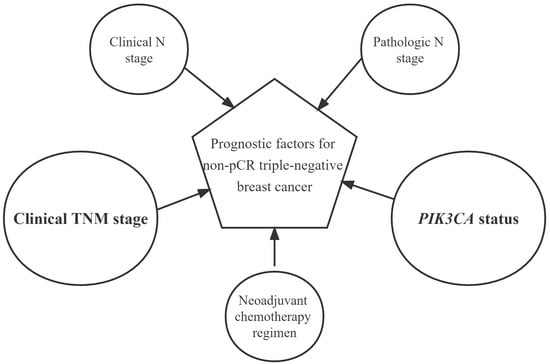
Prognostic Factors for Triple-Negative Breast Cancer with Residual Disease after Neoadjuvant Chemotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and Capture-Based Targeted DNA Sequencing
2.3. Sequence Data Analysis
2.4. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Overall Cohort and Treatment Subgroups
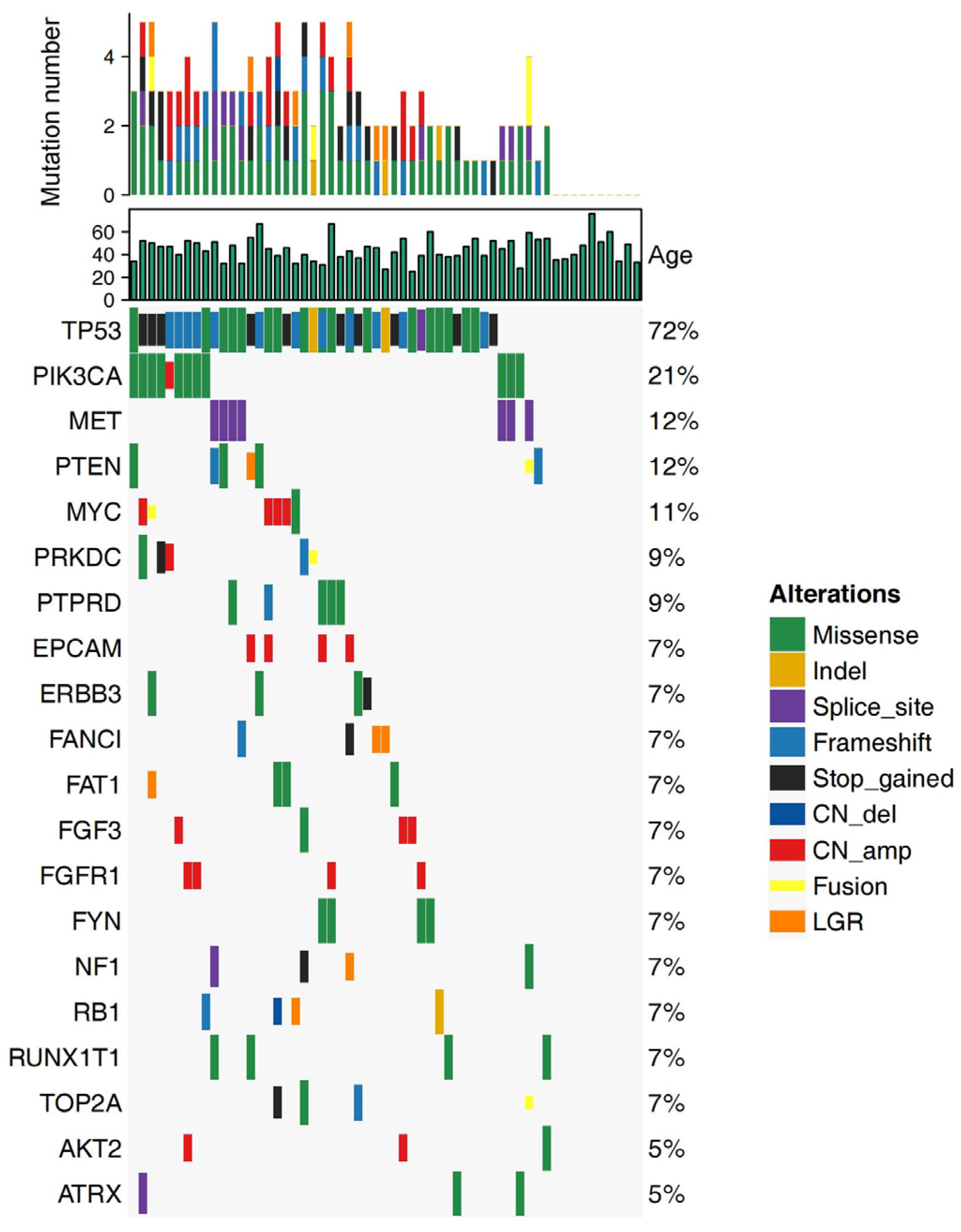
3.2. Alteration Landscape of the Cohort
3.3. Treatment Outcomes
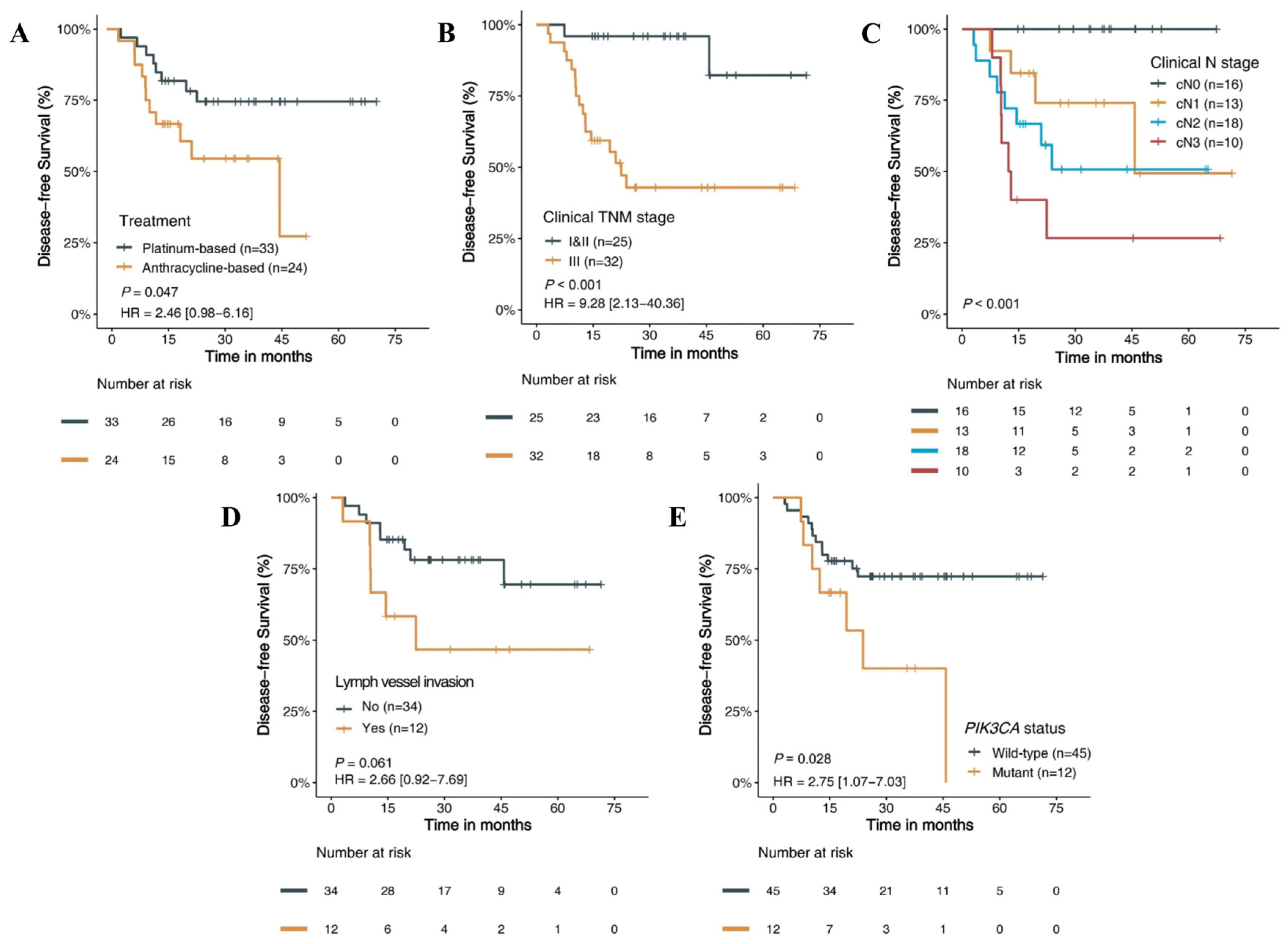
3.4. Clinical Survival Outcomes
3.4.1. Clinicopathology Features and DFS
3.4.2. Molecular Features and DFS
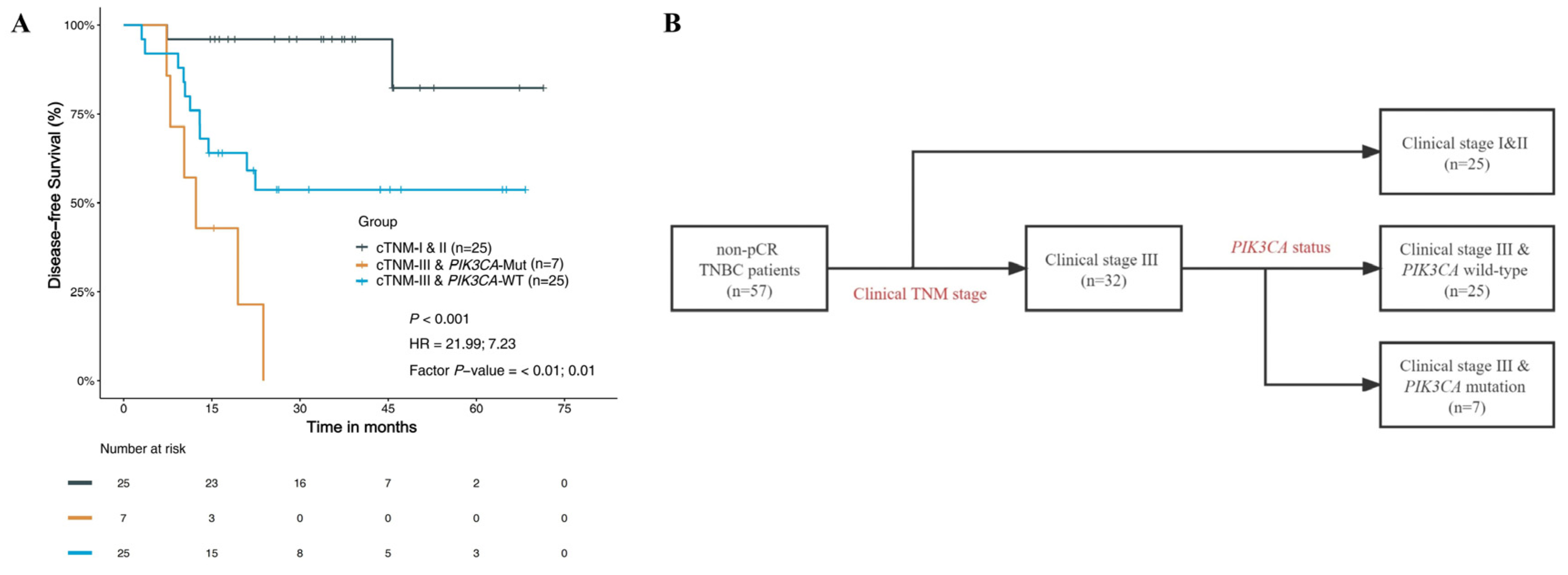
3.4.3. Prognostic Risk Factors and Stratified Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Alajez, N.M. Transcriptional landscape associated with TNBC resistance to neoadjuvant chemotherapy revealed by single-cell RNA-seq. Mol. Ther. Oncolytics 2021, 23, 151–162. [Google Scholar] [CrossRef]
- Singh, D.; Yadav, D. TNBC: Potential Targeting of Multiple Receptors for a Therapeutic Breakthrough, Nanomedicine, and Immunotherapy. Biomedicines 2021, 9, 876. [Google Scholar] [CrossRef]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin. Cancer Res. 2007, 13 Pt 1, 4429–4434. [Google Scholar] [CrossRef]
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Hwang, E.S.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Poggio, F.; Bruzzone, M.; Ceppi, M.; Pondé, N.F.; La Valle, G.; Del Mastro, L.; de Azambuja, E.; Lambertini, M. Platinum-based neoadjuvant chemotherapy in triple-negative breast cancer: A systematic review and meta-analysis. Ann. Oncol. 2018, 29, 1497–1508. [Google Scholar] [CrossRef]
