Pharmacogenetics of Long-Term Outcomes of Schizophrenia Spectrum Disorders: The Functional Role of CYP2D6 and CYP2C19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Outcome Measures
2.3. Covariables
2.4. Genotyping, Quality Control, and Imputation of Non-Genotyped Variants
2.5. Predicting Metabolizer Status
2.6. Polygenic Risk Scores
2.7. Data Analysis and Statistical Modeling
3. Results
3.1. Descriptive Statistics
3.2. Metabolizer Phenotype Status
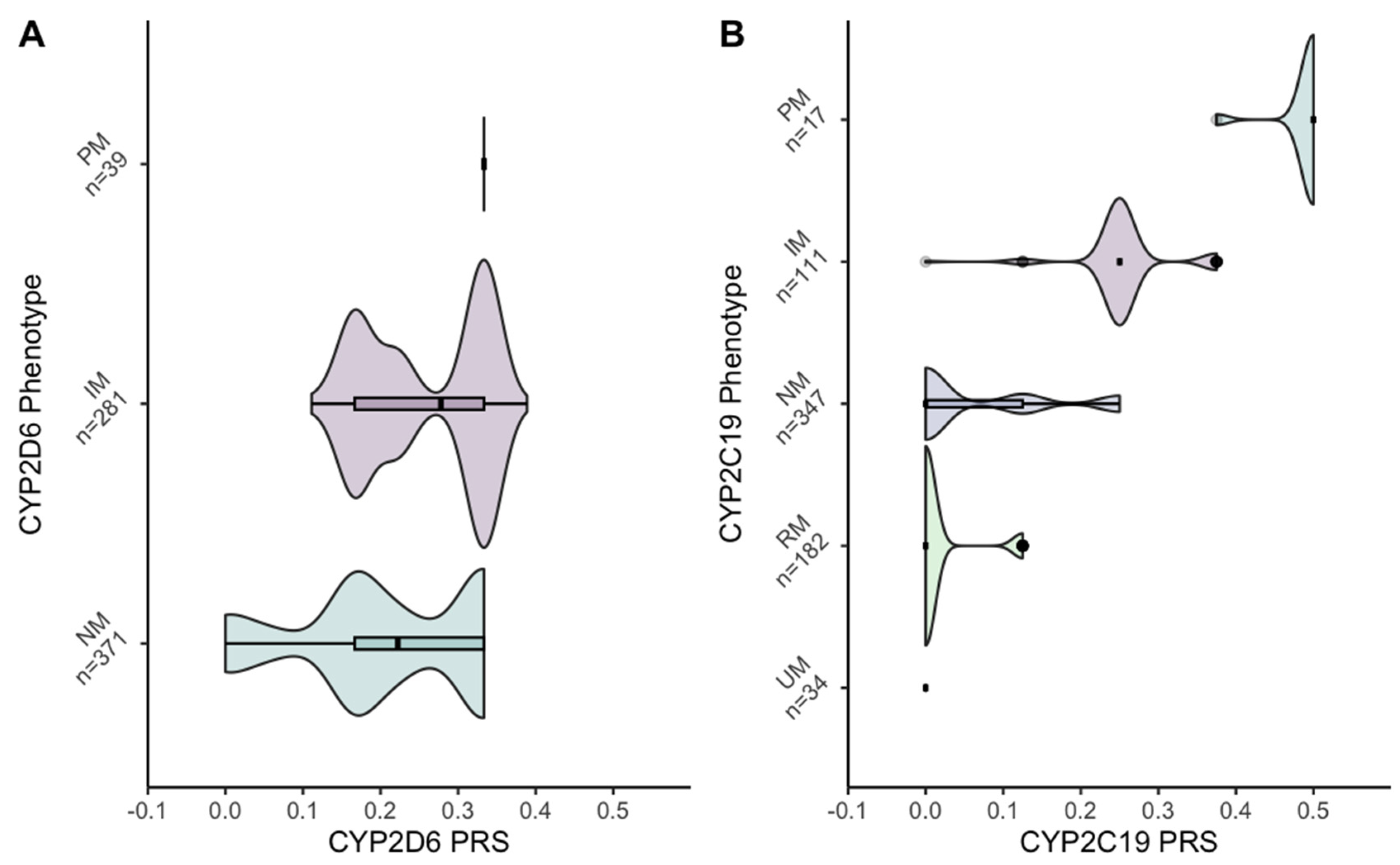
3.3. Polygenic Risk Scores
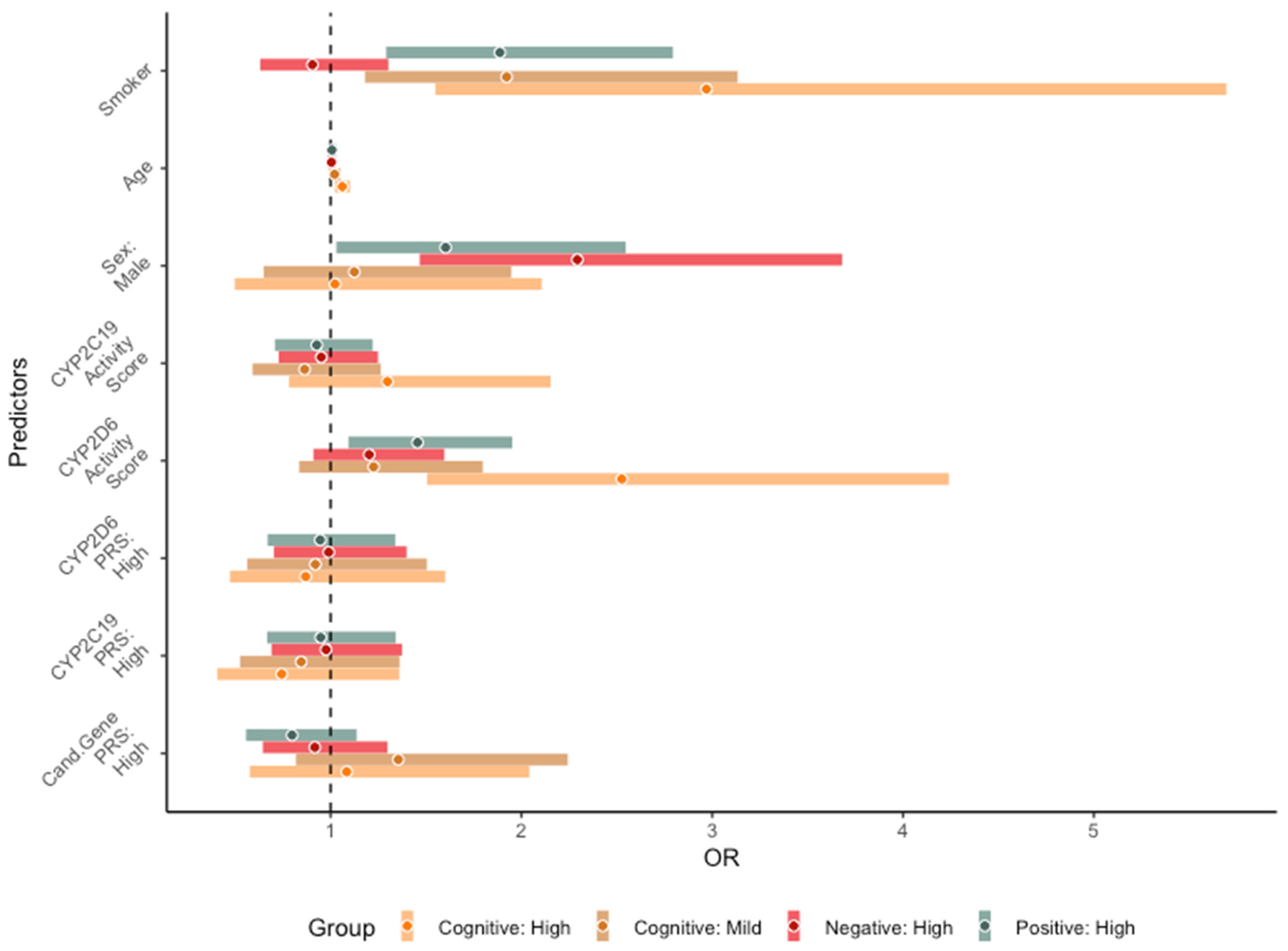
3.4. Long-Term Core Symptom Severity Trajectories
3.5. Cardiometabolic Variables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charlson, F.J.; Ferrari, A.J.; Santomauro, D.F.; Diminic, S.; Stockings, E.; Scott, J.G.; McGrath, J.J.; Whiteford, H.A. Global Epidemiology and Burden of Schizophrenia: Findings from the Global Burden of Disease Study 2016. Schizophr. Bull. 2018, 44, 1195–1203. [Google Scholar] [CrossRef]
- Malaspina, D.; Owen, M.J.; Heckers, S.; Tandon, R.; Bustillo, J.; Schultz, S.; Barch, D.M.; Gaebel, W.; Gur, R.E.; Tsuang, M.; et al. Schizoaffective Disorder in the DSM-5. Schizophr. Res. 2013, 150, 21–25. [Google Scholar] [CrossRef]
- Zhang, J.-P.; Malhotra, A.K. Pharmacogenetics and Antipsychotics: Therapeutic Efficacy and Side Effects Prediction. Expert Opin. Drug Metab. Toxicol. 2011, 7, 9–37. [Google Scholar] [CrossRef]
- Arranz, M.J.; Salazar, J.; Hernández, M.H. Pharmacogenetics of Antipsychotics: Clinical Utility and Implementation. Behav. Brain Res. 2021, 401, 113058. [Google Scholar] [CrossRef] [PubMed]
- Lisoway, A.J.; Chen, C.C.; Zai, C.C.; Tiwari, A.K.; Kennedy, J.L. Toward Personalized Medicine in Schizophrenia: Genetics and Epigenetics of Antipsychotic Treatment. Schizophr. Res. 2021, 232, 112–124. [Google Scholar] [CrossRef]
