Genetic and Clinical Insights into ALS/FTD: Profiling a Rare Cohort to Explore Spectrum Heterogeneity
Abstract
1. Introduction
2. Material and Methods
3. Results
3.1. ALS/FTD Cohort
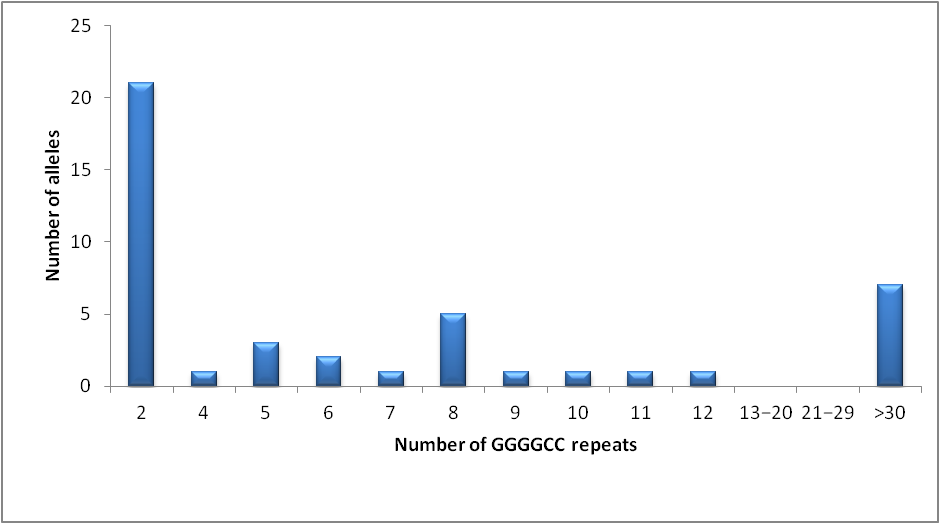
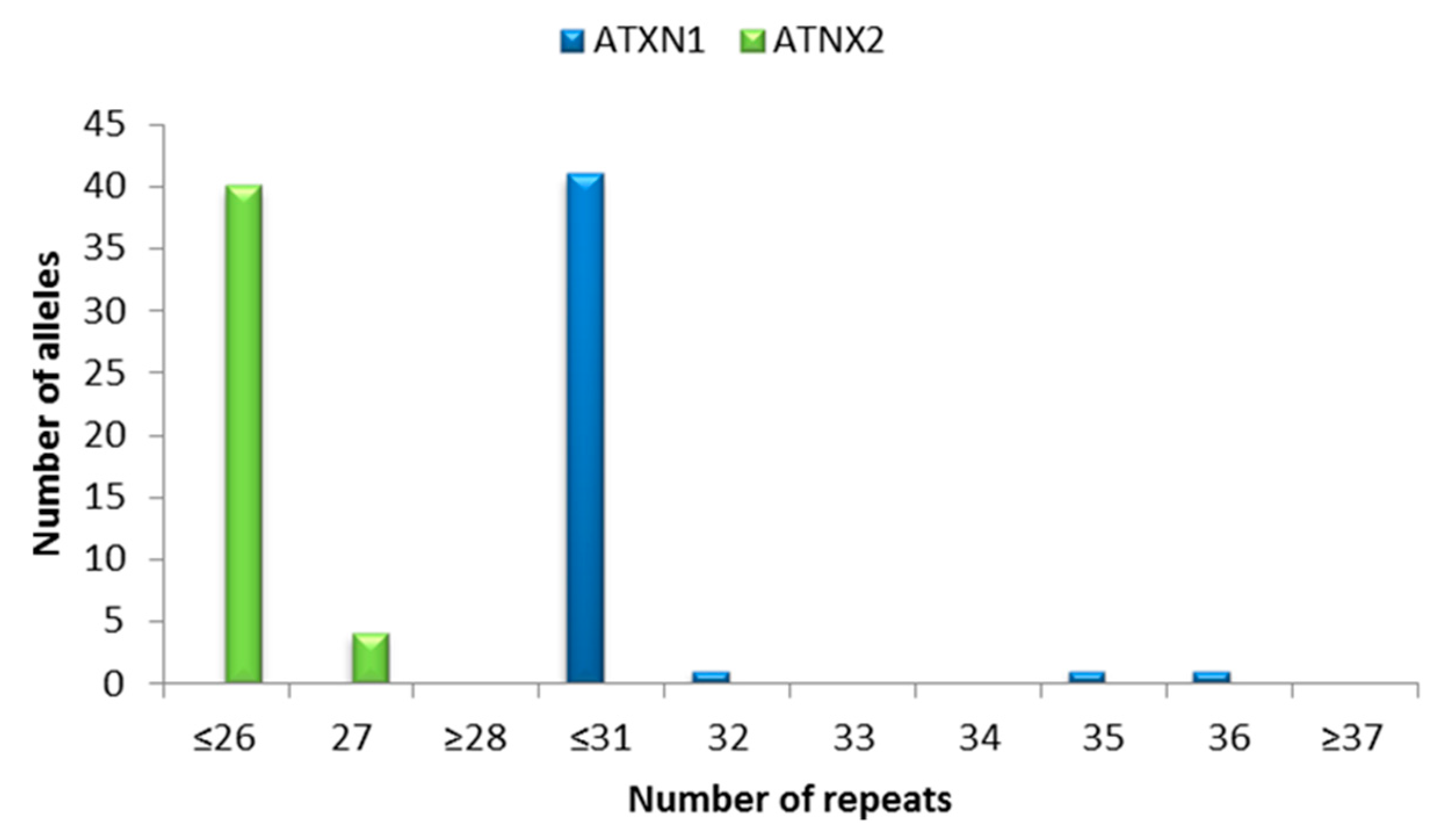
3.2. ALS/FTD Cohort vs. Control Group
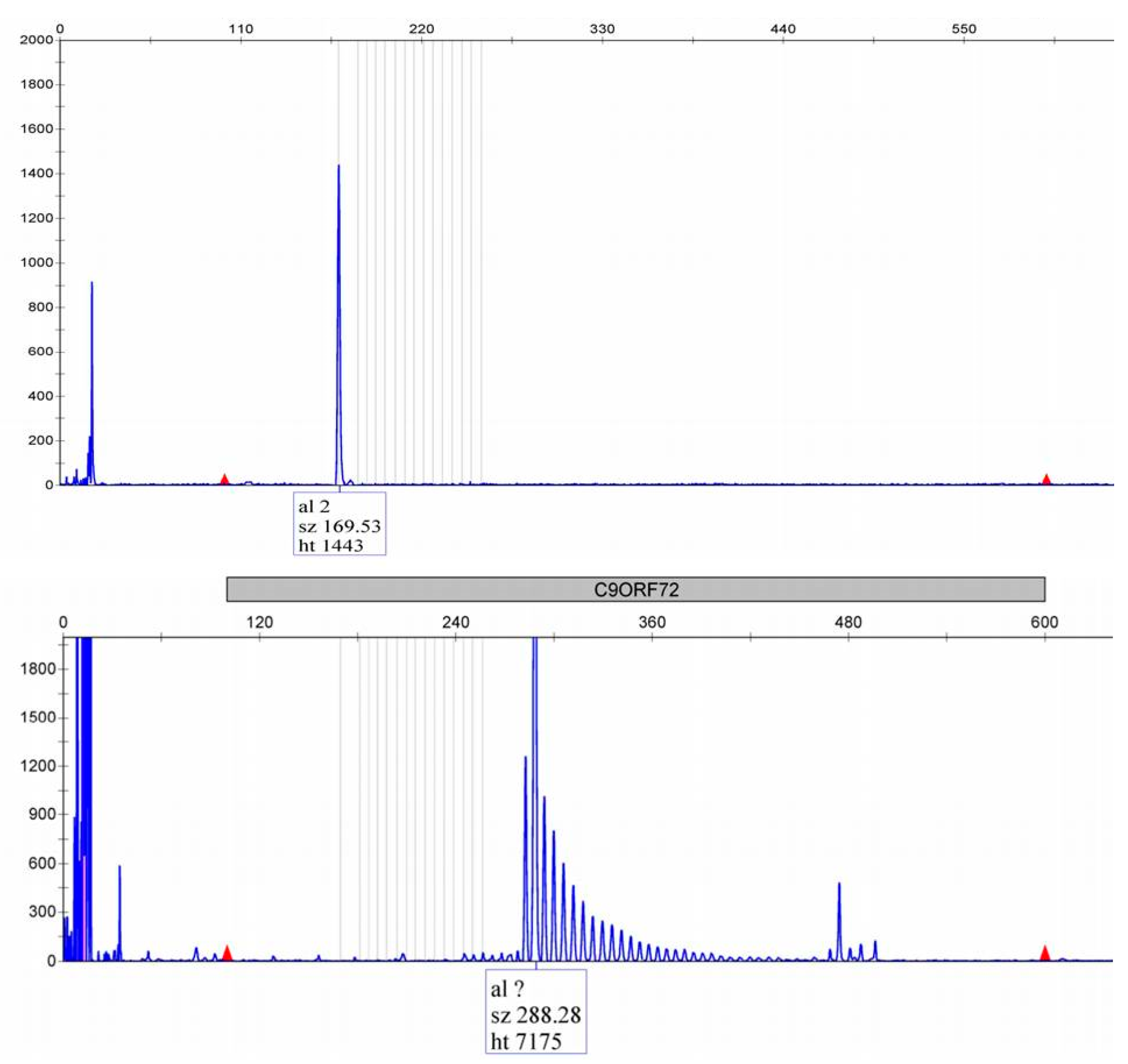
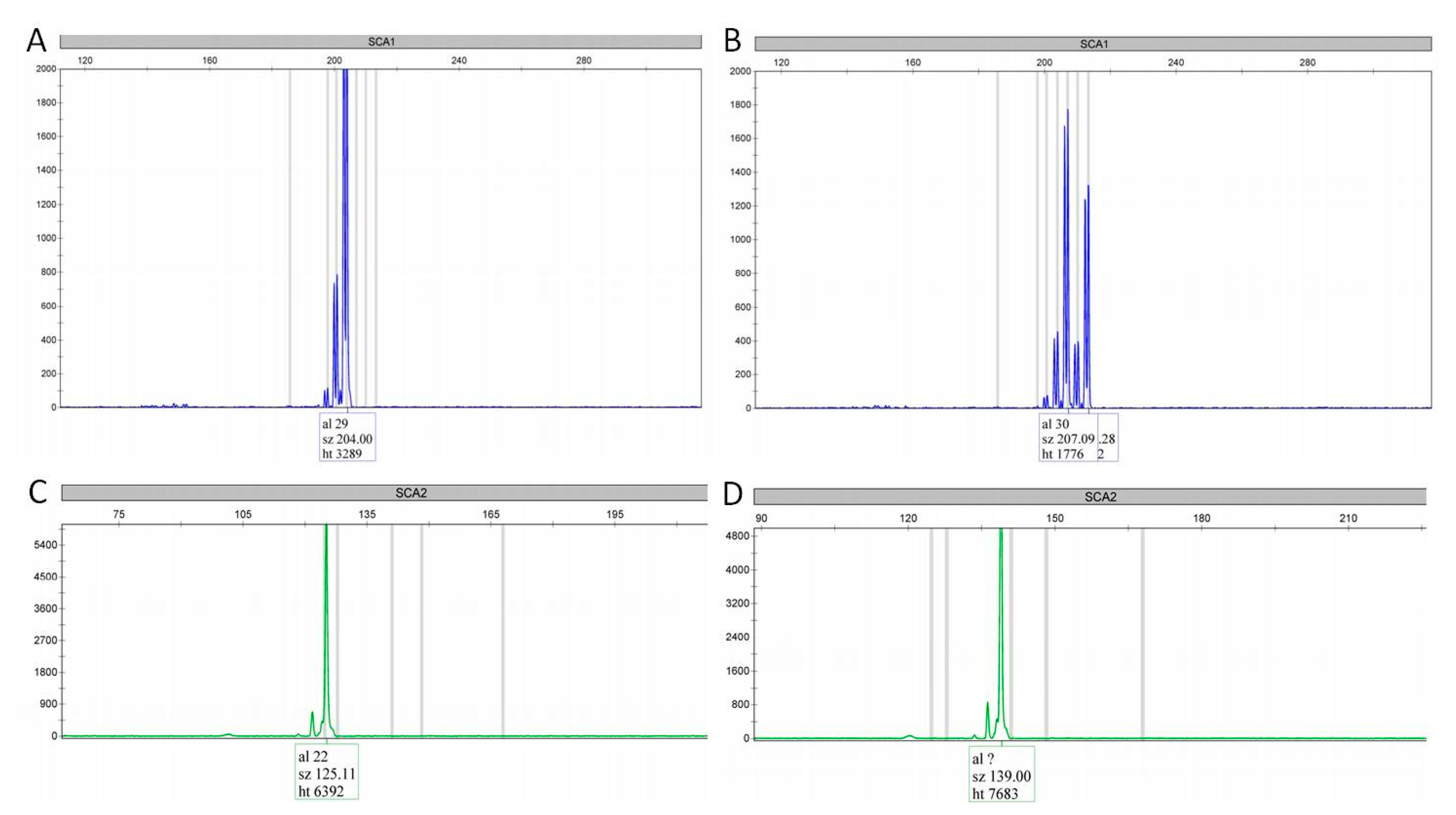
3.3. ALS/FTD C9orf72 Expansion Positive vs. C9orf72 Negative Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From Genes to Mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef]
- Antonioni, A.; Raho, E.M.; Lopriore, P.; Pace, A.P.; Latino, R.R.; Assogna, M.; Mancuso, M.; Gragnaniello, D.; Granieri, E.; Pugliatti, M.; et al. Frontotemporal Dementia, Where Do We Stand? A Narrative Review. Int. J. Mol. Sci. 2023, 24, 11732. [Google Scholar] [CrossRef]
- Erkkinen, M.G.; Kim, M.O.; Geschwind, M.D. Clinical Neurology and Epidemiology of the Major Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2018, 10, a033118. [Google Scholar] [CrossRef]
- Kumar, V.; Islam, A.; Hassan, M.I.; Ahmad, F. Delineating the Relationship between Amyotrophic Lateral Sclerosis and Frontotemporal Dementia: Sequence and Structure-Based Predictions. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 1742–1754. [Google Scholar] [CrossRef]
- Chiò, A.; Moglia, C.; Canosa, A.; Manera, U.; Vasta, R.; Brunetti, M.; Barberis, M.; Corrado, L.; D’Alfonso, S.; Bersano, E.; et al. Cognitive Impairment across ALS Clinical Stages in a Population-Based Cohort. Neurology 2019, 93, e984–e994. [Google Scholar] [CrossRef]
- Phukan, J.; Elamin, M.; Bede, P.; Jordan, N.; Gallagher, L.; Byrne, S.; Lynch, C.; Pender, N.; Hardiman, O. The Syndrome of Cognitive Impairment in Amyotrophic Lateral Sclerosis: A Population-Based Study. J. Neurol. Neurosurg. Psychiatry 2012, 83, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Lomen-Hoerth, C.; Anderson, T.; Miller, B. The Overlap of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Neurology 2002, 59, 1077–1079. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L.; World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Gorno-Tempini, M.L.; Hillis, A.E.; Weintraub, S.; Kertesz, A.; Mendez, M.; Cappa, S.F.; Ogar, J.M.; Rohrer, J.D.; Black, S.; Boeve, B.F.; et al. Classification of Primary Progressive Aphasia and Its Variants. Neurology 2011, 76, 1006–1014. [Google Scholar] [CrossRef]
