Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making
Abstract
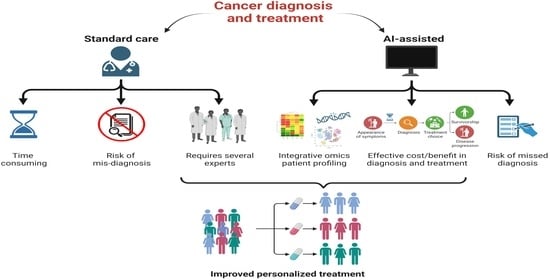
1. Introduction
Artificial Intelligence in Medicine at Glance
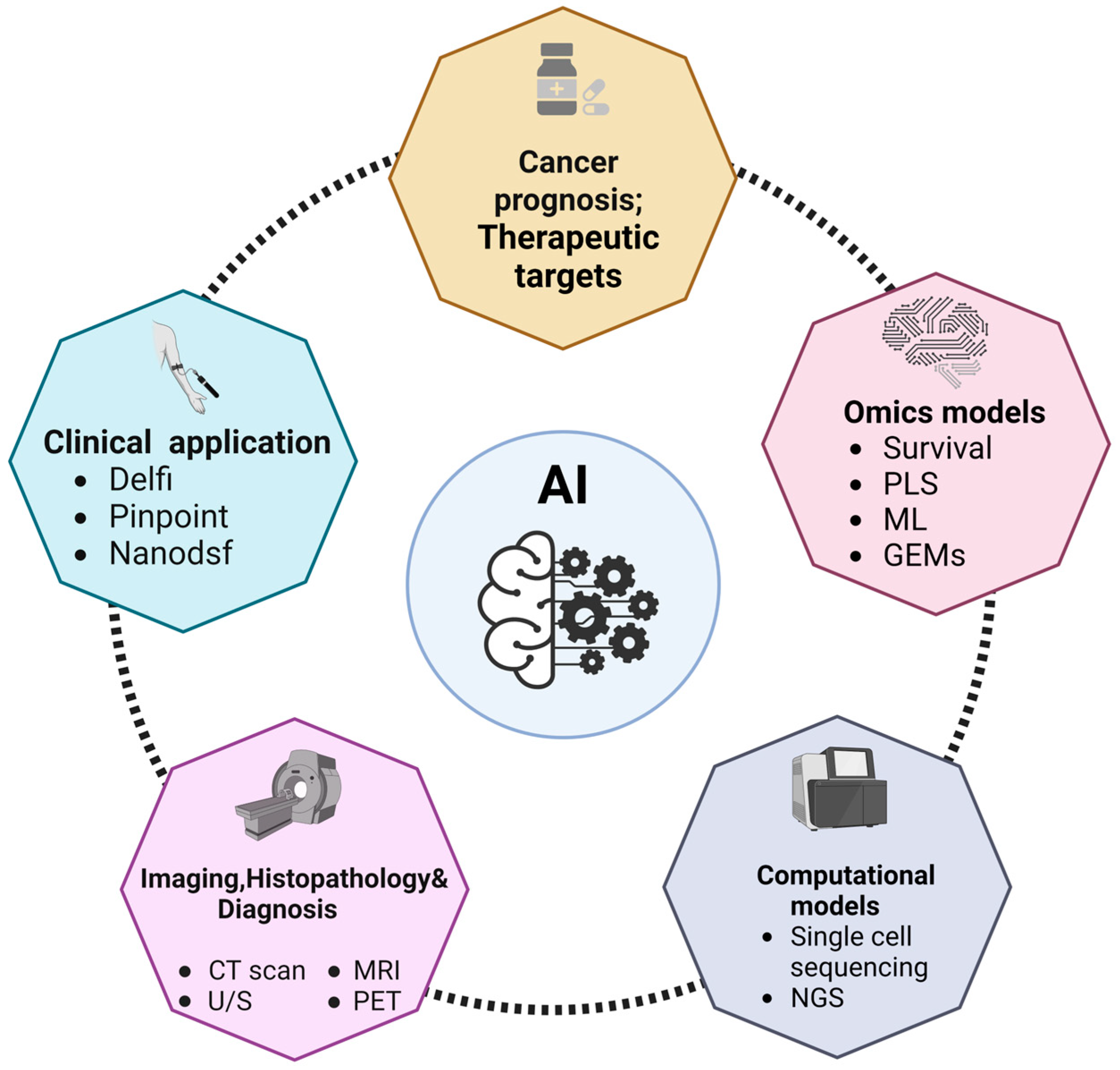
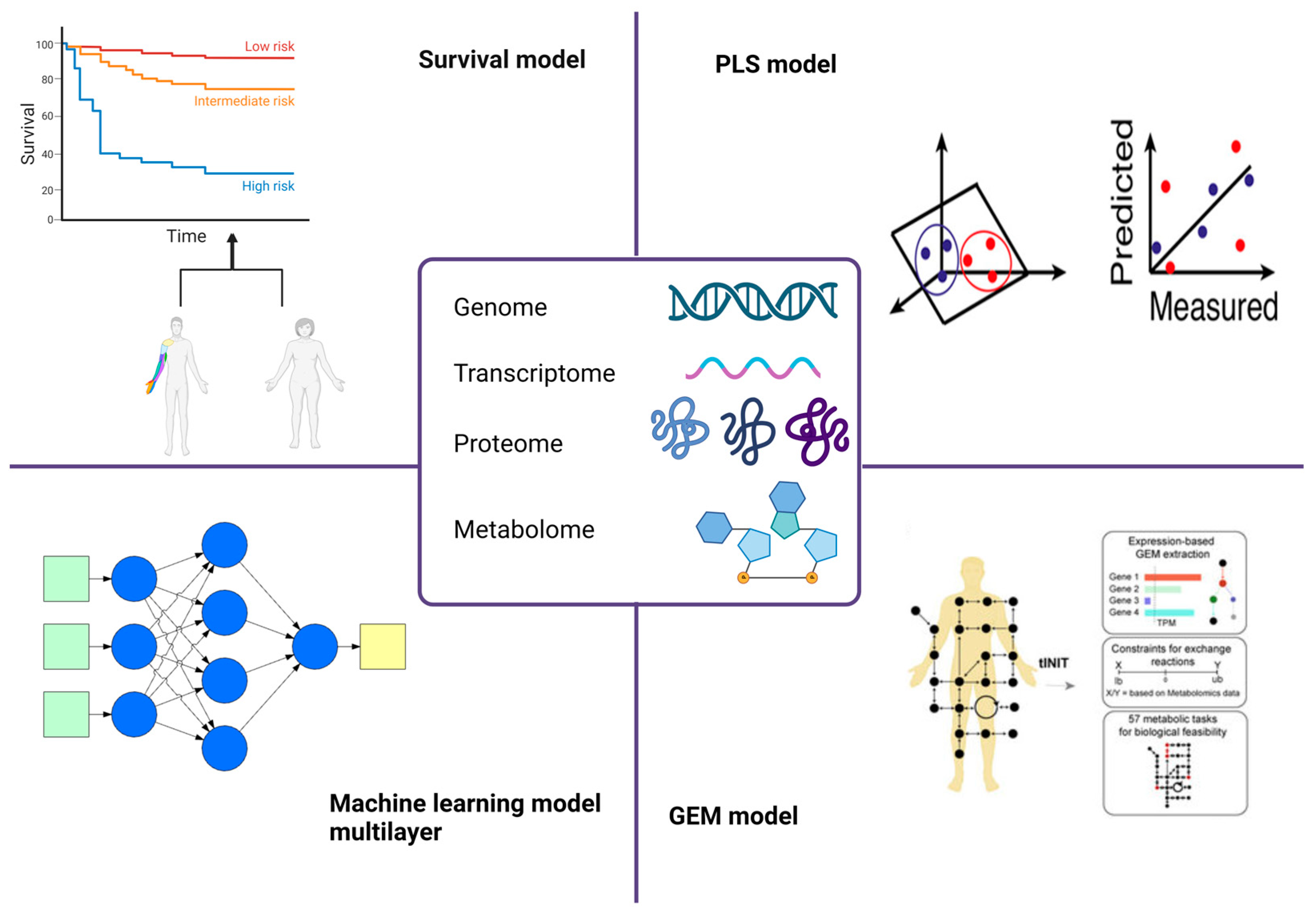
2. Omics Data for Identifying Cancer Metabolic Biomarkers
2.1. Survival Models
2.2. PLS Models
2.3. Genome-Scale Metabolic Models
2.4. Machine Learning Models
2.5. Deep Neural Networks (DNNs)
2.6. Graph Neural Networks (GNNs)
3. Computational Models for the Prediction of Cancer Metabolic Biomarkers
4. AI in Cancer Prognosis