- Gamucci, T.; Pizzuti, L.; Sperduti, I.; Mentuccia, L.; Vaccaro, A.; Moscetti, L.; Marchetti, P.; Carbognin, L.; Michelotti, A.; Iezzi, L.; et al. Neoadjuvant chemotherapy in triple-negative breast cancer: A multicentric retrospective observational study in real-life setting. J. Cell. Physiol. 2017, 233, 2313–2323. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Tovey, H.; Cheang, M.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P.; et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Sikov, W.M.; Berry, D.A.; Perou, C.M.; Singh, B.; Cirrincione, C.T.; Tolaney, S.M.; Kuzma, C.S.; Pluard, T.J.; Somlo, G.; Port, E.R.; et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J. Clin. Oncol. 2015, 33, 13–21. [Google Scholar] [PubMed]
- Ando, M.; Yamauchi, H.; Aogi, K.; Shimizu, S.; Iwata, H.; Masuda, N.; Yamamoto, N.; Inoue, K.; Ohono, S.; Kuroi, K.; et al. Randomized phase II study of weekly paclitaxel with and without carboplatin followed by cyclophosphamide/epirubicin/5-fluorouracil as neoadjuvant chemotherapy for stage II/IIIA breast cancer without HER2 overexpression. Breast Cancer Res. Treat. 2014, 145, 401–409. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Schneeweiss, A.; Loibl, S.; Salat, C.; Denkert, C.; Rezai, M.; Blohmer, J.U.; Jackisch, C.; Paepke, S.; Gerber, B.; et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): A randomised phase 2 trial. Lancet Oncol. 2014, 15, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Alba, E.; Chacon, J.I.; Lluch, A.; Anton, A.; Estevez, L.; Cirauqui, B.; Carrasco, E.; Calvo, L.; Segui, M.A.; Ribelles, N.; et al. A randomized phase II trial of platinum salts in basal-like breast cancer patients in the neoadjuvant setting. Results from the GEICAM/2006-03, multicenter study. Breast Cancer Res. Treat. 