- Rascovsky, K.; Hodges, J.R.; Knopman, D.; Mendez, M.F.; Kramer, J.H.; Neuhaus, J.; van Swieten, J.C.; Seelaar, H.; Dopper, E.G.; Onyike, C.U.; et al. Sensitivity of Revised Diagnostic Criteria for the Behavioural Variant of Frontotemporal Dementia. Brain J. Neurol. 2011, 134, 2456–2477. [Google Scholar] [CrossRef]
- Strong, M.J.; Abrahams, S.; Goldstein, L.H.; Woolley, S.; McLaughlin, P.; Snowden, J.; Mioshi, E.; Roberts-South, A.; Benatar, M.; HortobaGyi, T.; et al. Amyotrophic Lateral Sclerosis—Frontotemporal Spectrum Disorder (ALS-FTSD): Revised Diagnostic Criteria. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 153–174. [Google Scholar] [CrossRef]
- Logroscino, G.; Piccininni, M.; Graff, C.; Hardiman, O.; Ludolph, A.C.; Moreno, F.; Otto, M.; Remes, A.M.; Rowe, J.B.; Seelaar, H.; et al. Incidence of Syndromes Associated With Frontotemporal Lobar Degeneration in 9 European Countries. JAMA Neurol. 2023, 80, 279. [Google Scholar] [CrossRef]
- Wolfson, C.; Gauvin, D.E.; Ishola, F.; Oskoui, M. Global Prevalence and Incidence of Amyotrophic Lateral Sclerosis. Neurology 2023, 101, e613–e623. [Google Scholar] [CrossRef] [PubMed]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Sanchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Marogianni, C.; Rikos, D.; Provatas, A.; Dadouli, K.; Ntellas, P.; Tsitsi, P.; Patrinos, G.; Dardiotis, E.; Hadjigeorgiou, G.; Xiromerisiou, G. The Role of C9orf72 in Neurodegenerative Disorders: A Systematic Review, an Updated Meta-Analysis, and the Creation of an Online Database. Neurobiol. Aging 2019, 84, 238.e25–238.e34. [Google Scholar] [CrossRef]
- Gargano, M.A.; Matentzoglu, N.; Coleman, B.; Addo-Lartey, E.B.; Anagnostopoulos, A.V.; Anderton, J.; Avillach, P.; Bagley, A.M.; Bakštein, E.; Balhoff, J.P.; et al. The Human Phenotype Ontology in 2024: Phenotypes around the World. Nucleic Acids Res. 2023, 52, D1333–D1346. [Google Scholar] [CrossRef]
- Stefanova, E.; Marjanović, A.; Dobričić, V.; Mandić-Stojmenović, G.; Stojković, T.; Branković, M.; Šarčević, M.; Novaković, I.; Kostić, V.S. Frequency of C9orf72, GRN, and MAPT Pathogenic Variants in Patients Recruited at the Belgrade Memory Center. Neurogenetics 2024, 25, 193–200. [Google Scholar] [CrossRef]