5. AI in the Identification of Therapeutic Targets
6. AI Clinical Application
7. AI imaging in Cancer Diagnosis
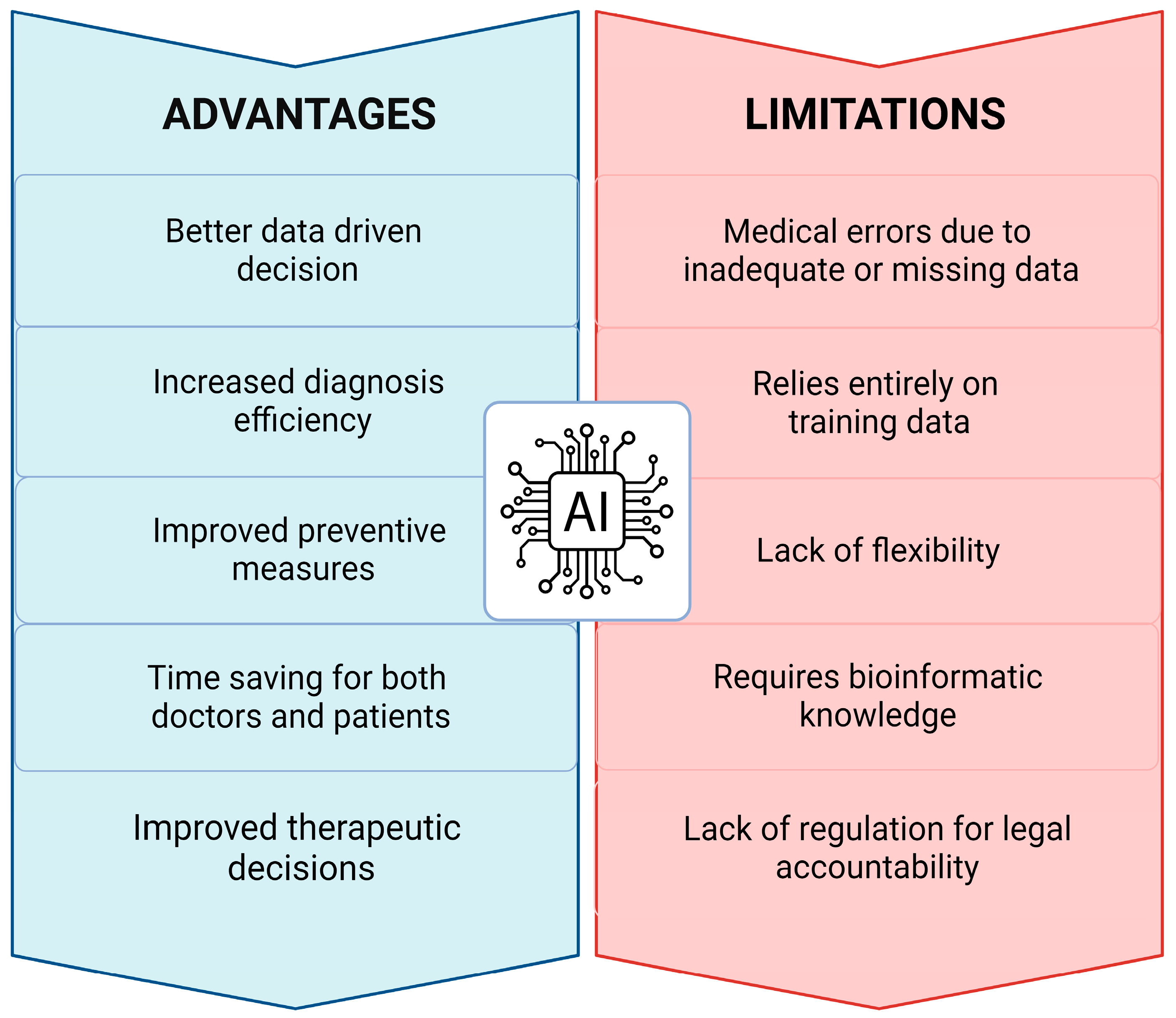
8. Critical Issues, Challenges, and Limitations
9. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Committee on Diagnostic Error in Health Care; Board on Health Care Services; Institute of Medicine; The National Academies of Sciences, Engineering, and Medicine. Improving Diagnosis in Health Care; Balogh, E.P., Miller, B.T., Ball, J.R., Eds.; National Academies Press: Washington, DC, USA, 2015. Available online: http://www.ncbi.nlm.nih.gov/books/NBK338596/ (accessed on 3 May 2023).
- Rodziewicz, T.L.; Houseman, B.; Hipskind, J.E. Medical Error Reduction and Prevention. In StatPearls; StatPearls Publishing: St. Petersburg, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK499956/ (accessed on 3 May 2023).
- Taylor, N. Duke Report Identifies Barriers to Adoption of AI Healthcare Systems. MedTech Dive. Available online: https://www.medtechdive.com/news/duke-report-identifies-barriers-to-adoption-of-ai-healthcare-systems/546739/ (accessed on 3 May 2023).
