Discovery of Biomarkers Related to Interstitial Fibrosis and Tubular Atrophy among Kidney Transplant Recipients by mRNA-Sequencing
Abstract
1. Introduction
2. Materials and Methods
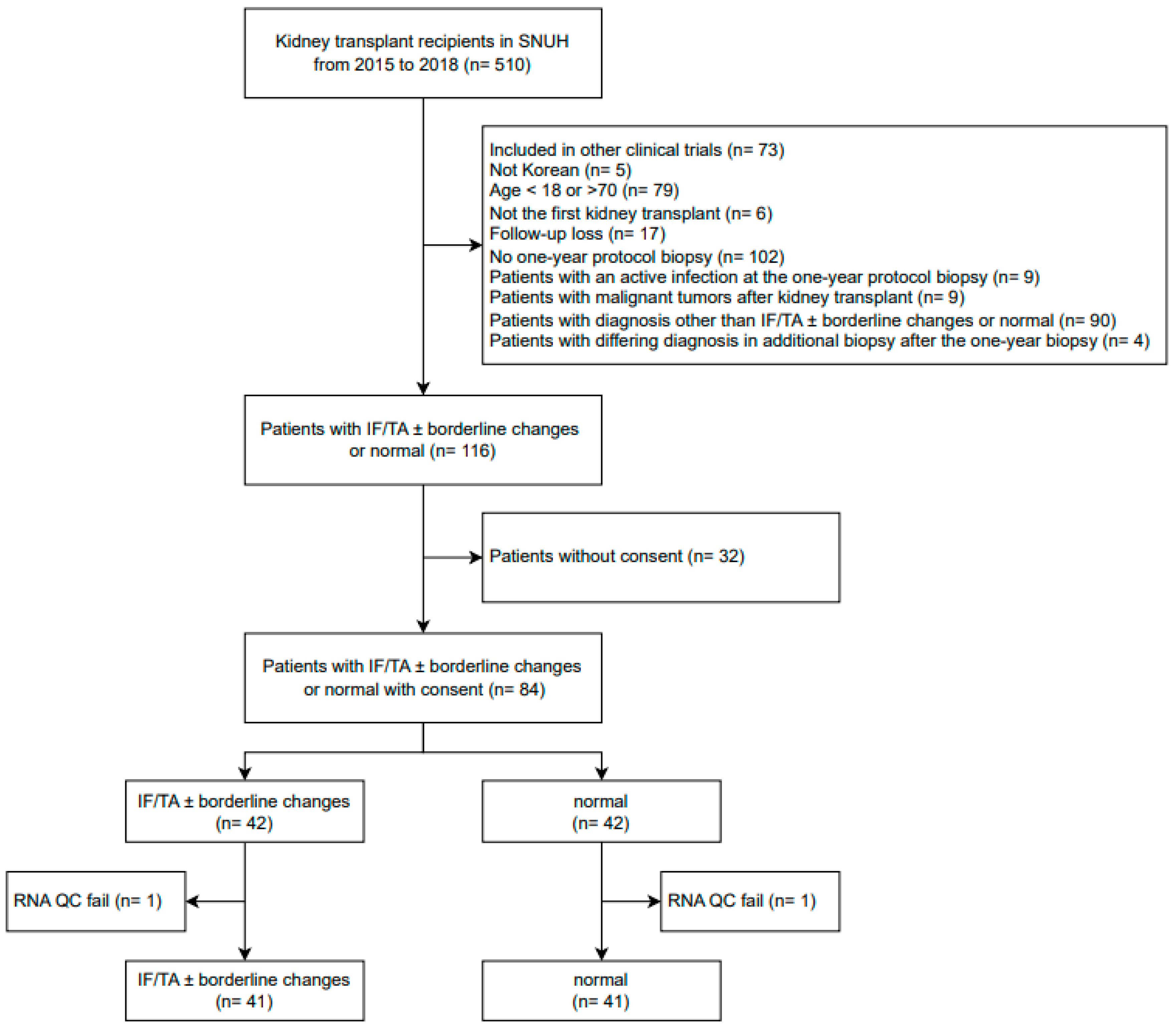
2.1. Study Population
2.2. Sample Preparation
2.3. mRNA-Sequencing
2.4. DEG Analysis
2.5. Ontology Analysis
2.6. Protein–Protein Interaction (PPI) Network Analysis
2.7. qPCR Validation of the Gene-Expression Profiles
2.8. Statistical Analysis
3. Results
3.1. Baseline Clinical Characteristics of Patients
3.2. DEGs Related to IF/TA
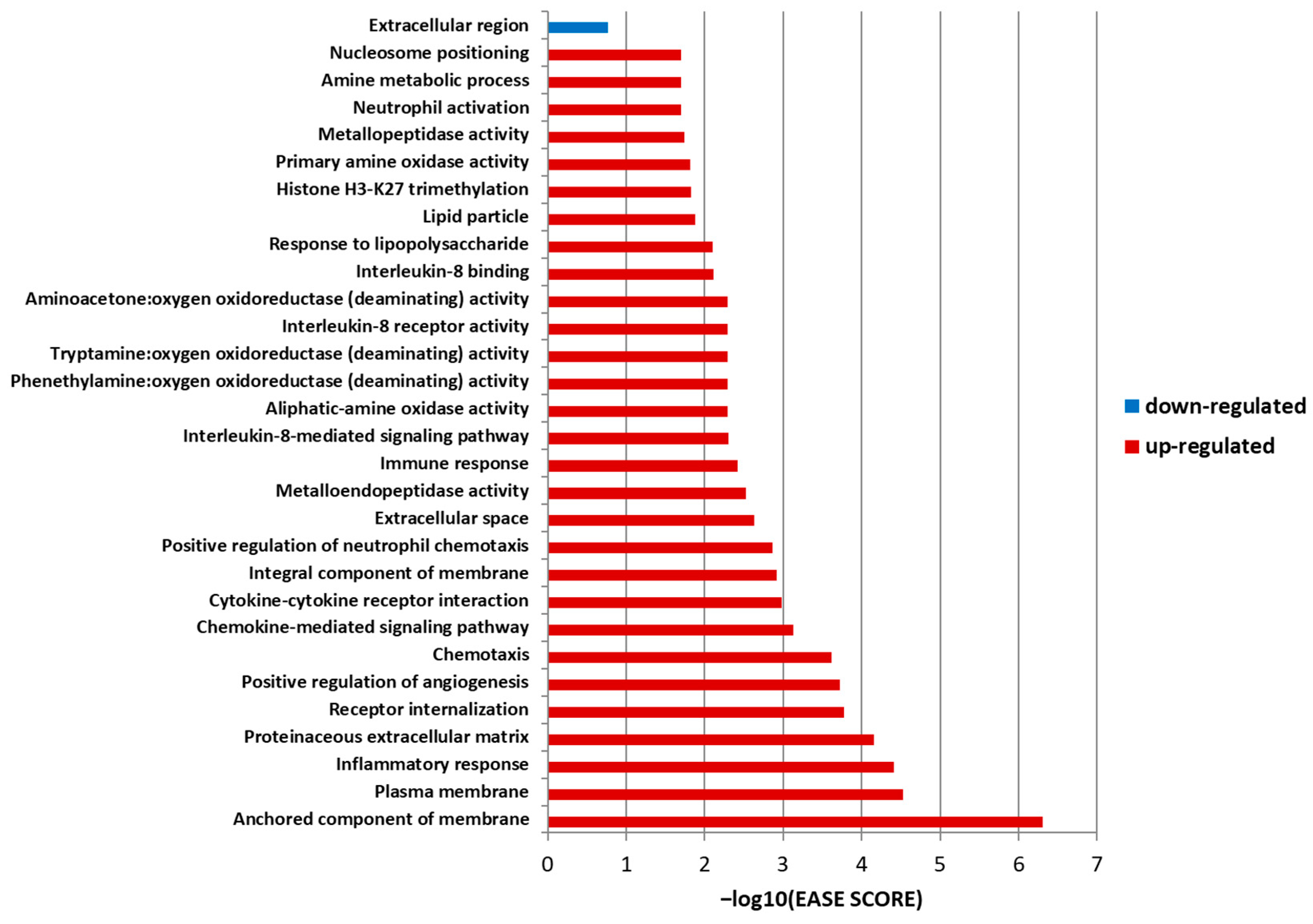
3.3. Ontology Analysis
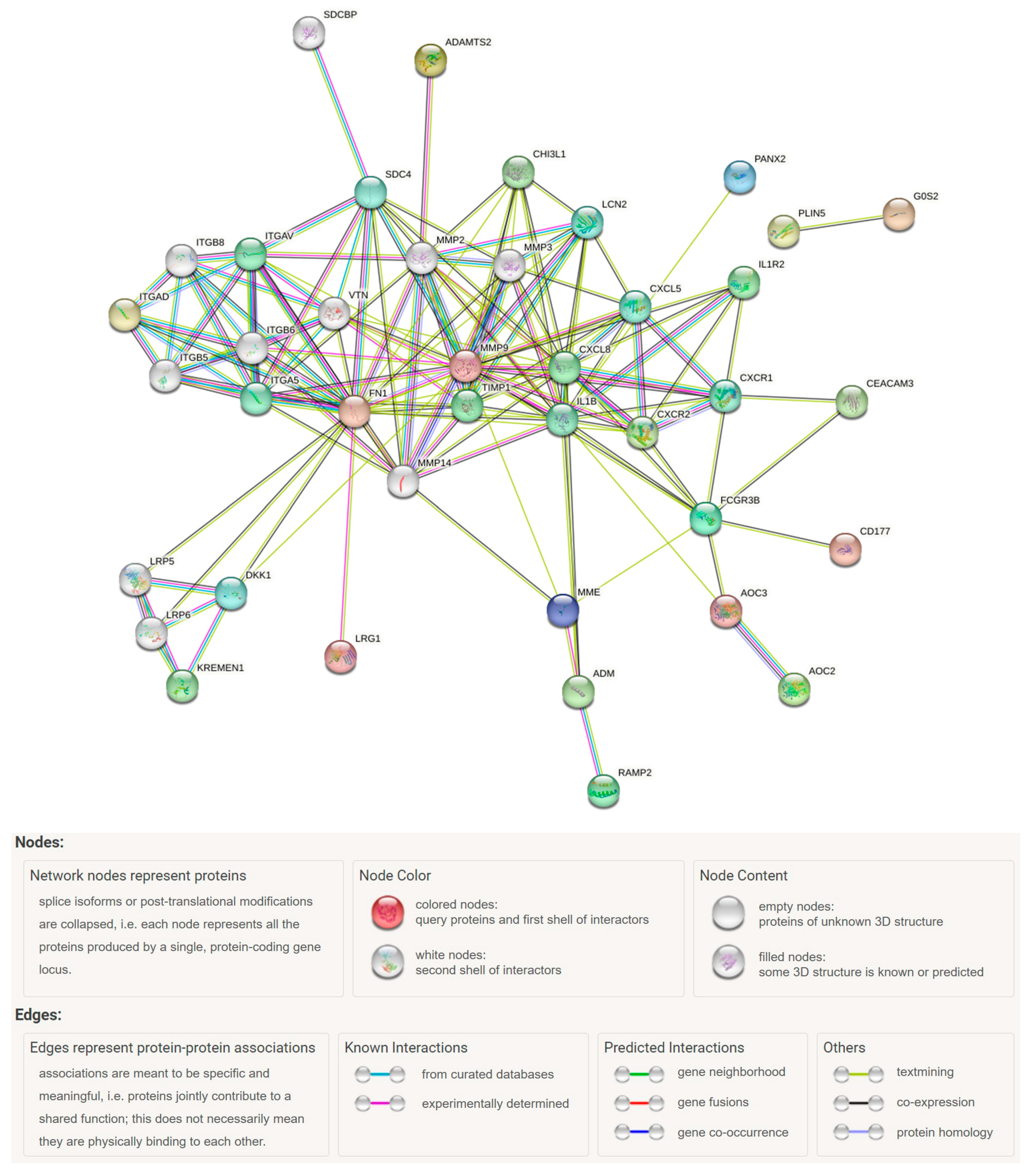
3.4. PPI Network Analysis
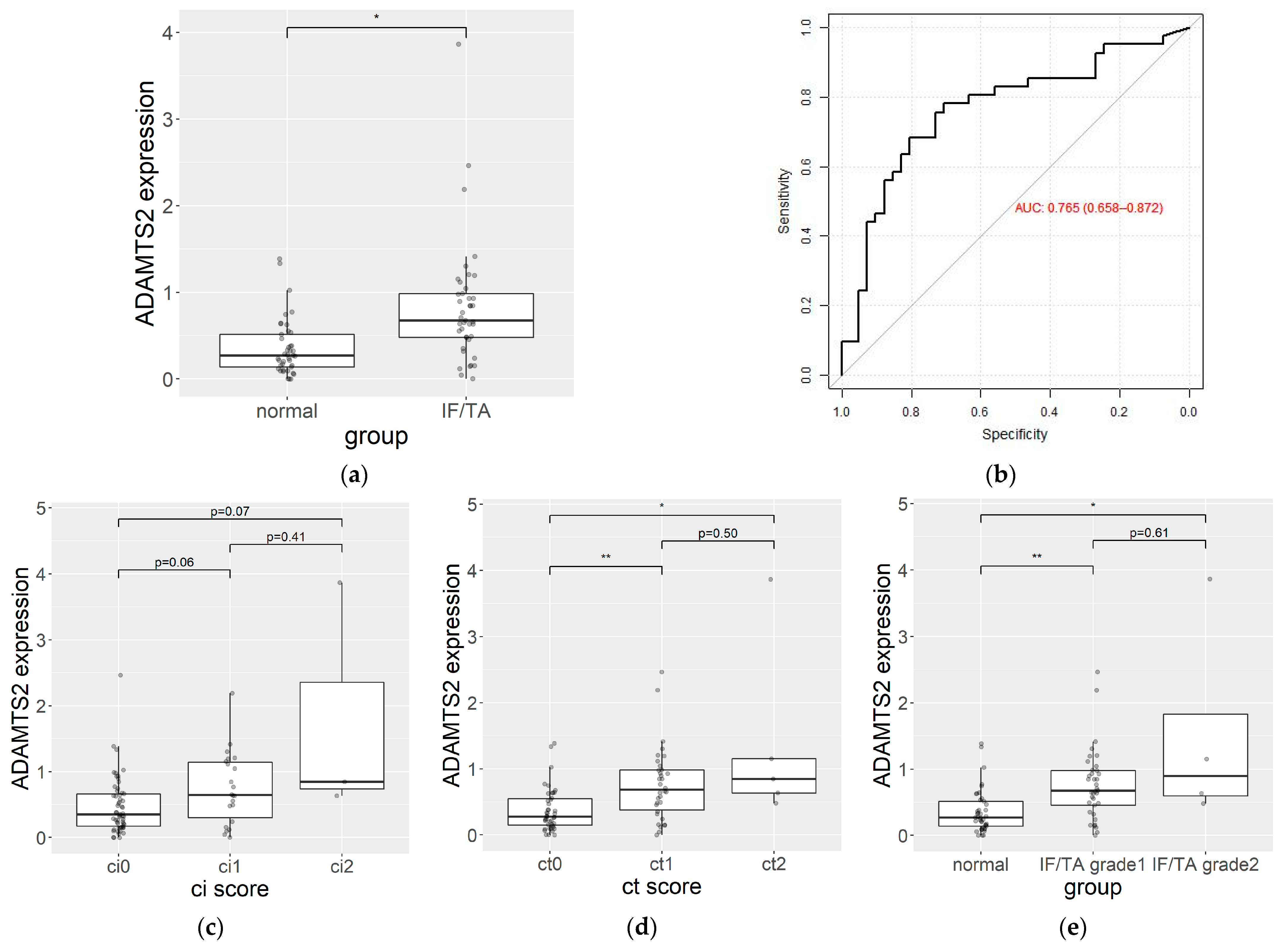
3.5. qPCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Solez, K.; Colvin, R.B.; Racusen, L.C.; Sis, B.; Halloran, P.F.; Birk, P.E.; Campbell, P.M.; Cascalho, M.; Collins, A.B.; Demetris, A.J.; et al. Banff ‘05 Meeting Report: Differential Diagnosis of Chronic Allograft Injury and Elimination of Chronic Allograft Nephropathy. Am. J. Transplant. 2007, 7, 518–526. [Google Scholar] [CrossRef]
- Nankivell, B.J.; Shingde, M.; Keung, K.L.; Fung, C.L.; Borrows, R.J.; O’Connell, P.J.; Chapman, J.R. The causes, significance and consequences of inflammatory fibrosis in kidney transplantation: The Banff i-IFTA lesion. Am. J. Transplant. 2018, 18, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Furness, P.N.; Philpott, C.M.; Chorbadjian, M.T.; Nicholson, M.L.; Bosmans, J.L.; Corthouts, B.L.; Bogers, J.J.; Schwarz, A.; Gwinner, W.; Haller, H.; et al. Protocol biopsy of the stable renal transplant: A multicenter study of methods and complication rates. Transplantation 2003, 76, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Kemmner, S.; Bachmann, Q.; Steiger, S.; Lorenz, G.; Honarpisheh, M.; Foresto-Neto, O.; Wang, S.; Carbajo-Lozoya, J.; Alt, V.; Schulte, C.; et al. STAT1 regulates macrophage number and phenotype and prevents renal fibrosis after ischemia-reperfusion injury. Am. J. Physiol.-Ren. Physiol. 2019, 316, F277–F291. [Google Scholar] [CrossRef]
- Heylen, L.; Thienpont, B.; Naesens, M.; Busschaert, P.; Depreeuw, J.; Smeets, D.; Jochmans, I.; Monbaliu, D.; Pirenne, J.; Lerut, E.; et al. Ischemia-Induced DNA Hypermethylation during Kidney Transplant Predicts Chronic Allograft Injury. J. Am. Soc. Nephrol. 2018, 29, 1566–1576. [Google Scholar] [CrossRef]