- Snowden, J.S.; Rollinson, S.; Thompson, J.C.; Harris, J.M.; Stopford, C.L.; Richardson, A.M.; Jones, M.; Gerhard, A.; Davidson, Y.S.; Robinson, A.; et al. Distinct Clinical and Pathological Characteristics of Frontotemporal Dementia Associated with C9ORF72 Mutations. Brain A J. Neurol. 2012, 135, 693–708. [Google Scholar] [CrossRef]
- Simon-Sanchez, J.; Dopper, E.G.; Cohn-Hokke, P.E.; Hukema, R.K.; Nicolaou, N.; Seelaar, H.; de Graaf, J.R.; de Koning, I.; van Schoor, N.M.; Deeg, D.J.; et al. The Clinical and Pathological Phenotype of C9ORF72 Hexanucleotide Repeat Expansions. Brain A J. Neurol. 2012, 135, 723–735. [Google Scholar] [CrossRef]
- Mehrabian, S.; Thonberg, H.; Raycheva, M.; Lilius, L.; Stoyanova, K.; Forsell, C.; Cavallin, L.; Nesheva, D.; Westman, E.; Toncheva, D.; et al. Phenotypic Variability and Neuropsychological Findings Associated with C9orf72 Repeat Expansions in a Bulgarian Dementia Cohort. PLoS ONE 2018, 13, e0208383. [Google Scholar] [CrossRef]
- Estevez-Fraga, C.; Magrinelli, F.; Hensman Moss, D.; Mulroy, E.; Di Lazzaro, G.; Latorre, A.; Mackenzie, M.; Houlden, H.; Tabrizi, S.J.; Bhatia, K.P. Expanding the Spectrum of Movement Disorders Associated With C9orf72 Hexanucleotide Expansions. Neurology. Genet. 2021, 7, e575. [Google Scholar] [CrossRef]
- Boeve, B.F.; Boylan, K.B.; Graff-Radford, N.R.; DeJesus-Hernandez, M.; Knopman, D.S.; Pedraza, O.; Vemuri, P.; Jones, D.; Lowe, V.; Murray, M.E.; et al. Characterization of Frontotemporal Dementia and/or Amyotrophic Lateral Sclerosis Associated with the GGGGCC Repeat Expansion in C9ORF72. Brain A J. Neurol. 2012, 135, 765–783. [Google Scholar] [CrossRef]
- Chiò, A.; Brunetti, M.; Barberis, M.; Iazzolino, B.; Montuschi, A.; Ilardi, A.; Cammarosano, S.; Canosa, A.; Moglia, C.; Calvo, A. The Role ofAPOEin the Occurrence of Frontotemporal Dementia in Amyotrophic Lateral Sclerosis. JAMA Neurol. 2016, 73, 425. [Google Scholar] [CrossRef] [PubMed]
- Lattante, S.; Pomponi, M.G.; Conte, A.; Marangi, G.; Bisogni, G.; Patanella, A.K.; Meleo, E.; Lunetta, C.; Riva, N.; Mosca, L.; et al. ATXN1 Intermediate-Length Polyglutamine Expansions Are Associated with Amyotrophic Lateral Sclerosis. Neurobiol. Aging 2018, 64, 157.e1–157.e5. [Google Scholar] [CrossRef]