2012, 136, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Iwase, M.; Ando, M.; Aogi, K.; Aruga, T.; Inoue, K.; Shimomura, A.; Tokunaga, E.; Masuda, N.; Yamauchi, H.; Yamashita, T.; et al. Long-term survival analysis of addition of carboplatin to neoadjuvant chemotherapy in HER2-negative breast cancer. Breast Cancer Res. Treat. 2020, 180, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Ha, R.; Chin, C.; Karcich, J.; Liu, M.Z.; Chang, P.; Mutasa, S.; Pascual Van Sant, E.; Wynn, R.T.; Connolly, E.; Jambawalikar, S. Prior to Initiation of Chemotherapy, Can We Predict Breast Tumor Response? Deep Learning Convolutional Neural Networks Approach Using a Breast MRI Tumor Dataset. J. Digit. Imaging 2019, 32, 693–701. [Google Scholar] [CrossRef]
- Liu, M.Z.; Mutasa, S.; Chang, P.; Siddique, M.; Jambawalikar, S.; Ha, R. A novel CNN algorithm for pathological complete response prediction using an I-SPY TRIAL breast MRI database. Magn. Reson. Imaging 2020, 73, 148–151. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Park, K.; Jung, H.H.; Lee, E.; Cho, E.Y.; Lee, K.H.; Bae, S.Y.; Lee, S.K.; Kim, S.W.; Lee, J.E.; et al. Association between Mutation and Expression of TP53 as a Potential Prognostic Marker of Triple-Negative Breast Cancer. Cancer Res. Treat. 2016, 48, 1338–1350. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Sporikova, Z.; Koudelakova, V.; Trojanec, R.; Hajduch, M. Genetic Markers in Triple-Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e841–e850. [Google Scholar] [CrossRef]
- Copson, E.R.; Maishman, T.C.; Tapper, W.J.; Cutress, R.I.; Greville-Heygate, S.; Altman, D.G.; Eccles, B.; Gerty, S.; Durcan, L.T.; Jones, L.; et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): A prospective cohort study. Lancet Oncol. 2018, 19, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Newman, A.; Bratman, S.; Stehr, H.; Lee, L.J.; Liu, C.L.; Diehn, M.; Alizadeh, A.A. FACTERA: A practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014, 30, 3390–3393. [Google Scholar] [CrossRef]