- Weston-Green, K. Antipsychotic Drug Development: From Historical Evidence to Fresh Perspectives. Front. Psychiatry 2022, 13, 903156. [Google Scholar] [CrossRef]
- Remington, G.; Hahn, M.K.; Agarwal, S.M.; Chintoh, A.; Agid, O. Schizophrenia: Antipsychotics and Drug Development. Behav. Brain Res. 2021, 414, 113507. [Google Scholar] [CrossRef]
- Keefe, R.S.E.; Bilder, R.M.; Davis, S.M.; Harvey, P.D.; Palmer, B.W.; Gold, J.M.; Meltzer, H.Y.; Green, M.F.; Capuano, G.; Stroup, T.S.; et al. Neurocognitive Effects of Antipsychotic Medications in Patients with Chronic Schizophrenia in the CATIE Trial. Arch. Gen. Psychiatry 2007, 64, 633–647. [Google Scholar] [CrossRef]
- McCutcheon, R.A.; Keefe, R.S.E.; McGuire, P.K. Cognitive Impairment in Schizophrenia: Aetiology, Pathophysiology, and Treatment. Mol. Psychiatry 2023, 1–17. [Google Scholar] [CrossRef]
- Zhang, J.-P.; Malhotra, A.K. Recent Progress in Pharmacogenomics of Antipsychotic Drug Response. Curr. Psychiatry Rep. 2018, 20, 24. [Google Scholar] [CrossRef]
- Butler, M.G. Pharmacogenetics and Psychiatric Care: A Review and Commentary. J. Ment. Health Clin. Psychol. 2018, 2, 17. [Google Scholar] [CrossRef]
- Teng, Y.; Sandhu, A.; Liemburg, E.J.; Naderi, E.; Alizadeh, B.Z. The Progress and Pitfalls of Pharmacogenetics-Based Precision Medicine in Schizophrenia Spectrum Disorders: A Systematic Review and Meta-Analysis. J. Pers. Med. 2023, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.C.F.; Wirgenes, K.V.; Haram, M.; Bettella, F.; Lunding, S.H.; Rødevand, L.; Hjell, G.; Agartz, I.; Djurovic, S.; Melle, I.; et al. Indicated Association between Polygenic Risk Score and Treatment-Resistance in a Naturalistic Sample of Patients with Schizophrenia Spectrum Disorders. Schizophr. Res. 2020, 218, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Arranz, M.; Munro, J.; Birkett, J.; Bolonna, A.; Mancama, D.; Sodhi, M.; Lesch, K.; Meyer, J.; Sham, P.; Collier, D.; et