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef]
- Nadhan, R.; Kashyap, S.; Ha, J.H.; Jayaraman, M.; Song, Y.S.; Isidoro, C.; Dhanasekaran, D.N. Targeting Oncometabolites in Peritoneal Cancers: Preclinical Insights and Therapeutic Strategies. Metabolites 2023, 13, 618. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.O.; Jeffries, C.; Sullivan, P. Expanding the ‘Central Dogma’: The Regulatory Role of Nonprotein Coding Genes and Implications for the Genetic Liability to Schizophrenia. Mol. Psychiatry 2005, 10, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Tsakiroglou, M.; Evans, A.; Pirmohamed, M. Leveraging Transcriptomics for Precision Diagnosis: Lessons Learned from Cancer and Sepsis. Front. Genet. 2023, 14, 1100352. [Google Scholar] [CrossRef] [PubMed]
- Haga, Y.; Minegishi, Y.; Ueda, K. Frontiers in Mass Spectrometry–Based Clinical Proteomics for Cancer Diagnosis and Treatment. Cancer Sci. 2023, 114, 1783–1791. [Google Scholar] [CrossRef]
- Janes, K.A.; Yaffe, M.B. Data-Driven Modelling of Signal-Transduction Networks. Nat. Rev. Mol. Cell Biol. 2006, 7, 820–828. [Google Scholar] [CrossRef]
- Luo, J.; Pan, M.; Mo, K.; Mao, Y.; Zou, D. Emerging Role of Artificial Intelligence in Diagnosis, Classification and Clinical Management of Glioma. Semin. Cancer Biol. 2023, 91, 110–123. [Google Scholar] [CrossRef]
- Wang, S.; Wang, S.; Wang, Z. A Survey on Multi-Omics-Based Cancer Diagnosis Using Machine Learning with the Potential Application in Gastrointestinal Cancer. Front. Med. 2023, 9, 1109365. [Google Scholar] [CrossRef]
- Liao, J.; Li, X.; Gan, Y.; Han, S.; Rong, P.; Wang, W.; Li, W.; Zhou, L. Artificial Intelligence Assists Precision Medicine in Cancer Treatment. Front. Oncol. 2023, 12, 998222. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, X.; Zuo, F.; Shi, H.; Jing, J. Artificial Intelligence-Based Multi-Omics Analysis Fuels Cancer Precision Medicine. Semin. Cancer Biol. 2023, 88, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Dembrower, K.; Wåhlin, E.; Liu, Y.; Salim, M.; Smith, K.; Lindholm, P.; Eklund, M.; Strand, F. Effect of Artificial Intelligence-Based Triaging of Breast Cancer Screening Mammograms on Cancer Detection and Radiologist Workload: A Retrospective Simulation Study. Lancet Digit. Health 2020, 2, e468–e474. [Google Scholar] [CrossRef] [PubMed]
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Bohr, A.; Memarzadeh, K. The rise of artificial intelligence in healthcare applications. In Artificial Intelligence in Healthcare; Elsevier: Amsterdam, The Netherlands, 2020; pp. 25–60. [Google Scholar] [CrossRef]
- Venkatesan, D.; Elangovan, A.; Winster, H.; Pasha, M.Y.; Abraham, K.S.; Satheeshkumar, J.; Sivaprakash, P.; Niraikulam, A.; Gopalakrishnan, A.V.; Narayanasamy, A.; et al. Diagnostic and therapeutic approach of artificial intelligence in neuro-oncological diseases. Biosens. Bioelectron. X 2022, 11, 100188. [Google Scholar] [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef]
- Mohammed, A.; Biegert, G.; Adamec, J.; Helikar, T. Identification of Potential Tissue-Specific Cancer Biomarkers and Development of Cancer versus Normal Genomic Classifiers. Oncotarget 2017, 8, 85692–85715. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, S.; Wang, Z.; Liu, Y.; Luo, H.; Li, B.; Zou, Q. Local Augmented Graph Neural Network for Multi-Omics Cancer Prognosis Prediction and Analysis. Methods 2023, 213, 1–9. [Google Scholar] [CrossRef]
- Peng, X.; Chen, Z.; Farshidfar, F.; Xu, X.; Lorenzi, P.L.; Wang, Y.; Cheng, F.; Tan, L.; Mojumdar, K.; Du, D.; et al. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018, 23, 255–269.e4. [Google Scholar] [CrossRef]
- Yokota, K.; Uchida, H.; Sakairi, M.; Abe, M.; Tanaka, Y.; Tainaka, T.; Shirota, C.; Sumida, W.; Oshima, K.; Makita, S.; et al. Identification of Novel Neuroblastoma Biomarkers in Urine Samples. Sci. Rep. 2021, 11, 4055. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA Cross Validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Partial Least Squares Discriminant Analysis: Taking the Magic Away: PLS-DA: Taking the Magic Away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A Tutorial Review: Metabolomics and Partial Least Squares-Discriminant Analysis—A Marriage of Convenience or a Shotgun Wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current Status and Applications of Genome-Scale Metabolic Models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef]