- Lattante, S.; Millecamps, S.; Stevanin, G.; Rivaud-Péchoux, S.; Moigneu, C.; Camuzat, A.; Da Barroca, S.; Mundwiller, E.; Couarch, P.; Salachas, F.; et al. Contribution of ATXN2 Intermediary polyQ Expansions in a Spectrum of Neurodegenerative Disorders. Neurology 2014, 83, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, J.P.N.; De Andrade, H.M.T.; Cintra, V.P.; Bonadia, L.C.; Leoni, T.B.; De Albuquerque, M.; Martins, M.P.; De Borba, F.C.; Couteiro, R.E.D.; De Oliveira, D.S.; et al. CAG Repeats ≥ 34 in Ataxin-1 Gene Are Associated with Amyotrophic Lateral Sclerosis in a Brazilian Cohort. J. Neurol. Sci. 2020, 414, 116842. [Google Scholar] [CrossRef] [PubMed]
- Kaivorinne, A.L.; Bode, M.K.; Paavola, L.; Tuominen, H.; Kallio, M.; Renton, A.E.; Traynor, B.J.; Moilanen, V.; Remes, A.M. Clinical Characteristics of C9ORF72-Linked Frontotemporal Lobar Degeneration. Dement. Geriatr. Cogn. Disord. Extra 2013, 3, 251–262. [Google Scholar] [CrossRef]
- Chio, A.; Mora, G.; Sabatelli, M.; Caponnetto, C.; Lunetta, C.; Traynor, B.J.; Johnson, J.O.; Nalls, M.A.; Calvo, A.; Moglia, C.; et al. ATNX2 Is Not a Regulatory Gene in Italian Amyotrophic Lateral Sclerosis Patients with C9ORF72 GGGGCC Expansion. Neurobiol. Aging 2016, 39, 218.e5–218.e8. [Google Scholar] [CrossRef][Green Version]
- Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.; George-Hyslop, P.H.; Pericak-Vance, M.A.; Joo, S.H.; Rosi, B.L.; Gusella, J.F.; Crapper-MacLachlan, D.R.; Alberts, M.J.; et al. Association of Apolipoprotein E Allele Epsilon 4 with Late-Onset Familial and Sporadic Alzheimer’s Disease. Neurology 1993, 43, 1467–1472. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Pulst, S.-M.; Nechiporuk, A.; Nechiporuk, T.; Gispert, S.; Chen, X.-N.; Lopes-Cendes, I.; Pearlman, S.; Starkman, S.; Orozco-Diaz, G.; Lunkes, A.; et al. Moderate Expansion of a Normally Biallelic Trinucleotide Repeat in Spinocerebellar Ataxia Type 2. Nat. Genet. 1996, 14, 269–276. [Google Scholar] [CrossRef]
- Marjanović, A.; Stojmenović, A.; Dobričić, V.; Milićević, O.; Branković, M.; Virić, V.; Drinić, A.; Mandić-Stojmenović, G.; Janković, M.; Basta, I.; et al. C9orf72 Genetic Screening in Amyotrophic Lateral Sclerosis Patients from Serbia. Genetika 2023, 55, 1–18. [Google Scholar] [CrossRef]
- Conforti, F.L.; Spataro, R.; Sproviero, W.; Mazzei, R.; Cavalcanti, F.; Condino, F.; Simone, I.L.; Logroscino, G.; Patitucci, A.; Magariello, A.; et al. Ataxin-1 and Ataxin-2 Intermediate-Length PolyQ Expansions in Amyotrophic Lateral Sclerosis. Neurology 2012, 79, 2315–2320. [Google Scholar] [CrossRef] [PubMed]