al. Pharmacogenetic Prediction of Clozapine Response. Lancet 2000, 355, 1615–1616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-P.; Lencz, T.; Zhang, R.X.; Nitta, M.; Maayan, L.; John, M.; Robinson, D.G.; Fleischhacker, W.W.; Kahn, R.S.; Ophoff, R.A.; et al. Pharmacogenetic Associations of Antipsychotic Drug-Related Weight Gain: A Systematic Review and Meta-Analysis. Schizophr. Bull. 2016, 42, 1418–1437. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.; Wilke, M.A.P.; Lyle, S.M.; Kowalec, K.; Jorgensen, A.; Wright, G.E.B.; Drögemöller, B.I. A Systematic Review and Analysis of the Use of Polygenic Scores in Pharmacogenomics. Clin. Pharmacol. Ther. 2022, 111, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yoshikawa, A.; Brennan, M.; Ramsey, T.; Meltzer, H.Y. Genetic Predictors of Antipsychotic Response to Lurasidone Identified in a Genome Wide Association Study and by Schizophrenia Risk Genes. Schizophr. Res. 2018, 192, 194–204. Available online: https://doi.org/10.1016/j.schres.2017.04.009 (accessed on 13 June 2022). [CrossRef] [PubMed]
- Zhang, J.-P.; Robinson, D.; Yu, J.; Gallego, J.; Fleischhacker, W.W.; Kahn, R.S.; Crespo-Facorro, B.; Vazquez-Bourgon, J.; Kane, J.M.; Malhotra, A.K.; et al. Schizophrenia Polygenic Risk Score as a Predictor of Antipsychotic Efficacy in First-Episode Psychosis. Am. J. Psychiatry 2019, 176, 21–28. Available online: https://ajp.psychiatryonline.org/doi/epub/10.1176/appi.ajp.2018.17121363 (accessed on 30 March 2022). [CrossRef]
- Hettige, N.C.; Cole, C.B.; Khalid, S.; De Luca, V. Polygenic Risk Score Prediction of Antipsychotic Dosage in Schizophrenia. Schizophr. Res. 2016, 170, 265–270. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Mkrtchian, S.; Zhou, Y.; Lauschke, V.M. Integrating Rare Genetic Variants into Pharmacogenetic Drug Response Predictions. Hum. Genom. 2018, 12, 26. [Google Scholar] [CrossRef]