- Shepherd, J.H.; Ballman, K.; Polley, M.-Y.C.; Campbell, J.D.; Fan, C.; Selitsky, S.; Fernandez-Martinez, A.; Parker, J.S.; Hoadley, K.A.; Hu, Z.; et al. CALGB 40603 (Alliance): Long-Term Outcomes and Genomic Correlates of Response and Survival After Neoadjuvant Chemotherapy with or Without Carboplatin and Bevacizumab in Triple-Negative Breast Cancer. J. Clin. Oncol. 2022, 40, 1323–1334. [Google Scholar] [CrossRef]
- Telli, M.L.; Timms, K.M.; Reid, J.; Hennessy, B.; Mills, G.B.; Jensen, K.C.; Szallasi, Z.; Barry, W.T.; Winer, E.P.; Tung, N.M.; et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016, 22, 3764–3773. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Yan, M.-D.; Wu, A.T.; Yuan, K.S.-P.; Liu, S.H. Brown Seaweed Fucoidan Inhibits Cancer Progression by Dual Regulation of mir-29c/ADAM12 and miR-17-5p/PTEN Axes in Human Breast Cancer Cells. J. Cancer 2016, 7, 2408–2419. [Google Scholar] [CrossRef]
- Uusitalo, E.; Kallionpää, R.A.; Kurki, S.; Rantanen, M.; Pitkäniemi, J.; Kronqvist, P.; Härkönen, P.; Huovinen, R.; Carpen, O.; Pöyhönen, M.; et al. Breast cancer in neurofibromatosis type 1: Overrepresentation of unfavourable prognostic factors. Br. J. Cancer 2017, 116, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhu, J.; Zhong, Y.; Geng, R.; Ji, Y.; Guan, Q.; Hong, C.; Wei, Y.; Min, N.; Qi, A.; et al. PIK3CA mutation confers resistance to chemotherapy in triple-negative breast cancer by inhibiting apoptosis and activating the PI3K/AKT/mTOR signaling pathway. Ann. Transl. Med. 2021, 9, 410. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Loibl, S.