- Fang, X.; Lloyd, C.J.; Palsson, B.O. Reconstructing Organisms in Silico: Genome-Scale Models and Their Emerging Applications. Nat. Rev. Microbiol. 2020, 18, 731–743. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.O. A Protocol for Generating a High-Quality Genome-Scale Metabolic Reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- O’Brien, J.E.; Monk, J.M.; Palsson, B.O. Using Genome-Scale Models to Predict Biological Capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef]
- Chand, S. A comparative study of breast cancer tumor classification by classical machine learning methods and deep learning method. Mach. Vis. Appl. 2020, 31, e270. [Google Scholar]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep Learning for Computational Biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef]
- Samiei, M.; Würfl, T.; Deleu, T.; Weiss, M.; Dutil, F.; Fevens, T.; Boucher, G.; Lemieux, S.; Cohen, J.P. The TCGA Meta-Dataset Clinical Benchmark. arXiv 2019, arXiv:1910.08636. [Google Scholar]
- Jin, S.; Zeng, X.; Xia, F.; Huang, W.; Liu, X. Application of Deep Learning Methods in Biological Networks. Brief. Bioinform. 2021, 22, 1902–1917. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.; Seo, S.; Kim, S. Hybrid Approach of Relation Network and Localized Graph Convolutional Filtering for Breast Cancer Subtype Classification. arXiv 2018, arXiv:1711.05859. [Google Scholar]
- Lee, S.; Lim, S.; Lee, T.; Sung, I.; Kim, S. Cancer Subtype Classification and Modeling by Pathway Attention and Propagation. Bioinformatics 2020, 36, 3818–3824. [Google Scholar] [CrossRef]
- Dai, H.; Li, H.; Tian, T.; Huang, X.; Wang, L.; Zhu, J.; Song, L. Adversarial Attack on Graph Structured Data. In Proceedings of the 35th International Conference on Machine Learning (ICML 2018), Stockholm, Sweden, 10–15 July 2018; Available online: https://proceedings.mlr.press/v80/dai18b.html (accessed on 3 May 2023).
- Zhang, X.; Zitnik, M. GNNGuard: Defending Graph Neural Networks against Adversarial Attacks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: New York, NY, USA, 2020; Volume 33, pp. 9263–9275. Available online: https://papers.nips.cc/paper/2020/hash/690d83983a63aa1818423fd6edd3bfdb-Abstract.html (accessed on 3 May 2023).
- Abdelaal, T.; Michielsen, L.; Cats, D.; Hoogduin, D.; Mei, H.; Reinders, M.J.T.; Mahfouz, A. A Comparison of Automatic Cell Identification Methods for Single-Cell RNA Sequencing Data. Genome Biol. 2019, 20, 194. [Google Scholar] [CrossRef]
- Tan, Y.; Cahan, P. SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data Across Platforms and Across Species. Cell Syst. 2019, 9, 207–213.e2. [Google Scholar] [CrossRef]
- Hu, J.; Li, X.; Hu, G.; Lyu, Y.; Susztak, K.; Li, M. Iterative Transfer Learning with Neural Network for Clustering and Cell Type Classification in Single-Cell RNA-Seq Analysis. Nat. Mach. Intell. 2020, 2, 607–618. [Google Scholar] [CrossRef]
- Andreatta, M.; Corria-Osorio, J.; Müller, S.; Cubas, R.; Coukos, G.; Carmona, S.J. Interpretation of T Cell States from Single-Cell Transcriptomics Data Using Reference Atlases. Nat. Commun. 2021, 12, 2965. [Google Scholar] [CrossRef]
- Michielsen, L.; Reinders, M.J.T.; Mahfouz, A. Hierarchical Progressive Learning of Cell Identities in Single-Cell Data. Nat. Commun. 2021, 12, 2799. [Google Scholar] [CrossRef]
- Ranjan, B.; Schmidt, F.; Sun, W.; Park, J.; Honardoost, M.A.; Tan, J.; Rayan, N.A.; Prabhakar, S. ScConsensus: Combining Supervised and Unsupervised Clustering for Cell Type Identification in Single-Cell RNA Sequencing Data. BMC Bioinform. 2021, 22, 186. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Creixell, P.; Reimand, J.; Haider, S.; Wu, G.; Shibata, T.; Vazquez, M.; Mustonen, V.; Gonzalez-Perez, A.; Pearson, J.; Sander, C.; et al. Pathway and Network Analysis of Cancer Genomes. Nat. Methods 2015, 12, 615–621. [Google Scholar] [PubMed]