- Wannasuphoprasit, Y.; Andersen, S.E.; Arranz, M.J.; Catalan, R.; Jurgens, G.; Kloosterboer, S.M.; Rasmussen, H.B.; Bhat, A.; Irizar, H.; Koller, D.; et al. CYP2D6 Genetic Variation and Antipsychotic-Induced Weight Gain: A Systematic Review and Meta-Analysis. Front. Psychol. 2022, 12, 768748. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.E.; Fonseka, T.M.; Bousman, C.A.; Müller, D.J. Gene-Drug Pairings for Antidepressants and Antipsychotics: Level of Evidence and Clinical Application. Mol. Psychiatry 2022, 27, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Korver, N.; Quee, P.J.; Boos, H.B.M.; Simons, C.J.P.; de Haan, L. Genetic Risk and Outcome of Psychosis (GROUP), a Multi Site Longitudinal Cohort Study Focused on Gene–Environment Interaction: Objectives, Sample Characteristics, Recruitment and Assessment Methods. Int. J. Methods Psychiatr. Res. 2012, 21, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.R.; Fiszbein, A.; Opler, L.A. The Positive and Negative Syndrome Scale (PANSS) for Schizophrenia. Schizophr. Bull. 1987, 13, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Galderisi, S.; Mucci, A.; Dollfus, S.; Nordentoft, M.; Falkai, P.; Kaiser, S.; Giordano, G.M.; Vandevelde, A.; Nielsen, M.Ø.; Glenthøj, L.B.; et al. EPA Guidance on Assessment of Negative Symptoms in Schizophrenia. Eur. Psychiatry 2021, 64, e23. [Google Scholar] [CrossRef]
- Nuechterlein, K.H.; Barch, D.M.; Gold, J.M.; Goldberg, T.E.; Green, M.F.; Heaton, R.K. Identification of Separable Cognitive Factors in Schizophrenia. Schizophr. Res. 2004, 72, 29–39. [Google Scholar] [CrossRef]
- Nuechterlein, K.H.; Green, M.F.; Kern, R.S.; Baade, L.E.; Barch, D.M.; Cohen, J.D.; Essock, S.; Fenton, W.S.; Frese, F.J.; Gold, J.M.; et al. The MATRICS Consensus Cognitive Battery, Part 1: Test Selection, Reliability, and Validity. Am. J. Psychiatry 2008, 165, 203–213. [Google Scholar] [CrossRef]