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Denkert, C. PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane-Based Neoadjuvant Chemotherapy. Cancer Res. Treat. 2020, 52, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, L.; Yao, L.; Kuang, X.Y.; Zuo, W.J.; Li, S.; Qiao, F.; Liu, Y.R.; Cao, Z.G.; Zhou, S.L.; et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat. Commun. 2018, 9, 1357. [Google Scholar] [CrossRef] [PubMed]

| Variable | Overall (n = 57) | Platinum–Paclitaxel Regimen (n = 33) | Anthracycline–Paclitaxel Regimen (n = 24) | p-Value |
|---|---|---|---|---|
| Age | 0.312 | |||
| Median [range] | 45.0 [38.0, 52.0] | 43.0 [36.0, 51.0] | 46.5 [38.0, 54.0] | |
| Menopausal status | 0.765 | |||
| Premenopause | 42 (73.7%) | 25 (75.8%) | 17 (70.8%) | |
| Postmenopause | 15 (26.3%) | 8 (24.2%) | 7 (29.2%) | |
| Family history of cancer | 0.584 | |||
| No | 38 (66.7%) | 23 (69.7%) | 15 (62.5%) | |
| Yes | 19 (33.3%) | 10 (30.3%) | 9 (37.5%) | |
| Ki-67(%) | 0.416 | |||
| Median [range] | 55.0 [21.3, 70.0] | 50.0 [10.0, 70.0] | 60.0 [30.0, 75.0] | |
| Missing | 1 (1.8%) | 0 (0.0%) | 1 (4.2%) | |
| Clinical T stage | 0.417 | |||
| cT1 | 6 (10.5%) | 2 (6.1%) | 4 (16.7%) | |
| cT2 | 36 (63.2%) | 23 (69.7%) | 13 (54.2%) | |
| cT3 | 12 (21.1%) | 7 (21.2%) | 5 (20.8%) | |
| cT4 | 3 (5.3%) | 1 (3.0%) | 2 (8.3%) | |
| Clinical N stage | 0.836 | |||
| cN0 | 16 (28.1%) | 9 (27.3%) | 7 (29.2%) | |
| cN1 | 13 (22.8%) | 9 (27.3%) | 4 (16.7%) | |
| cN2 | 18 (31.6%) | 10 (30.3%) | 8 (33.3%) | |
| cN3 | 10 (17.5%) | 5 (15.2%) | 5 (20.8%) | |
| Clinical TNM stage | 0.794 | |||
| I and II | 25 (43.9%) | 15 (45.5%) | 10 (41.7%) | |
| III | 32 (56.4%) | 18 (54.5%) | 14 (58.3%) | |
| Pathologic T stage | 1.000 | |||