- Ngiam, K.Y.; Khor, I.W. Big Data and Machine Learning Algorithms for Health-Care Delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef] [PubMed]
- Reyna, M.A.; Haan, D.; Paczkowska, M.; Verbeke, L.P.C.; Vazquez, M.; Kahraman, A.; Pulido-Tamayo, S.; Barenboim, J.; Wadi, L.; Dhingra, P.; et al. Pathway and Network Analysis of More than 2500 Whole Cancer Genomes. Nat. Commun. 2020, 11, 729. [Google Scholar] [CrossRef]
- Luo, P.; Ding, Y.; Lei, X.; Wu, F.-X. DeepDriver: Predicting Cancer Driver Genes Based on Somatic Mutations Using Deep Convolutional Neural Networks. Front. Genet. 2019, 10, 13. [Google Scholar] [CrossRef]
- Jiao, W.; Atwal, G.; Polak, P.; Karlic, R.; Cuppen, E.; PCAWG Tumor Subtypes and Clinical Translation Working Group; Danyi, A.; de Ridder, J.; van Herpen, C.; Lolkema, M.P.; et al. A Deep Learning System Accurately Classifies Primary and Metastatic Cancers Using Passenger Mutation Patterns. Nat. Commun. 2020, 11, 728. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef]
- Gao, F.; Wang, W.; Tan, M.; Zhu, L.; Zhang, Y.; Fessler, E.; Vermeulen, L.; Wang, X. DeepCC: A Novel Deep Learning-Based Framework for Cancer Molecular Subtype Classification. Oncogenesis 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhu, S.; Liu, X.; Zhou, Y.; Nussinov, R.; Cheng, F. DeepDR: A Network-Based Deep Learning Approach to in Silico Drug Repositioning. Bioinformatics 2019, 35, 5191–5198. [Google Scholar] [CrossRef] [PubMed]
- Issa, N.T.; Stathias, V.; Schürer, S.; Dakshanamurthy, S. Machine and Deep Learning Approaches for Cancer Drug Repurposing. Semin. Cancer Biol. 2021, 68, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Heider, D.; Hauschild, A.-C. Integrative Analysis of Next-Generation Sequencing for Next-Generation Cancer Research toward Artificial Intelligence. Cancers 2021, 13, 3148. [Google Scholar] [CrossRef]
- Zhang, L.; Lv, C.; Jin, Y.; Cheng, G.; Fu, Y.; Yuan, D.; Tao, Y.; Guo, Y.; Ni, X.; Shi, T. Deep Learning-Based Multi-Omics Data Integration Reveals Two Prognostic Subtypes in High-Risk Neuroblastoma. Front. Genet. 2018, 9, 477. [Google Scholar] [CrossRef]
- Francescatto, M.; Chierici, M.; Dezfooli, S.R.; Zandonà, A.; Jurman, G.; Furlanello, C. Multi-Omics Integration for Neuroblastoma Clinical Endpoint Prediction. Biol. Direct 2018, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhan, X.; Xiang, S.; Johnson, T.S.; Helm, B.; Yu, C.Y.; Zhang, J.; Salama, P.; Rizkalla, M.; Han, Z.; et al. SALMON: Survival Analysis Learning with Multi-Omics Neural Networks on Breast Cancer. Front. Genet. 2019, 10, 166. [Google Scholar] [CrossRef]
- Xie, G.; Dong, C.; Kong, Y.; Zhong, J.F.; Li, M.; Wang, K. Group Lasso Regularized Deep Learning for Cancer Prognosis from Multi-Omics and Clinical Features. Genes 2019, 10, 240. [Google Scholar] [CrossRef]
- Chen, L.; Wu, J. Bio-Network Medicine. J. Mol. Cell Biol. 2015, 7, 185–186. [Google Scholar] [CrossRef][Green Version]
- Song, H.; Chen, L.; Cui, Y.; Li, Q.; Wang, Q.; Fan, J.; Yang, J.; Zhang, L. Denoising of MR and CT Images Using Cascaded Multi-Supervision Convolutional Neural Networks with Progressive Training. Neurocomputing 2022, 469, 354–365. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Guo, Y.; Xiao, M.; Feng, L.; Yang, C.; Wang, G.; Ouyang, L. MCDB: A Comprehensive Curated Mitotic Catastrophe Database for Retrieval, Protein Sequence Alignment, and Target Prediction. Acta Pharm. Sin. B 2021, 11, 3092–3104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial Intelligence in COVID-19 Drug Repurposing. Lancet Digit. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef] [PubMed]
- Suhail, Y.; Cain, M.P.; Vanaja, K.; Kurywchak, P.A.; Levchenko, A.; Kalluri, R.; Kshitiz. Systems Biology of Cancer Metastasis. Cell Syst. 2019, 9, 109–127. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Ali, M.; Khan, S.A.; Wennerberg, K.; Aittokallio, T. Global Proteomics Profiling Improves Drug Sensitivity Prediction: Results from a Multi-Omics, Pan-Cancer Modeling Approach. Bioinformatics 2018, 34, 1353–1362. [Google Scholar] [CrossRef]
