Interface Alters the Response to Particle Exposure. Part. Fibre Toxicol. 2013, 10, 25. [Google Scholar] [CrossRef]
- Whitcutt, M.J.; Adler, K.B.; Wu, R. A Biphasic Chamber System for Maintaining Polarity of Differentiation of Cultured Respiratory Tract Epithelial Cells. Vitr. Cell. Dev. Biol. J. Tissue Cult. Assoc. 1988, 24, 420–428. [Google Scholar] [CrossRef]
- Jiang, D.; Schaefer, N.; Chu, H.W. Air-Liquid Interface Culture of Human and Mouse Airway Epithelial Cells. Methods Mol. Biol. 2018, 1809, 91–109. [Google Scholar] [CrossRef]
- Wu, R.; Zhao, Y.H.; Chang, M.M. Growth and Differentiation of Conducting Airway Epithelial Cells in Culture. Eur. Respir. J. 1997, 10, 2398–2403. [Google Scholar] [CrossRef]
- Kesimer, M.; Kirkham, S.; Pickles, R.J.; Henderson, A.G.; Alexis, N.E.; Demaria, G.; Knight, D.; Thornton, D.J.; Sheehan, J.K. Tracheobronchial Air-Liquid Interface Cell Culture: A Model for Innate Mucosal Defense of the Upper Airways? Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L92–L100. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Eo, E.-Y.; Kim, B.; Lee, H.; Kim, J.; Koo, B.-K.; Kim, H.-J.; Cho, S.; Kim, J.; Cho, Y.-J. Transcriptomic Analysis of Air-Liquid Interface Culture in Human Lung Organoids Reveals Regulators of Epithelial Differentiation. Cells 2024, 13, 1991. [Google Scholar] [CrossRef]
- Usui, T.; Sakurai, M.; Umata, K.; Yamawaki, H.; Ohama, T.; Sato, K. Preparation of Human Primary Colon Tissue-Derived Organoid Using Air Liquid Interface Culture. Curr. Protoc. Toxicol. 2018, 75, 22.6.1–22.6.7. [Google Scholar] [CrossRef]
- Sekiya, S.; Kikuchi, T.; Shimizu, T. Perfusion Culture Maintained with an Air-Liquid Interface to Stimulate Epithelial Cell Organization in Renal Organoids in Vitro. BMC Biomed. Eng. 2019, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Ivancic, D.Z.; Naved, B.A.; Wertheim, J.A.; Oxburgh, L. An Efficient Method to Generate Kidney Organoids at the Air-Liquid Interface. J. Biol. Methods 2021, 8, e150. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, I.; Nakai, T.; Iwao, T.; Matsunaga, T. Air-Liquid Interface Culture Combined with Differentiation Factors Reproducing Intestinal Cell Structure Formation in Vitro. Biol. Open 2025, 14, bio061612. [Google Scholar] [CrossRef]
- King, J.A.; Miller, W.M. Bioreactor Development for Stem Cell Expansion and Controlled Differentiation. Curr. Opin. Chem. Biol. 2007, 11, 394–398. [Google Scholar] [CrossRef]
- dos Santos, F.F.; Andrade, P.Z.; da Silva, C.L.; Cabral, J.M.S. Bioreactor Design for Clinical-Grade Expansion of Stem Cells. Biotechnol. J. 2013, 8, 644–654. [Google Scholar] [CrossRef]
- Avena, P.; Zavaglia, L.; Casaburi, I.; Pezzi, V. Perfusion Bioreactor Technology for Organoid and Tissue Culture: A Mini Review. Onco 2025, 5, 17. [Google Scholar] [CrossRef]
- Licata, J.P.; Schwab, K.H.; Har-El, Y.-E.; Gerstenhaber, J.A.; Lelkes, P.I. Bioreactor Technologies for Enhanced Organoid Culture. Int. J. Mol. Sci. 2023, 24, 1427. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-Bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell Diversity and Network Dynamics in Photosensitive Human Brain Organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef]
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G. Generation of Human Brain Region–specific Organoids Using a Miniaturized Spinning Bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef]
- Przepiorski, A.; Sander, V.; Tran, T.; Hollywood, J.A.; Sorrenson, B.; Shih, J.-H.; Wolvetang, E.J.; McMahon, A.P.; Holm, T.M.; Davidson, A.J. A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Rep. 2018, 11, 470–484. [Google Scholar] [CrossRef]
- Schneeberger, K.; Sánchez-Romero, N.; Ye, S.; van Steenbeek, F.G.; Oosterhoff, L.A.; Pla Palacin, I.; Chen, C.; van Wolferen, M.E.; van Tienderen, G.; Lieshout, R.; et al. Large-Scale Production of LGR5-Positive Bipotential Human Liver Stem Cells. Hepatology 2020, 72, 257–270. [Google Scholar] [CrossRef]