| ypT1 | 39 (68.4%) | 23 (69.7%) | 16 (66.7%) | |
| ypT2 | 18 (31.6%) | 10 (30.3%) | 8 (33.3%) | |
| Pathologic N stage | 0.497 | |||
| ypN0 | 26 (45.6%) | 14 (42.4%) | 12 (50.0%) | |
| ypN1 | 18 (31.6%) | 13 (39.4%) | 5 (20.8%) | |
| ypN2 | 6 (10.5%) | 3 (9.1%) | 3 (12.5%) | |
| ypN3 | 7 (12.3%) | 3 (9.1%) | 4 (16.7%) | |
| Pathologic TNM stage | 0.376 | |||
| I | 21 (36.8%) | 11 (33.3%) | 10 (41.7%) | |
| II | 23 (40.4%) | 16 (48.5%) | 7 (29.2%) | |
| III | 13 (22.8%) | 6 (18.2%) | 7 (29.2%) | |
| Miller–Payne | 0.565 | |||
| G1 and G2 | 18 (31.6%) | 9 (27.3%) | 9 (37.5%) | |
| G3 and G4 | 39 (68.4%) | 24 (72.7%) | 15 (62.5%) | |
| Histological grade | 0.732 | |||
| Grade II | 11 (19.30%) | 7 (21.21%) | 4 (16.67%) | |
| Grade III | 35 (61.40%) | 19 (57.58%) | 16 (66.67%) | |
| Missing | 11 (19.30%) | 7 (21.21%) | 4 (16.67%) | |
| Lymph vessel invasion | 0.738 | |||
| No | 34 (59.7%) | 20 (60.6%) | 14 (58.3%) | |
| Yes | 12 (21.1%) | 6 (18.2%) | 6 (25.0%) | |
| Missing | 11 (19.3%) | 7 (21.2%) | 4 (16.7%) | |
| Adjuvant chemotherapy | 0.787 | |||
| No | 24 (42.1%) | 13 (39.4%) | 11 (45.8%) | |
| Yes | 33 (57.9%) | 20 (60.6%) | 13 (54.2%) | |
| Adjuvant radiotherapy | 1.000 | |||
| No | 9 (15.8%) | 5 (15.2%) | 4 (16.7%) | |
| Yes | 48 (84.2%) | 28 (84.9%) | 20 (83.3%) | |
| Initial metastatic sites | 0.168 | |||
| Bone/soft tissue only | 12 (21.1%) | 6 (18.2%) | 6 (25.0%) | |
| Visceral | 7 (12.3%) | 2 (6.1%) | 5 (20.8%) | |
| None | 38 (66.7%) | 25 (75.8%) | 13 (54.2%) | |
| Number of metastatic sites | 1.000 | |||
| 1 | 12 (21.1%) | 5 (15.2%) | 7 (29.2%) | |
| 2 | 7 (12.3%) | 3 (9.1%) | 4 (16.7%) | |
| None | 38 (66.7%) | 25 (75.8%) | 13 (54.2%) | |
| BRCA1/2 status | 0.073 | |||
| Mutant | 6 (10.5%) | 1 (3.0%) | 5 (20.8%) | |