- Bontha, S.; Maluf, D.; Archer, K.; Dumur, C.; Dozmorov, M.; King, A.; Akalin, E.; Mueller, T.; Gallon, L.; Mas, V. Effects of DNA Methylation on Progression to Interstitial Fibrosis and Tubular Atrophy in Renal Allograft Biopsies: A Multi-Omics Approach. Am. J. Transplant. 2017, 17, 3060–3075. [Google Scholar] [CrossRef] [PubMed]
- Modena, B.D.; Kurian, S.M.; Gaber, L.W.; Waalen, J.; Su, A.I.; Gelbart, T.; Mondala, T.S.; Head, S.R.; Papp, S.; Heilman, R.; et al. Gene Expression in Biopsies of Acute Rejection and Interstitial Fibrosis/Tubular Atrophy Reveals Highly Shared Mechanisms That Correlate with Worse Long-Term Outcomes. Am. J. Transplant. 2016, 16, 1982–1998. [Google Scholar] [CrossRef]
- Scian, M.J.; Maluf, D.G.; Archer, K.J.; Suh, J.L.; Massey, D.; Fassnacht, R.C.; Whitehill, B.; Sharma, A.; King, A.; Gehr, T.; et al. Gene Expression Changes Are Associated With Loss of Kidney Graft Function and Interstitial Fibrosis and Tubular Atrophy: Diagnosis Versus Prediction. Transplantation 2011, 91, 657–665. [Google Scholar] [CrossRef]
- Park, W.D.; Griffin, M.D.; Cornell, L.D.; Cosio, F.G.; Stegall, M.D. Fibrosis with Inflammation at One Year Predicts Transplant Functional Decline. J. Am. Soc. Nephrol. 2010, 21, 1987–1997. [Google Scholar] [CrossRef]
- Scherer, A.; Gwinner, W.; Mengel, M.; Kirsch, T.; Raulf, F.; Szustakowski, J.D.; Hartmann, N.; Staedtler, F.; Engel, G.; Klupp, J.; et al. Transcriptome Changes in Renal Allograft Protocol Biopsies at 3 Months Precede the Onset of Interstitial Fibrosis/Tubular Atrophy (IF/TA) at 6 Months. Nephrol. Dial. Transplant. 2009, 24, 2567–2575. [Google Scholar] [CrossRef]
- Rödder, S.; Scherer, A.; Raulf, F.; Berthier, C.C.; Hertig, A.; Couzi, L.; Durrbach, A.; Rondeau, E.; Marti, H.-P. Renal Allografts with IF/TA Display Distinct Expression Profiles of Metzincins and Related Genes. Am. J. Transplant. 2009, 9, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Nogare, A.L.; Veronese, F.V.; Carpio, V.N.; Montenegro, R.M.; Pedroso, J.A.; Pegas, K.L.; Gonçalves, L.F.; Manfro, R.C. Kidney injury molecule-1 expression in human kidney transplants with interstitial fibrosis and tubular atrophy. BMC Nephrol. 2015, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Maluf, D.G.; Mas, V.R.; Archer, K.J.; Yanek, K.; Gibney, E.M.; King, A.L.; Cotterell, A.; Fisher, R.A.; Posner, M.P. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol. Med. 2008, 14, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Nakorchevsky, A.; Hewel, J.A.; Kurian, S.M.; Mondala, T.S.; Campbell, D.; Head, S.R.; Marsh, C.L.; Yates, J.R., 3rd; Salomon, D.R. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J. Am. Soc. Nephrol. 2010, 21, 362–373. [Google Scholar] [CrossRef]