- Sharifi-Noghabi, H.; Zolotareva, O.; Collins, C.C.; Ester, M. MOLI: Multi-Omics Late Integration with Deep Neural Networks for Drug Response Prediction. Bioinformatics 2019, 35, i501–i509. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Kim, Y.; Lee, S.; Namkung, J.; Yun, T.; Yi, S.G.; Han, S.; Kang, M.; Kim, S.W.; Jang, J.-Y.; et al. Integrative Analysis of Multi-Omics Data for Identifying Multi-Markers for Diagnosing Pancreatic Cancer. BMC Genom. 2015, 16, S4. [Google Scholar] [CrossRef]
- Gautam, P.; Jaiswal, A.; Aittokallio, T.; Al-Ali, H.; Wennerberg, K. Phenotypic Screening Combined with Machine Learning for Efficient Identification of Breast Cancer-Selective Therapeutic Targets. Cell Chem. Biol. 2019, 26, 970–979.e4. [Google Scholar] [CrossRef]
- Peng, C.; Zheng, Y.; Huang, D.-S. Capsule Network Based Modeling of Multi-Omics Data for Discovery of Breast Cancer-Related Genes. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1605–1612. [Google Scholar] [CrossRef]
- Mazzone, P.J.; Sears, C.R.; Arenberg, D.A.; Gaga, M.; Gould, M.K.; Massion, P.P.; Nair, V.S.; Powell, C.A.; Silvestri, G.A.; Vachani, A.; et al. Evaluating Molecular Biomarkers for the Early Detection of Lung Cancer: When Is a Biomarker Ready for Clinical Use? An Official American Thoracic Society Policy Statement. Am. J. Respir. Crit. Care Med. 2017, 196, e15–e29. [Google Scholar] [CrossRef] [PubMed]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2019, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.O.; Eyraud, R.; Ayache, S.; Bougaev, A.A.; Malesinski, S.; Benazha, H.; Gorokhova, S.; Buffat, C.; Dehais, C.; Sanson, M.; et al. An AI-Powered Blood Test to Detect Cancer Using NanoDSF. Cancers 2021, 13, 1294. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Lu, T.; Li, J. Clinical Applications of Urinary Cell-Free DNA in Cancer: Current Insights and Promising Future. Am. J. Cancer Res. 2017, 7, 2318–2332. [Google Scholar] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and Future Perspectives of Liquid Biopsies in Genomics-Driven Oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Savage, R.; Messenger, M.; Neal, R.D.; Ferguson, R.; Johnston, C.; Lloyd, K.L.; Neal, M.D.; Sansom, N.; Selby, P.; Sharma, N.; et al. Development and Validation of Multivariable Machine Learning Algorithms to Predict Risk of Cancer in Symptomatic Patients Referred Urgently from Primary Care: A Diagnostic Accuracy Study. BMJ Open 2022, 12, e053590. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Cree, I.A.; Uttley, L.; Woods, H.B.; Kikuchi, H.; Reiman, A.; Harnan, S.; Whiteman, B.L.; Philips, S.T.; Messenger, M.; Cox, A.; et al. The Evidence Base for Circulating Tumour DNA Blood-Based Biomarkers for the Early Detection of Cancer: A Systematic Mapping Review. BMC Cancer 2017, 17, 697. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Chan, H.-P.; Hadjiiski, L.; Zhou, C.; Sahiner, B. Computer-aided diagnosis of lung cancer and pulmonary embolism in computed tomography-a review. Acad. Radiol. 2008, 15, 535–555. [Google Scholar] [CrossRef] [PubMed]
- Rasch, C.; Barillot, I.; Remeijer, P.; Touw, A.; van Herk, M.; Lebesque, J.V. Definition of the Prostate in CT and MRI: A Multi-Observer Study. Int. J. Radiat. Oncol. Biol. Phys. 1999, 43, 57–66. [Google Scholar] [CrossRef]
- Chen, W.; Giger, M.L.; Li, H.; Bick, U.; Newstead, G.M. Volumetric Texture Analysis of Breast Lesions on Contrast-Enhanced Magnetic Resonance Images. Magn. Reson. Med. 2007, 58, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Song, N.; Shen, R.; Arora, A.; Machiela, M.J.; Song, L.; Landi, M.T.; Ghosh, D.; Chatterjee, N.; Baladandayuthapani, V.; et al. Integrating Clinical and Multiple Omics Data for Prognostic Assessment across Human Cancers. Sci. Rep. 2017, 7, 16954. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, H.; Feng, Y.; Li, W. Prostate Cancer Diagnosis Using Deep Learning with 3D Multiparametric MRI. arXiv 2017, arXiv:1703.04078. [Google Scholar]
- Chen, Q.; Xu, X.; Hu, S.; Li, X.; Zou, Q.; Li, Y. A Transfer Learning Approach for Classification of Clinical Significant Prostate Cancers from MpMRI Scans. Proc. SPIE 2017, 10134, 101344F. [Google Scholar]