- Kern, R.S.; Nuechterlein, K.H.; Green, M.F.; Baade, L.E.; Fenton, W.S.; Gold, J.M.; Keefe, R.S.E.; Mesholam-Gately, R.; Mintz, J.; Seidman, L.J.; et al. The MATRICS Consensus Cognitive Battery, Part 2: Co-Norming and Standardization. Am. J. Psychiatry 2008, 165, 214–220. [Google Scholar] [CrossRef]
- Habtewold, T.D.; Liemburg, E.J.; Islam, M.A.; de Zwarte, S.M.C.; Boezen, H.M.; Bruggeman, R.; Alizadeh, B.Z.; Luykx, J.J.; Rutten, B.P.F.; van Winkel, R.; et al. Association of Schizophrenia Polygenic Risk Score with Data-Driven Cognitive Subtypes: A Six-Year Longitudinal Study in Patients, Siblings and Controls. Schizophr. Res. 2020, 223, 135–147. [Google Scholar] [CrossRef]
- Austin-Zimmerman, I.; Wronska, M.; Wang, B.; Irizar, H.; Thygesen, J.H.; Bhat, A.; Denaxas, S.; Fatemifar, G.; Finan, C.; Harju-Seppänen, J.; et al. The Influence of CYP2D6 and CYP2C19 Genetic Variation on Diabetes Mellitus Risk in People Taking Antidepressants and Antipsychotics. Genes 2021, 12, 1758. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wheeler, M.M.; Thummel, K.E.; Nickerson, D.A. Calling Star Alleles with Stargazer in 28 Pharmacogenes with Whole Genome Sequences. Clin. Pharmacol. Ther. 2019, 106, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- PharmVar. Gene Info:CYP2D6. 2022. Available online: https://www.pharmvar.org/gene/CYP2D6 (accessed on 8 February 2023).
- Pratt, V.M.; Cavallari, L.H.; Del Tredici, A.L.; Gaedigk, A.; Hachad, H.; Ji, Y.; Kalman, L.V.; Ly, R.C.; Moyer, A.M.; Scott, S.A.; et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection. J. Mol. Diagn. 2021, 23, 1047–1064. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.r-project.org/ (accessed on 23 March 2023).