- Flechner, S.M.; Kurian, S.M.; Solez, K.; Cook, D.J.; Burke, J.T.; Rollin, H.; Hammond, J.A.; Whisenant, T.; Lanigan, C.M.; Head, S.R.; et al. De novo kidney transplantation without use of calcineurin inhibitors preserves renal structure and function at two years. Am. J. Transplant. 2004, 4, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Mas, V.; Maluf, D.; Archer, K.; Yanek, K.; Mas, L.; King, A.; Gibney, E.; Massey, D.; Cotterell, A.; Fisher, R.; et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation 2007, 83, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Ventura, C.G.; Whisenant, T.; Gelbart, T.; David, D.S.R.; Agena, F.; Salomon, D.R.; David-Neto, E.; Kurian, S.M. Discovery and cross-validation of peripheral blood and renal biopsy gene expression signatures from ethnically diverse kidney transplant populations. Am. J. Transplant. 2019, 19, 3356–3366. [Google Scholar] [CrossRef]
- Kurian, S.M.; Heilman, R.; Mondala, T.S.; Nakorchevsky, A.; Hewel, J.A.; Campbell, D.; Robison, E.H.; Wang, L.; Lin, W.; Gaber, L.; et al. Biomarkers for Early and Late Stage Chronic Allograft Nephropathy by Proteogenomic Profiling of Peripheral Blood. PLoS ONE 2009, 4, e6212. [Google Scholar] [CrossRef]
- Matz, M.; Lorkowski, C.; Fabritius, K.; Wu, K.; Rudolph, B.; Frischbutter, S.; Brakemeier, S.; Gaedeke, J.; Neumayer, H.-H.; Mashreghi, M.-F.; et al. The selective biomarker IL-8 identifies IFTA after kidney transplantation in blood cells. Transpl. Immunol. 2016, 39, 18–24. [Google Scholar] [CrossRef]
- Roufosse, C.; Simmonds, N.; Clahsen-van Groningen, M.; Haas, M.; Henriksen, K.J.; Horsfield, C.; Loupy, A.; Mengel, M.; Perkowska-Ptasińska, A.; Rabant, M.; et al. A 2018 Reference Guide to the Banff Classification of Renal Allograft Pathology. Transplantation 2018, 102, 1795–1814. [Google Scholar] [CrossRef]
- Tophat. Available online: http://ccb.jhu.edu/software/tophat/index.shtml (accessed on 28 September 2020).
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Cuffdiff. Available online: http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/ (accessed on 28 September 2020).
- DAVID. Available online: https://david.ncifcrf.gov/ (accessed on 28 September 2020).