- Elden, A.C.; Kim, H.J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 Intermediate-Length Polyglutamine Expansions Are Associated with Increased Risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef]
- Byrne, S.; Elamin, M.; Bede, P.; Shatunov, A.; Walsh, C.; Corr, B.; Heverin, M.; Jordan, N.; Kenna, K.; Lynch, C.; et al. Cognitive and Clinical Characteristics of Patients with Amyotrophic Lateral Sclerosis Carrying a C9orf72 Repeat Expansion: A Population-Based Cohort Study. Lancet Neurol. 2012, 11, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Gijselinck, I.; Van Langenhove, T.; van der Zee, J.; Sleegers, K.; Philtjens, S.; Kleinberger, G.; Janssens, J.; Bettens, K.; Van Cauwenberghe, C.; Pereson, S.; et al. A C9orf72 Promoter Repeat Expansion in a Flanders-Belgian Cohort with Disorders of the Frontotemporal Lobar Degeneration-Amyotrophic Lateral Sclerosis Spectrum: A Gene Identification Study. Lancet Neurol. 2012, 11, 54–65. [Google Scholar] [CrossRef]
- Dobson-Stone, C.; Hallupp, M.; Bartley, L.; Shepherd, C.E.; Halliday, G.M.; Schofield, P.R.; Hodges, J.R.; Kwok, J.B. C9ORF72 Repeat Expansion in Clinical and Neuropathologic Frontotemporal Dementia Cohorts. Neurology 2012, 79, 995–1001. [Google Scholar] [CrossRef]
- Ratti, A.; Corrado, L.; Castellotti, B.; Del Bo, R.; Fogh, I.; Cereda, C.; Tiloca, C.; D’Ascenzo, C.; Bagarotti, A.; Pensato, V.; et al. C9ORF72 Repeat Expansion in a Large Italian ALS Cohort: Evidence of a Founder Effect. Neurobiol. Aging 2012, 33, 2528.e7–2528.e14. [Google Scholar] [CrossRef]
- Borrego-Écija, S.; Turon-Sans, J.; Ximelis, T.; Aldecoa, I.; Molina-Porcel, L.; Povedano, M.; Rubio, M.A.; Gámez, J.; Cano, A.; Paré-Curell, M.; et al. Cognitive Decline in Amyotrophic Lateral Sclerosis: Neuropathological Substrate and Genetic Determinants. Brain Pathol. 2021, 31, e12942. [Google Scholar] [CrossRef] [PubMed]
- Canosa, A.; Pagani, M.; Brunetti, M.; Barberis, M.; Iazzolino, B.; Ilardi, A.; Cammarosano, S.; Manera, U.; Moglia, C.; Calvo, A.; et al. Correlation between Apolipoprotein E Genotype and Brain Metabolism in Amyotrophic Lateral Sclerosis. Eur. J. Neurol. 2019, 26, 306–312. [Google Scholar] [CrossRef]
- König, T.; Wurm, R.; Parvizi, T.; Silvaieh, S.; Hotzy, C.; Cetin, H.; Klotz, S.; Gelpi, E.; Bancher, C.; Benke, T.; et al. C9orf72 Repeat Length Might Influence Clinical Sub-Phenotypes in Dementia Patients. Neurobiol. Dis. 2022, 175, 105927. [Google Scholar] [CrossRef]