- Ovando-Roche, P.; West, E.L.; Branch, M.J.; Sampson, R.D.; Fernando, M.; Munro, P.; Georgiadis, A.; Rizzi, M.; Kloc, M.; Naeem, A.; et al. Use of Bioreactors for Culturing Human Retinal Organoids Improves Photoreceptor Yields. Stem Cell Res. Ther. 2018, 9, 156. [Google Scholar] [CrossRef]
- Ye, S.; Marsee, A.; van Tienderen, G.S.; Rezaeimoghaddam, M.; Sheikh, H.; Samsom, R.-A.; de Koning, E.J.P.; Fuchs, S.; Verstegen, M.M.A.; van der Laan, L.J.W.; et al. Accelerated Production of Human Epithelial Organoids in a Miniaturized Spinning Bioreactor. Cell Rep. Methods 2024, 4, 100903. [Google Scholar] [CrossRef] [PubMed]
| System or Body District | Organ | Number of Samples | Country | Diagnosis | Primary or Metastatic | Main Experimental PDOs Validation | Main Translational Applications | References | |
|---|---|---|---|---|---|---|---|---|---|
| Tumor | Paired Healthy | ||||||||
| Digestive | Colorectal | 22 | 19 | The Netherlands | Colorectal carcinoma | Primary | WGS; RNA-seq | High-throughput screening (in vitro) | [17] |
| Digestive | Colorectal | 55 | 41 | Japan | Colorectal carcinoma | Primary and metastatic | Histology, WGS, RNA microarray, | Disease modeling | [18] |
| Digestive | Colorectal | 32 | 18 | China | Early-onset colorectal carcinoma | Primary | MSI analysis, WES, WGS, RNA-seq, sc-RNA-seq, gene editing | Disease modeling | [19] |
| Digestive | Rectal | 96 | 0 | China | Rectal carcinoma | Primary | Histology, WES | Drug/radiation response prediction | [20] |
| Digestive | Colorectal | 151 | 0 | China | Colorectal carcinoma | Primary and metastatic | Histology, RNA-seq | Drug response prediction | [21] |
| Digestive | Colorectal | 94 | 0 | The Netherlands | Colorectal carcinoma | Primary and metastatic | RNA-seq | Disease modeling | [22] |
| Digestive | Colorectal | 58 | 0 | China | Colorectal carcinoma | Primary and metastatic | Histology, WES, RNA-seq, sc-RNA-seq | Drug response prediction | [23] |
| Digestive | Colorectal | 34 | 21 | China | Colorectal carcinoma | Primary | Histology, WES | Drug response prediction | [24] |
| Digestive | Colorectal | 77 | 31 | The Netherlands | Colorectal carcinoma | Primary and metastatic | WES | High-throughput screening (in vitro/in vivo) | [25] |
| Digestive | Colorectal | 106 | 0 | Germany | Colorectal carcinoma | Primary and metastatic | WGS, WES, RNA-seq | High-throughput screening, gene–drug response correlation | [26] |
| Digestive | Colorectal, gastroesophagus, bile ducts | 110 | 0 | The United Kingdom | Colorectal, gastroesophageal cancers and cholangiocarcinoma carcinoma | Metastatic | Histology, WGS, NGS, RNA-seq, | High-throughput screening (in vitro/in vivo) | [27] |
| Digestive | Stomach | 46 | 17 | China | Gastric tumor | Primary and metastatic | Histology, WES, RNA-seq | High-throughput screening, drug response prediction | [28] |
| Digestive | Liver | 11 | 0 | Switzerland | Hepatocellular carcinoma | Primary and metastatic | Histology, WES | Disease modeling, drug response prediction | [29] |
| Digestive | Pancreas | 13 | 13 | USA | Intraductal papillary mucinous neoplasms | - | Histology, WGS | Disease modeling | [30] |
| Digestive | Pancreas | 31 | 0 | Switzerland | Pancreatic carcinoma | Primary and metastatic | Histology, WGS, WES, RNA-seq | Disease modeling, high-throughput screening, gene–drug response correlation | [31] |
| Digestive | Pancreas | 77 | 0 | USA | Pancreatic ductal adenocarcinoma | Primary and metastatic | Histology | Drug response prediction | [32] |
| Digestive | Pancreas | 10 | 7 | USA | Intraductal papillary mucinous neoplasms | - | Histology, WGS, WES, RNA-seq | Disease modeling | [33] |
| Digestive | Pancreas | 30 | 5 | The Netherlands | Pancreatic ductal adenocarcinoma and distal cholangiocarcinomas | Primary and metastatic | Histology, WGS, RNA-seq | High-throughput screening | [34] |
| Digestive | Pancreas | 10 | 0 | The Netherlands | Pancreatic carcinoma | Not specified | Histology, WGS, RNA-seq | Disease modeling | [35] |
| Digestive | Pancreas, gallbladder, duodenum | 25 | 0 | Japan | Gastroenteropancreatic neuroendocrine neoplasms | Primary and metastatic | Histology, WGS, WES, RNA-seq, | Disease modeling | [36] |