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Hosack, D.A.; Dennis, G.; Sherman, B.T.; Lane, H.C.; Lempicki, R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003, 4, R70. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Bekhouche, M.; Colige, A. The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. 2015, 44–46, 46–53. [Google Scholar] [CrossRef]
- Wang, X.; Chen, W.; Zhang, J.; Khan, A.; Li, L.; Huang, F.; Qiu, Z.; Wang, L.; Chen, X. Critical Role of ADAMTS2 (A Disintegrin and Metalloproteinase With Thrombospondin Motifs 2) in Cardiac Hypertrophy Induced by Pressure Overload. Hypertension 2017, 69, 1060–1069. [Google Scholar] [CrossRef]
- Yang, C.Y.; Chanalaris, A.; Troeberg, L. ADAMTS and ADAM metalloproteinases in osteoarthritis—Looking beyond the ‘usual suspects’. Osteoarthr. Cartil. 2017, 25, 1000–1009. [Google Scholar] [CrossRef]
- Li, G.; Li, X.; Yang, M.; Xu, L.; Deng, S.; Ran, L. Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci. Rep. 2017, 7, 42105. [Google Scholar] [CrossRef]
- Kimmel, A.R.; Sztalryd, C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr. Opin. Lipidol. 2014, 25, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Jin, Y.; Wang, Q.; Huang, J.; Wu, X.; Ren, Z. Perilipin 5 Protects against Cellular Oxidative Stress by Enhancing Mitochondrial Function in HepG2 Cells. Cells 2019, 8, 1241. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhuang, S. Recent advances in renal interstitial fibrosis and tubular atrophy after kidney transplantation. Fibrogenesis Tissue Repair 2014, 7, 15. [Google Scholar] [CrossRef]
- Djamali, A. Oxidative stress as a common pathway to chronic tubulointerstitial injury in kidney allografts. Am. J. Physiol. Renal. Physiol. 2007, 293, F445–F455. [Google Scholar] [CrossRef]
- Abuazza, G.; Becker, A.; Williams, S.S.; Chakravarty, S.; Truong, H.-T.; Lin, F.; Baum, M. Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am. J. Physiol.-Ren. Physiol. 2006, 291, F1132–F1141. [Google Scholar] [CrossRef]
- Schlingmann, K.P.; Renigunta, A.; Hoorn, E.J.; Forst, A.-L.; Renigunta, V.; Atanasov, V.; Mahendran, S.; Barakat, T.S.; Gillion, V.; Godefroid, N.; et al. Defects in KCNJ16 Cause a Novel Tubulopathy with Hypokalemia, Salt Wasting, Disturbed Acid-Base Homeostasis, and Sensorineural Deafness. J. Am. Soc. Nephrol. 2021, 32, 1498–1512. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Iwasaki, N.; Doi, K.; Noiri, E.; Iwamoto, Y.; Uchigata, Y.; Fujita, T.; Tokunaga, K. Inhibition of Glucose-Stimulated Insulin Secretion by KCNJ15, a Newly Identified Susceptibility Gene for Type 2 Diabetes. Diabetes 2012, 61, 1734–1741. [Google Scholar] [CrossRef]