- Kohli, M.A.; John-Williams, K.; Rajbhandary, R.; Naj, A.; Whitehead, P.; Hamilton, K.; Carney, R.M.; Wright, C.; Crocco, E.; Gwirtzman, H.E.; et al. Repeat Expansions in the C9ORF72 Gene Contribute to Alzheimer’s Disease in Caucasians. Neurobiol. Aging 2013, 34, 1519.e5–1519.e12. [Google Scholar] [CrossRef]
- Mui, S.; Rebeck, G.W.; McKenna-Yasek, D.; Hyman, B.T.; Brown, R.H. Apolipoprotein E Epsilon 4 Allele Is Not Associated with Earlier Age at Onset in Amyotrophic Lateral Sclerosis. Ann. Neurol. 1995, 38, 460–463. [Google Scholar] [CrossRef]
- Geschwind, D.; Karrim, J.; Nelson, S.F.; Miller, B. The Apolipoprotein E Epsilon4 Allele Is Not a Significant Risk Factor for Frontotemporal Dementia. Ann. Neurol. 1998, 44, 134–138. [Google Scholar] [CrossRef]
- Rubino, E.; Vacca, A.; Govone, F.; De Martino, P.; Pinessi, L.; Rainero, I. Apolipoprotein E Polymorphisms in Frontotemporal Lobar Degeneration: A Meta-Analysis. Alzheimers Dement. 2013, 9, 706–713. [Google Scholar] [CrossRef]
- Rosas, I.; Martinez, C.; Coto, E.; Clarimon, J.; Lleo, A.; Illan-Gala, I.; Dols-Icardo, O.; Borroni, B.; Almeida, M.R.; van der Zee, J.; et al. Genetic Variation in APOE, GRN, and TP53 Are Phenotype Modifiers in Frontotemporal Dementia. Neurobiol. Aging 2021, 99, 99.e15–99.e22. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, S.L.; Boogaard, M.W.; Trompet, S.; de Mutsert, R.; Rosendaal, F.R.; Gussekloo, J.; Jukema, J.W.; Roos, R.A.C.; Aziz, N.A. Prevalence of Carriers of Intermediate and Pathological Polyglutamine Disease–Associated Alleles Among Large Population-Based Cohorts. JAMA Neurol. 2019, 76, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, C.; Sprovieri, T.; Morello, G.; Perrone, B.; Spampinato, A.G.; Simone, I.L.; Trojsi, F.; Monsurro, M.R.; Spataro, R.; La Bella, V.; et al. Genetic Investigation of Amyotrophic Lateral Sclerosis Patients in South Italy: A Two-Decade Analysis. Neurobiol. Aging 2021, 99, 99.e7–99.e14. [Google Scholar] [CrossRef] [PubMed]
- Rubino, E.; Mancini, C.; Boschi, S.; Ferrero, P.; Ferrone, M.; Bianca, S.; Zucca, M.; Orsi, L.; Pinessi, L.; Govone, F.; et al. ATXN2 Intermediate Repeat Expansions Influence the Clinical Phenotype in Frontotemporal Dementia. Neurobiol. Aging 2019, 73, 231.e7–231.e9. [Google Scholar] [CrossRef]

| Clinical Domain | Main Clinical Features with HP Terms [17] | Comments/Distinctions | References |
|---|---|---|---|