- Champely, S.; Ekstrom, C.; Dalgaard, P.; Gill, J.; Weibelzahl, S.; Anandkumar, A.; Ford, C.; Volcic, R.; De Rosario, H. pwr: Basic Functions for Power Analysis. 2017. Available online: https://cran.r-project.org/web/packages/pwr/ (accessed on 15 August 2022).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Okhuijsen-Pfeifer, C.; van der Horst, M.Z.; Bousman, C.A.; Lin, B.; van Eijk, K.R.; Ripke, S.; Ayhan, Y.; Babaoglu, M.O.; Bak, M.; Alink, W.; et al. Genome-Wide Association Analyses of Symptom Severity among Clozapine-Treated Patients with Schizophrenia Spectrum Disorders. Transl. Psychiatry 2022, 12, 145. [Google Scholar] [CrossRef]
- Lu, J.; Yang, Y.; Lu, J.; Wang, Z.; He, Y.; Yan, Y.; Fu, K.; Jiang, W.; Xu, Y.; Wu, R.; et al. Effect of CYP2D6 Polymorphisms on Plasma Concentration and Therapeutic Effect of Risperidone. BMC Psychiatry 2021, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Lesche, D.; Mostafa, S.; Everall, I.; Pantelis, C.; Bousman, C.A. Impact of CYP1A2, CYP2C19, and CYP2D6 Genotype- and Phenoconversion-Predicted Enzyme Activity on Clozapine Exposure and Symptom Severity. Pharmacogenomics J. 2020, 20, 192–201. [Google Scholar] [CrossRef]
- Paribello, P.; Manchia, M.; Pinna, F.; Isayeva, U.; Squassina, A.; Pisanu, C.; Balderi, L.; Contu, M.; Pinna, M.; Carpiniello, B. Pharmacokinetic Markers of Clinical Outcomes in Severe Mental Illness: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 4776. [Google Scholar] [CrossRef]
- Viviani, R.; Messina, I.; Bosch, J.E.; Dommes, L.; Paul, A.; Schneider, K.L.; Scholl, C.; Stingl, J.C. Effects of Genetic Variability of CYP2D6 on Neural Substrates of Sustained Attention during On-Task Activity. Transl. Psychiatry 2020, 10, 338. [Google Scholar] [CrossRef]
- Ma, L.; Shcherbina, A.; Chetty, S. Variations and Expression Features of CYP2D6 Contribute to Schizophrenia Risk. Mol. Psychiatry 2021, 26, 2605–2615. [Google Scholar] [CrossRef]
- Morozova, A.; Zorkina, Y.; Abramova, O.; Pavlova, O.; Pavlov, K.; Soloveva, K.; Volkova, M.; Alekseeva, P.; Andryshchenko, A.; Kostyuk, G.; et al. Neurobiological Highlights of Cognitive Impairment in Psychiatric Disorders. Int. J. Mol. Sci. 2022, 23, 1217. [Google Scholar] [CrossRef] [PubMed]
- Ohi, K.; Sumiyoshi, C.; Fujino, H.; Yasuda, Y.; Yamamori, H.; Fujimoto, M.; Shiino, T.; Sumiyoshi, T.; Hashimoto, R. Genetic Overlap between General Cognitive Function and Schizophrenia: A Review of Cognitive GWASs. Int. J. Mol. Sci. 2018, 19, 3822. [Google Scholar] [CrossRef]
- Sosin, D.; Ivashchenko, D.; Sozaeva, Z.; Ryzhikova, K.; Fadeeva, V.; Chomskaya, V.; Sheidakov, R.; Yanushko, M.; Otmakhov, A.; Grishina, E.; et al. Cognitive Impairment in Patients with Treatment Resistant Schizophrenia: Associations with DRD2, DRD3, HTR2A, BDNF and CYP2D6 Genetic Polymorphisms. Neurol. Psychiatry Brain Res. 2019, 33, 48–55. [Google Scholar] [CrossRef]