| Wild-type | 51 (89.5%) | 32 (97.0%) | 19 (79.2%) |
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Treatment | 0.047 | |||
| Platinum–Paclitaxel (n = 33) | Ref. | |||
| Anthracycline–Paclitaxel (n = 24) | 2.46 (0.98–6.16) | |||
| Clinical N stage | <0.001 | |||
| cN0 and cN1 (n = 29) | Ref. | |||
| cN2 (n = 18) | 4.41 (1.32–14.72) | |||
| cN3 (n = 10) | 8.07 (2.33–27.98) | |||
| Clinical TNM stage | <0.001 | <0.001 | ||
| I and II (n = 25) | Ref. | Ref. | ||
| III (n = 32) | 9.28 (2.13–40.36) | 11.02 (3.02–40.2) | ||
| Pathologic N stage | <0.001 | |||
| pN0& and N1 (n = 44) | Ref. | |||
| pN2 (n = 6) | 0.68 (0.09–5.25) | |||
| pN3 (n = 7) | 8.58 (2.78–26.51) | |||
| PIK3CA status | 0.028 | 0.03 | ||
| Wild-type (n = 45) | Ref. | Ref. | ||
| Mutant (n = 12) | 2.75 (1.07–7.03) | 2.70 (1.10–6.6) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Han, Y.; Wang, J.; Xu, B. Prognostic Factors for Triple-Negative Breast Cancer with Residual Disease after Neoadjuvant Chemotherapy. J. Pers. Med. 2023, 13, 190. https://doi.org/10.3390/jpm13020190
Li Z, Han Y, Wang J, Xu B. Prognostic Factors for Triple-Negative Breast Cancer with Residual Disease after Neoadjuvant Chemotherapy. Journal of Personalized Medicine. 2023; 13(2):190. https://doi.org/10.3390/jpm13020190
Chicago/Turabian StyleLi, Zhijun, Yiqun Han, Jiayu Wang, and Binghe Xu. 2023. "Prognostic Factors for Triple-Negative Breast Cancer with Residual Disease after Neoadjuvant Chemotherapy" Journal of Personalized Medicine 13, no. 2: 190. https://doi.org/10.3390/jpm13020190
APA StyleLi, Z., Han, Y., Wang, J., & Xu, B. (2023). Prognostic Factors for Triple-Negative Breast Cancer with Residual Disease after Neoadjuvant Chemotherapy. Journal of Personalized Medicine, 13(2), 190. https://doi.org/10.3390/jpm13020190