| ALS | Limb- or bulbar-onset weakness (HP:0003690; HP:0001283), UMN and LMN signs (HP:0002127; HP:0002366), early respiratory involvement (HP:0002098) | Most common phenotype; often positive family history | [14,15] |
| FTD | Executive dysfunction (HP:0033051), frontal memory disorder (HP:0100543), language impairment (HP:0002463), changes in behavior (HP:0000708) aphasia (HP:0002381), apathy (HP:0000741), disinhibition (HP:0000734), loss of empathy (HP:5200037), compulsive behaviors (HP:0000722) | bvFTD is the most frequent dementia subtype non-fluent/agrammatic PPA is less common than bvFTD | [18,19,20] |
| Overlap phenotypes | ALS/FTD—concomitant motor and cognitive/behavioral impairment (HP:0100543; HP:0000708) FTD/PSP—behavioral an cognitive changes (HP:0100543; HP:0000708) accompanied with motor and oculomotor deficits (HP:000605) | ALS/FTD reported in up to ~40–50% of carriers | [14,15,18,21] |
| Psychiatric features | Delusions (HP:0000746), psychosis (HP:0000709), hallucinations(HP:0000738), mood disorders (HP:0000712), paranoid schizophrenia (HP:0100753) | May precede neurological signs by years | [18,19] |
| Movement disorders | Parkinsonism (HP:0001300), motor stereotypes (HP:0000733), chorea (HP:0002072), oromandibular dyskinesia (HP:0012048), ataxia (HP:0001251), dystonia (HP:0001332), myoclonus (HP:0001336), tremor (HP:0001337) | Occasionally isolated or combined with ALS and/or FTD | [18,22,23] |
| Other dementias | Alzheimer’s disease, Unspecified dementia, Corticobasal degeneration, Lewy body dementia, sporadic Creutzfeld-Jacobs disease | Reported at a very low frequency | [16,18] |
| Primer Sequences | Reference | |
|---|---|---|
| C9orf72 | C9F: FAM-GAAACAACCGCAGCCTGTAG C9R: GCCTCCTCACTCACCCACT | in-house |
| C9orf72 repeat-primed PCR | C9orf72F: FAM-AGTCGCTAGAGGCGAAAGC C9orf72R: TACGCATCCCAGTTTGAGACGGGGGCCGGGGCCGGGGCCGGGG C9orf72A: TACGCATCCCAGTTTGAGACG | [14] |
| MRX-F: NED-TGTAAAACGACGGCCAGTCAAGGAGGGAAACAACCGCAGCC MRX-M13R: CAGGAAACAGCTATGACC MRX-R1:CAGGAAACAGCTATGACCGGGCCCGCCCCGACCACGCCCCGGCCCCGGCCCCGG | [15] | |
| ATXN1 | SCA1F: CCAACATGGGCAGTCTGAG SCA1R: FAM-TGGACGTACTGGTTCTGCTG | in-house |
| ATXN2 | SCA2F: GGGCCCCTCACCATGTCG SCA2R: VIC-CGGGCTTGCGGACATTGG | [32] |
| Characteristic | ALS/FTD (n = 22) | C9orf72+ (n = 6) | C9orf72– (n = 15) |
|---|---|---|---|
| Demographics and Genetic Features | |||
| Gender (male %) | 90% | 100% | 85.7% |
| GGGGCC expansion | 3 (30%) | – | – |