- Kanasaki, K.; Taduri, G.; Koya, D. Diabetic nephropathy: The role of inflammation in fibroblast activation and kidney fibrosis. Front. Endocrinol. Lausanne 2013, 4, 7. [Google Scholar] [CrossRef]
- Wesson, D.E.; Buysse, J.M.; Bushinsky, D.A. Mechanisms of Metabolic Acidosis–Induced Kidney Injury in Chronic Kidney Disease. J. Am. Soc. Nephrol. 2020, 31, 469–482. [Google Scholar] [CrossRef]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur. J. Med. Chem. 2020, 194, 112260. [Google Scholar] [CrossRef]
- Žlajpah, M.; Hauptman, N.; Boštjančič, E.; Zidar, N. Differential expression of extracellular matrix-related genes DCN, EPHA4, FN1, SPARC, SPON2 and SPP1 in colorectal carcinogenesis. Oncol. Rep. 2019, 42, 1539–1548. [Google Scholar] [CrossRef]
- Liu, Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 2011, 7, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Von Toerne, C.; Schmidt, C.; Adams, J.; Kiss, E.; Bedke, J.; Porubsky, S.; Gretz, N.; Lindenmeyer, M.T.; Cohen, C.D.; Gröne, H.-J.; et al. Wnt Pathway Regulation in Chronic Renal Allograft Damage. Am. J. Transplant. 2009, 9, 2223–2239. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.-L.; Lin, Q.-Y.; Liu, Y.; Guan, X.-M.; Ma, X.-L.; Cao, H.-J.; Liu, Y.; Bai, J.; Xia, Y.-L.; et al. CXCL1–CXCR2 axis mediates angiotensin II-induced cardiac hypertrophy and remodelling through regulation of monocyte infiltration. Eur. Heart J. 2018, 39, 1818–1831. [Google Scholar] [CrossRef] [PubMed]
- Thézénas, M.-L.; De Leo, B.; Laux-Biehlmann, A.; Bafligil, C.; Elger, B.; Tapmeier, T.; Morten, K.; Rahmioglu, N.; Dakin, S.G.; Charles, P.; et al. Amine oxidase 3 is a novel pro-inflammatory marker of oxidative stress in peritoneal endometriosis lesions. Sci. Rep. 2020, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Willcocks, L.C.; Lyons, P.A.; Clatworthy, M.R.; Robinson, J.I.; Yang, W.; Newland, S.A.; Plagnol, V.; McGovern, N.N.; Condliffe, A.M.; Chilvers, E.R.; et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J. Exp. Med. 2008, 205, 1573–1582. [Google Scholar] [CrossRef]
- Peters, V.A.; Joesting, J.J.; Freund, G.G. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immun. 2013, 32, 1–8. [Google Scholar] [CrossRef]
- Bonsignore, P.; Kuiper, J.W.P.; Adrian, J.; Goob, G.; Hauck, C.R. CEACAM3—A Prim(at)e Invention for Opsonin-Independent Phagocytosis of Bacteria. Front. Immunol. 2020, 10, 3160. [Google Scholar] [CrossRef]