- Richards-Belle, A.; Austin-Zimmerman, I.; Wang, B.; Zartaloudi, E.; Cotic, M.; Gracie, C.; Saadullah Khani, N.; Wannasuphoprasit, Y.; Wronska, M.; Dawda, Y.; et al. Associations of Antidepressants and Antipsychotics with Lipid Parameters: Do CYP2C19/CYP2D6 Genes Play a Role? A UK Population-Based Study. J. Psychopharmacol. 2023, 37, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Pillinger, T.; McCutcheon, R.A.; Vano, L.; Mizuno, Y.; Arumuham, A.; Hindley, G.; Beck, K.; Natesan, S.; Efthimiou, O.; Cipriani, A.; et al. Comparative Effects of 18 Antipsychotics on Metabolic Function in Patients with Schizophrenia, Predictors of Metabolic Dysregulation, and Association with Psychopathology: A Systematic Review and Network Meta-Analysis. Lancet Psychiatry 2020, 7, 64–77. [Google Scholar] [CrossRef]
- Klomp, S.D.; Manson, M.L.; Guchelaar, H.-J.; Swen, J.J. Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. J. Clin. Med. 2020, 9, 2890. [Google Scholar] [CrossRef]
- Huang, H.; Dong, M.; Zhang, L.; Zhong, B.-L.; Ng, C.H.; Ungvari, G.S.; Yuan, Z.; Meng, X.; Xiang, Y.-T. Psychopathology and Extrapyramidal Side Effects in Smoking and Non-Smoking Patients with Schizophrenia: Systematic Review and Meta-Analysis of Comparative Studies. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 92, 476–482. [Google Scholar] [CrossRef]
- Coustals, N.; Martelli, C.; Brunet-Lecomte, M.; Petillion, A.; Romeo, B.; Benyamina, A. Chronic Smoking and Cognition in Patients with Schizophrenia: A Meta-Analysis. Schizophr. Res. 2020, 222, 113–121. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Cui, C.; Liu, Z.; Lu, J. Gray Matter Morphological Anomalies in the Cerebellar Vermis in First-Episode Schizophrenia Patients with Cognitive Deficits. BMC Psychiatry 2017, 17, 374. [Google Scholar] [CrossRef]
- Sagud, M.; Mihaljevic Peles, A.; Pivac, N. Smoking in Schizophrenia: Recent Findings about an Old Problem. Curr. Opin. Psychiatry 2019, 32, 402. [Google Scholar] [CrossRef]
- Quik, M.; Boyd, J.T.; Bordia, T.; Perez, X. Potential Therapeutic Application for Nicotinic Receptor Drugs in Movement Disorders. Nicotine Tob. Res. 2018, 21, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Terry, A.V.; Callahan, P.M. A7 Nicotinic Acetylcholine Receptors as Therapeutic Targets in Schizophrenia: Update on Animal and Clinical Studies and Strategies for the Future. Neuropharmacology 2020, 170, 108053. [Google Scholar] [CrossRef] [PubMed]
- Koromina, M.; Koutsilieri, S.; Patrinos, G.P. Delineating Significant Genome-Wide Associations of Variants with Antipsychotic and Antidepressant Treatment Response: Implications for Clinical Pharmacogenomics. Hum. Genom. 2020, 14, 4. [Google Scholar] [CrossRef]
- Allen, J.D.; Bishop, J.R. A Systematic Review of Genome-Wide Association Studies of Antipsychotic Response. Pharmacogenomics 2019, 20, 291–306. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping Genomic Loci Implicates Genes and Synaptic Biology in Schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Siemens, A.; Anderson, S.J.; Rassekh, S.R.; Ross, C.J.D.; Carleton, B.C. A Systematic Review of Polygenic Models for Predicting Drug Outcomes. J. Pers. Med. 2022, 12, 1394. [Google Scholar] [CrossRef]