| Clinical Features | |||
| Limb weakness (HP:0003690) | 10 (45.5%) | 3 (42.9%) | 7 (46.7%) |
| Upper extremity (UE, HP:0003484) | 9 (90%) | 2 (66.7%) | 7 (100%) |
| Left | 1 (11.1%) | 1 (50%) | – |
| Right | 3 (33.3%) | 1 (50%) | 2 (28.6%) |
| Both | 5 (55.6%) | – | 5 (71.4%) |
| Lower extremity (LE, HP:0007340) | 1 (10%) | 1 (33.3%) | – |
| Left | 1 (100%) | 1 (100%) | – |
| Bulbar weakness (HP:0001283) | 11 (50%) | 3 (42.9%) | 8 (53.3%) |
| C9orf72 Expansion Positive | C9orf72 Expansion Negative | p Value | |
|---|---|---|---|
| Gender | 0.867 | ||
| male | 57.14% | 53.33% | |
| female | 42.86% | 46.67% | |
| Onset | 0.890 | ||
| spinal | 50% | 46.67% | |
| bulbar | 50% | 53.33% | |
| Positive family history | 50% | 30% | 0.424 |
| Age of onset | 0.264 | ||
| Mean ± SD | 56.43 ± 9 | 61.07 ± 8.75 | |
| (95% CI) | (48.11–64.75) | (56.22–65.91) | |
| Range:min-max | 42.0–67.0 | 46.0–74.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marjanovic, A.; Stefanova, E.; Viric, V.; Palibrk, A.; Mandić Stojmenović, G.; Stojković, T.; Stojadinovic, L.; Basta, I.; Novakovic, I.; Stević, Z.; et al. Genetic and Clinical Insights into ALS/FTD: Profiling a Rare Cohort to Explore Spectrum Heterogeneity. J. Pers. Med. 2025, 15, 451. https://doi.org/10.3390/jpm15100451
Marjanovic A, Stefanova E, Viric V, Palibrk A, Mandić Stojmenović G, Stojković T, Stojadinovic L, Basta I, Novakovic I, Stević Z, et al. Genetic and Clinical Insights into ALS/FTD: Profiling a Rare Cohort to Explore Spectrum Heterogeneity. Journal of Personalized Medicine. 2025; 15(10):451. https://doi.org/10.3390/jpm15100451
Chicago/Turabian StyleMarjanovic, Ana, Elka Stefanova, Vanja Viric, Aleksa Palibrk, Gorana Mandić Stojmenović, Tanja Stojković, Lenka Stojadinovic, Ivana Basta, Ivana Novakovic, Zorica Stević, and et al. 2025. "Genetic and Clinical Insights into ALS/FTD: Profiling a Rare Cohort to Explore Spectrum Heterogeneity" Journal of Personalized Medicine 15, no. 10: 451. https://doi.org/10.3390/jpm15100451
APA StyleMarjanovic, A., Stefanova, E., Viric, V., Palibrk, A., Mandić Stojmenović, G., Stojković, T., Stojadinovic, L., Basta, I., Novakovic, I., Stević, Z., & Jankovic, M. (2025). Genetic and Clinical Insights into ALS/FTD: Profiling a Rare Cohort to Explore Spectrum Heterogeneity. Journal of Personalized Medicine, 15(10), 451. https://doi.org/10.3390/jpm15100451