| Reproductive | Mammary gland | 13 | 14 | The Netherlands | Breast carcinoma (TNBC, ER+/PR+, Her2+) | Primary and metastatic | Histology, imaging | Disease modeling | [37] |
| Reproductive | Mammary gland | 168 | 0 | The Netherlands | Breast carcinoma (TNBC, ER+/PR+ or ER+/PR-, Her2+) | Primary and metastatic | Histology, WGS, RNA-seq | Drug response prediction | [38] |
| Reproductive | Mammary gland | 33 | 0 | Italy | Invasive ductal and lobular breast carcinoma (TNBC, Her2+, Her2-) | Primary | Histology | Disease modeling | [39] |
| Reproductive | Mammary gland | 11 | 0 | China | Breast carcinoma (TNBC, Her2+, Luminal B) | Primary and metastatic | Histology | Drug response prediction | [40] |
| Reproductive | Mammary gland | 38 | 0 | The Republic of Korea | Breast carcinoma (TNBC) | Primary and metastatic | RNA-Seq | Disease modeling, high-throughput screening gene–drug response correlation | [41] |
| Reproductive | Mammary gland | 87 | 0 | USA | Invasive ductal and lobular breast carcinoma (TNBC) | Primary and metastatic | Histology, WGS, RNA-seq, sc-RNA-seq | Disease modeling | [42] |
| Reproductive | Cervix | 12 | 6 | The Netherlands | Cervical tumors | Primary | Histology, WES, RNA-seq | Disease modeling, high-throughput screening | [43] |
| Reproductive | Ovaries | 76 | 0 | The United Kingdom | High-grade serous ovarian carcinoma | Primary and metastatic | Histology, WES, RNA-seq | Disease modeling, drug response prediction | [44] |
| Urinary | Kidney | 54 | 47 | The Netherlands | Kidney tumors | Primary and metastatic | Histology, WGS, sc-RNA-seq | Disease modeling | [45] |
| Urinary | Bladder | 22 | 0 | USA | Urothelial carcinoma | Primary | Histology, WGS | Disease modeling, gene-drug response correlation | [46] |
| Urinary | Bladder | 16 | 0 | The Netherlands | Urothelial carcinoma | Primary | Histology, WGS | Disease modeling, gene-drug response correlation | [47] |
| Urinary | Bladder | 53 | 0 | The Netherlands | Urothelial carcinoma | Primary | Histology | Disease modeling, high-throughput screening | [48] |
| Urinary | Prostate | 4 | 0 | USA | Prostate cancer | Metastatic | Histology, WGS, WES, RNA-seq | Gene–drug response correlation, drug response prediction | [49] |
| Nervous | Brain | 70 | 0 | USA | Glioblastoma | Primary | Histology, WES, RNA-seq, sc-RNA-seq | Disease modeling | [50] |
| Nervous | Brain | 33 | 0 | USA | Glioma | Primary | Histology, gene sequencing | Disease modeling | [51] |
| Head and Neck | Nasopharynx | 62 | 15 | China | Nasopharyngeal carcinoma | Primary | Histology | Disease modeling | [52] |
| Respiratory | Lungs | 84 | 0 | The Republic of Korea | Lung adenocarcinoma | Primary | Histology, WES, RNA-seq | Gene–drug response correlation | [53] |
| Respiratory | Lungs | 14 | 0 | China | NSCLC | Primary | Histology, gene sequencing | High-throughput screening | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Paola, F.J.; Calafato, G.; Piccaluga, P.P.; Tallini, G.; Rhoden, K.J. Patient-Derived Organoid Biobanks for Translational Research and Precision Medicine: Challenges and Future Perspectives. J. Pers. Med. 2025, 15, 394. https://doi.org/10.3390/jpm15080394
Di Paola FJ, Calafato G, Piccaluga PP, Tallini G, Rhoden KJ. Patient-Derived Organoid Biobanks for Translational Research and Precision Medicine: Challenges and Future Perspectives. Journal of Personalized Medicine. 2025; 15(8):394. https://doi.org/10.3390/jpm15080394
Chicago/Turabian StyleDi Paola, Floriana Jessica, Giulia Calafato, Pier Paolo Piccaluga, Giovanni Tallini, and Kerry Jane Rhoden. 2025. "Patient-Derived Organoid Biobanks for Translational Research and Precision Medicine: Challenges and Future Perspectives" Journal of Personalized Medicine 15, no. 8: 394. https://doi.org/10.3390/jpm15080394
APA StyleDi Paola, F. J., Calafato, G., Piccaluga, P. P., Tallini, G., & Rhoden, K. J. (2025). Patient-Derived Organoid Biobanks for Translational Research and Precision Medicine: Challenges and Future Perspectives. Journal of Personalized Medicine, 15(8), 394. https://doi.org/10.3390/jpm15080394