- Desai, B.; Mattson, J.; Paintal, H.; Nathan, M.; Shen, F.; Beaumont, M.; Malinao, M.-C.; Li, Y.; Canfield, J.; Basham, B.; et al. Differential expression of monocyte/macrophage-selective markers in human idiopathic pulmonary fibrosis. Exp. Lung Res. 2011, 37, 227–238. [Google Scholar] [CrossRef]
- Puthumana, J.; Thiessen-Philbrook, H.; Xu, L.; Coca, S.G.; Garg, A.X.; Himmelfarb, J.; Bhatraju, P.K.; Ikizler, T.A.; Siew, E.D.; Ware, L.B.; et al. Biomarkers of inflammation and repair in kidney disease progression. J. Clin. Investig. 2021, 131, e139927. [Google Scholar] [CrossRef]
- Zhu, T.; Yang, J.; Liu, X.; Zhang, L.; Zhang, J.; Wang, Y.; Ma, H.; Ren, Z. Hypoxia-Inducible Adrenomedullin Ameliorates the Epithelial-To-Mesenchymal Transition in Human Proximal Tubular Epithelial Cells. Mol. Med. Rep. 2015, 11, 3760–3766. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, H.; Fang, X.; Zhu, H.; Li, S.; He, J.; Gu, P.; Fan, D.; Han, F.; Zeng, Y.; Yu, X.; et al. Gene expression profile analysis for different idiopathic interstitial pneumonias subtypes. Exp. Lung Res. 2014, 40, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Jeong, H.; Roh, J.S.; Lee, B.; Lee, D.; Han, M.-E.; Oh, S.-O.; Sohn, D.H.; Kim, Y.H. VNN3 is a potential novel biomarker for predicting prognosis in clear cell renal cell carcinoma. Anim. Cells Syst. 2019, 23, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [PubMed]

| IF/TA (n = 41) | Control (n = 41) | p | |
|---|---|---|---|
| Age (y) at transplant date, median (range) | 50 (27–67) | 52 (20–68) | 0.94 |
| Male, n (%) | 21 (51.2) | 25 (61.0) | 0.50 |
| BMI (kg/m2, mean ± SD) at transplant date | 23.3 ± 4.0 | 23.4 ± 3.2 | 0.84 |
| Dialysis type, n (%) | 0.60 | ||
| No dialysis | 9 (22.0) | 10 (24.4) | |
| Hemodialysis | 27 (65.9) | 29 (70.7) | |
| Peritoneal dialysis | 5 (12.2) | 2 (4.9) | |
| Original disease, n (%) | 0.75 | ||
| Diabetes | 8 (19.5) | 9 (22.0) | |
| Glomerular nephritis | 13 (31.7) | 14 (34.1) | |
| Hypertension | 2 (4.9) | 2 (4.9) | |
| Polycystic kidney disease | 7 (17.1) | 3 (7.3) | |
| Others | 3 (7.3) | 6 (14.6) | |
| Unknown | 8 (19.5) | 7 (17.1) | |
| Donor type, n (%) | 0.06 | ||
| Deceased | 18 (43.9) | 8 (19.5) | |
| Living/Related | 15 (36.6) | 22 (53.7) | |
| Living/Unrelated | 8 (19.5) | 11 (26.8) | |
| Donor age (y, mean ± SD) | 49.8 ± 12.3 | 46.1 ± 13.3 | 0.20 |
| Missing, n (%) | 3 (7.3) | 0 (0) | |