- Seah, J.C.Y.; Tang, J.S.N.; Kitchen, A. Detection of Prostate Cancer on Multiparametric MRI. In Medical Imaging 2017: Computer-Aided Diagnosis; Armato, S.G., Petrick, N.A., Eds.; SPIE: Orlando, FL, USA, 2017; Volume 10134, p. 1013429. [Google Scholar]
- Mehrtash, A.; Sedghi, A.; Ghafoorian, M.; Taghipour, M.; Tempany, C.M.; Wells, W.M.; Kapur, T.; Mousavi, P.; Abolmaesumi, P.; Fedorov, A. Classification of Clinical Significance of MRI Prostate Findings Using 3D Convolutional Neural Networks. Proc. SPIE Int. Soc. Opt. Eng. 2017, 10134, 101342A. [Google Scholar]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Nir, G.; Kwak, J.T.; Xu, S.; Wilson, S.; Iczkowski, K.A.; Lucia, M.S.; et al. Detection and Grading of Prostate Cancer Using Temporal Enhanced Ultrasound: Combining Deep Neural Networks and Tissue Mimicking Simulations. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1293–1305. [Google Scholar] [CrossRef]
- Al Bakir, M.; Huebner, A.; Martínez-Ruiz, C.; Grigoriadis, K.; Watkins, T.B.K.; Pich, O.; Moore, D.A.; Veeriah, S.; Ward, S.; Laycock, J.; et al. The Evolution of Non-Small Cell Lung Cancer Metastases in TRACERx. Nature 2023, 616, 534–542. [Google Scholar] [CrossRef]
- Martínez-Ruiz, C.; Black, J.R.M.; Puttick, C.; Hill, M.S.; Demeulemeester, J.; Cadieux, E.L.; Thol, K.; Jones, T.P.; Veeriah, S.; Naceur-Lombardelli, C.; et al. Genomic–Transcriptomic Evolution in Lung Cancer and Metastasis. Nature 2023, 616, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Ding, Y.; Wang, L.; Wang, S.; Wang, H.; Qin, Y. DNA Methylation: From Cancer Biology to Clinical Perspectives. Front. Biosci. Landmark 2022, 27, 326. [Google Scholar] [CrossRef] [PubMed]
- Olaronke, I.; Oluwaseun, O. Big Data in Healthcare: Prospects, Challenges and Resolutions. In Proceedings of the 2016 Future Technologies Conference (FTC), San Francisco, CA, USA, 6–7 December 2016; pp. 1152–1157. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
| Advantages | Limitations |
|---|---|
| Ability to robustly handle more descriptor variables | Higher risk of overlooking ‘real’ correlations |
| Provide more predictive accuracy | Sensitivity to the relative scaling of the descriptor variables |
| Low risk of chance correlation |
| Advantages | Limitations |
|---|---|
| Explore metabolism in multiple cell types | Uncertainties in the estimated parameters regarding quantitative flux predictions |
| Validating or discovering biomarkers for screening, diagnostics, prognostics, and/or patient stratification | Ambiguous normalization of experimentally quantified fluxes |
| Identify cancer-specific metabolic features that constitute generic potential drug targets for cancer treatment |
| Advantages | Limitations |
|---|---|
| Ability to handle complex data and relationships | Massive data requirement |
| Effective at producing high-quality results | High processing and computational power |
| Extremely scalable because of its capacity to analyze large volumes of data | Black box problem making them hard to debug and understand how they make decisions |
| Advantages | Limitations |
|---|---|
| Rapid processing of massive data | Limited to a fixed number of points |
| Reliable performance in mining deep-level topological information | Time and space complexity are higher |
| Extracting text relationship and reasoning the structure of graphics and images | Less handling of edges of graphs based on their types and relations |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fawaz, A.; Ferraresi, A.; Isidoro, C. Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making. J. Pers. Med. 2023, 13, 1590. https://doi.org/10.3390/jpm13111590
Fawaz A, Ferraresi A, Isidoro C. Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making. Journal of Personalized Medicine. 2023; 13(11):1590. https://doi.org/10.3390/jpm13111590
Chicago/Turabian StyleFawaz, Alaa, Alessandra Ferraresi, and Ciro Isidoro. 2023. "Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making" Journal of Personalized Medicine 13, no. 11: 1590. https://doi.org/10.3390/jpm13111590
APA StyleFawaz, A., Ferraresi, A., & Isidoro, C. (2023). Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making. Journal of Personalized Medicine, 13(11), 1590. https://doi.org/10.3390/jpm13111590