- Alchakee, A.; Ahmed, M.; Eldohaji, L.; Alhaj, H.; Saber-Ayad, M. Pharmacogenomics in Psychiatry Practice: The Value and the Challenges. Int. J. Mol. Sci. 2022, 23, 13485. [Google Scholar] [CrossRef]
- Wolff, J.; Hefner, G.; Normann, C.; Kaier, K.; Binder, H.; Hiemke, C.; Toto, S.; Domschke, K.; Marschollek, M.; Klimke, A. Polypharmacy and the Risk of Drug–Drug Interactions and Potentially Inappropriate Medications in Hospital Psychiatry. Pharmacoepidemiol. Drug Saf. 2021, 30, 1258–1268. [Google Scholar] [CrossRef]

| Baseline Measures | Longitudinal Measures (6 Year Follow-Up) | ||
|---|---|---|---|
| Age (years), mean (SE) | 27.71 (0.27) | Antipsychotic use N (%): | |
| Sex (male), N (%) | 525 (75.98) | AAP use | 485 (70.19) |
| Education (years), mean (SE) | 12.46 (0.15) | TAP use | 42 (6.08) |
| Smokers N (%) | 449 (64.98) | Mixed (AAP & TAP) | 57 (8.25) |
| Age onset illness (years), mean (SE) | 23.19 (0.23) | Unknown | 107 (15.48) |
| Duration of Illness (years), mean (SE) | 4.97 (0.17) | Positive symptoms N (%): | |
| Psychotic episodes, mean (SE) | 1.72 (1.17) | Low | 491 (71.06) |
| Systolic BP (mmHg), mean (SD) | 127.35 (15.68) | High | 200 (28.94) |
| Diastolic BP (mmHg), mean (SD) | 79.38 (11.36) | Negative symptoms N (%): | |
| Pulse rate (beat/min), mean (SD) | 75.30 (15.80) | Low | 489 (70.77) |
| Triglycerides (mmol/L), median (IQR) | 1.84 (1.30) | High | 202 (29.23) |
| HDL (mmol/L), mean (SD) | 1.27 (0.74) | Cognitive impairments N (%): | |
| LDL (mmol/L), mean (SD) | 3.12 (0.94) | None | 84 (12.16) |
| HbA1c (mmol/mol), mean (SD) | 35.01 (6.05) | Mild | 500 (72.36) |
| BMI (kg/m2), mean (SD) | 26.01 (4.72) | High | 107 (15.48) |
| Waist circumference (cm), mean (SD) | 95.08 (14.13) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandhu, A.K.; Naderi, E.; Wijninga, M.J.; Liemburg, E.J.; GROUP Investigators; Cath, D.; Bruggeman, R.; Alizadeh, B.Z. Pharmacogenetics of Long-Term Outcomes of Schizophrenia Spectrum Disorders: The Functional Role of CYP2D6 and CYP2C19. J. Pers. Med. 2023, 13, 1354. https://doi.org/10.3390/jpm13091354
Sandhu AK, Naderi E, Wijninga MJ, Liemburg EJ, GROUP Investigators, Cath D, Bruggeman R, Alizadeh BZ. Pharmacogenetics of Long-Term Outcomes of Schizophrenia Spectrum Disorders: The Functional Role of CYP2D6 and CYP2C19. Journal of Personalized Medicine. 2023; 13(9):1354. https://doi.org/10.3390/jpm13091354
Chicago/Turabian StyleSandhu, Amrit K., Elnaz Naderi, Morenika J. Wijninga, Edith J. Liemburg, GROUP Investigators, Danielle Cath, Richard Bruggeman, and Behrooz Z. Alizadeh. 2023. "Pharmacogenetics of Long-Term Outcomes of Schizophrenia Spectrum Disorders: The Functional Role of CYP2D6 and CYP2C19" Journal of Personalized Medicine 13, no. 9: 1354. https://doi.org/10.3390/jpm13091354
APA StyleSandhu, A. K., Naderi, E., Wijninga, M. J., Liemburg, E. J., GROUP Investigators, Cath, D., Bruggeman, R., & Alizadeh, B. Z. (2023). Pharmacogenetics of Long-Term Outcomes of Schizophrenia Spectrum Disorders: The Functional Role of CYP2D6 and CYP2C19. Journal of Personalized Medicine, 13(9), 1354. https://doi.org/10.3390/jpm13091354