| Postoperative days, median (range) | 814 (428–1951) | 1103 (508–2008) | 0.14 |
| Immunosuppressants at protocol biopsy, n (%) | 0.48 | ||
| Tac/MMF or MPS/steroid | 37 (90.2) | 35 (85.4) | |
| Tac/mizoribine/steroid | 3 (7.3) | 6 (14.6) | |
| Tac/steroid | 1 (2.4) | 0 (0) | |
| IF/TA grade, n (%) | N/A | N/A | |
| Grade 1 | 37 (90.2) | ||
| Grade 2 | 4 (9.8) | ||
| Banff score ci, n (%) | < 0.001 | ||
| 0 | 17 (41.5) | 40 (97.6) | |
| 1 | 21 (51.2) | 1 (2.4) | |
| 2 | 3 (7.3) | 0 (0) | |
| Banff score ct, n (%) | < 0.001 | ||
| 0 | 2 (4.9) | 41 (100.0) | |
| 1 | 34 (82.9) | 0 (0) | |
| 2 | 5 (12.2) | 0 (0) | |
| Previous biopsy-proven acute rejection, n (%) | 3 (7.3) | 1 (2.4) | 0.61 |
| Delayed graft function, n (%) | 2 (4.9) | 0 (0) | 0.47 |
| Renal artery stenosis, n (%) | 1 (2.4) | 0 (0) | 1.0 |
| eGFR (mL/min/1.73 m2, mean ± SD) | |||
| On one-year protocol biopsy day | 61.8 ± 13.3 | 65.7 ± 11.7 | 0.17 |
| On blood collection day | 62.4 ± 13.5 | 65.26 ± 10.8 | 0.30 |
| Gene ID | AUC (95% CI) | log2( Fold Change) | Absolute Fold Change | p | q |
|---|---|---|---|---|---|
| ADAMTS2 | 0.77 (0.66–0.87) | 1.82 | 3.52 | 0.00005 | 0.00625 |
| FN1 | 0.69 (0.58–0.81) | 1.08 | 2.12 | 0.00005 | 0.00625 |
| PLIN5 | 0.67 (0.55–0.79) | 1.84 | 3.57 | 0.00005 | 0.00625 |
| CLDN9 | 0.66 (0.55–0.78) | 1.57 | 2.97 | 0.00005 | 0.00625 |
| IL1R2 | 0.66 (0.54–0.78) | 1.42 | 2.68 | 0.00005 | 0.00625 |
| KREMEN1 | 0.64 (0.52–0.77) | 1.29 | 2.44 | 0.00005 | 0.00625 |
| LRG1 | 0.64 (0.52–0.76) | 1.13 | 2.19 | 0.00005 | 0.00625 |
| KCNJ15 | 0.63 (0.51–0.76) | 1.85 | 3.60 | 0.00005 | 0.00625 |
| CEACAM3 | 0.63 (0.51–0.75) | 1.36 | 2.56 | 0.00005 | 0.00625 |
| PANX2 | 0.62 (0.49–0.74) | 1.26 | 2.40 | 0.00005 | 0.00625 |
| LUCAT1 | 0.61 (0.49–0.73) | 1.21 | 2.31 | 0.00050 | 0.03779 |
| MANSC1 | 0.61 (0.48–0.73) | 1.13 | 2.19 | 0.00005 | 0.00625 |
| AOC2 | 0.60 (0.48–0.73) | 1.25 | 2.39 | 0.00005 | 0.00625 |
| LINC01002 | 0.59 (0.47–0.72) | 1.65 | 3.13 | 0.00005 | 0.00625 |
| MMP9 | 0.58 (0.45–0.70) | 1.80 | 3.48 | 0.00005 | 0.00625 |
| CXCR2 | 0.58 (0.46–0.71) | 1.57 | 2.96 | 0.00005 | 0.00625 |
| ADM | 0.58 (0.46–0.71) | 1.28 | 2.43 | 0.00005 | 0.00625 |
| VNN3 | 0.58 (0.45–0.70) | 1.07 | 2.10 | 0.00005 | 0.00625 |
| KRT23 | 0.57 (0.44–0.70) | 1.59 | 3.01 | 0.00005 | 0.00625 |
| CXCL8 | 0.57 (0.44–0.69) | 1.32 | 2.50 | 0.00005 | 0.00625 |
| BMX | 0.57 (0.44–0.70) | 1.30 | 2.45 | 0.00005 | 0.00625 |
| G0S2 | 0.56 (0.44–0.69) | 2.50 | 5.65 | 0.00155 | 0.08566 |
| CD177 | 0.56 (0.43–0.69) | 1.08 | 2.11 | 0.00025 | 0.023000 |
| CYP4F3 | 0.55 (0.43–0.68) | 1.42 | 2.68 | 0.00005 | 0.00625 |
| CHI3L1 | 0.55 (0.43–0.68) | 1.08 | 2.12 | 0.00005 | 0.00625 |
| ITGAD | 0.55 (0.42–0.68) | 1.01 | 2.01 | 0.00005 | 0.00625 |
| ALPL | 0.54 (0.41–0.67) | 2.83 | 7.09 | 0.00005 | 0.00625 |
| MME | 0.54 (0.41–0.67) | 1.97 | 3.92 | 0.00005 | 0.00625 |
| CMTM2 | 0.54 (0.41–0.67) | 1.69 | 3.24 | 0.00005 | 0.00625 |
| MGAM | 0.54 (0.41–0.66) | 1.11 | 2.16 | 0.00005 | 0.00625 |
| FCGR3B | 0.53 (0.40–0.66) | 2.35 | 5.11 | 0.00005 | 0.00625 |
| AOC3 | 0.53 (0.40–0.66) | 1.58 | 2.99 | 0.00005 | 0.00625 |
| ADGRG3 | 0.53 (0.40–0.66) | 1.51 | 2.85 | 0.00005 | 0.00625 |
| HCAR3 | 0.53 (0.41–0.66) | 1.34 | 2.53 | 0.00005 | 0.00625 |
| Models | AUC (95% Confidence Interval) |
|---|---|
| A | 0.765 (0.658–0.872) |
| A + P + C + K | 0.721 (0.609–0.833) |
| A + P + C | 0.782 (0.682–0.881) |
| A + P | 0.776 (0.674–0.879) |
| A + C | 0.786 (0.687–0.886) |
| A + K | 0.717 (0.605–0.830) |
| A + recipient age | 0.799 (0.701–0.897) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.K.; Jung, N.H.; Lee, D.E.; Lee, H.; Yang, J.; Kim, Y.S.; Han, S.S.; Han, N.; Kim, I.-W.; Oh, J.M. Discovery of Biomarkers Related to Interstitial Fibrosis and Tubular Atrophy among Kidney Transplant Recipients by mRNA-Sequencing. J. Pers. Med. 2023, 13, 1242. https://doi.org/10.3390/jpm13081242
Lee HK, Jung NH, Lee DE, Lee H, Yang J, Kim YS, Han SS, Han N, Kim I-W, Oh JM. Discovery of Biomarkers Related to Interstitial Fibrosis and Tubular Atrophy among Kidney Transplant Recipients by mRNA-Sequencing. Journal of Personalized Medicine. 2023; 13(8):1242. https://doi.org/10.3390/jpm13081242
Chicago/Turabian StyleLee, Hyun Kyung, Na Hyun Jung, Da Eun Lee, Hajeong Lee, Jaeseok Yang, Yon Su Kim, Seung Seok Han, Nayoung Han, In-Wha Kim, and Jung Mi Oh. 2023. "Discovery of Biomarkers Related to Interstitial Fibrosis and Tubular Atrophy among Kidney Transplant Recipients by mRNA-Sequencing" Journal of Personalized Medicine 13, no. 8: 1242. https://doi.org/10.3390/jpm13081242
APA StyleLee, H. K., Jung, N. H., Lee, D. E., Lee, H., Yang, J., Kim, Y. S., Han, S. S., Han, N., Kim, I.-W., & Oh, J. M. (2023). Discovery of Biomarkers Related to Interstitial Fibrosis and Tubular Atrophy among Kidney Transplant Recipients by mRNA-Sequencing. Journal of Personalized Medicine, 13(8), 1242. https://doi.org/10.3390/jpm13081242







