The Association of HLA-B*35 and GSTT1 Genotypes and Hepatotoxicity in Thai People Living with HIV
Abstract
:1. Introduction
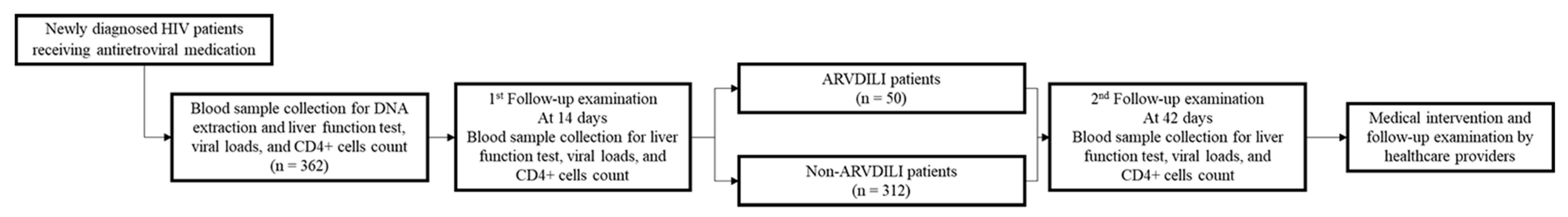
2. Materials and Methods
2.1. Study Subjects
2.2. DNA Sample Retrieval and DNA Quantification
2.3. Genetic Genotyping and Data Retrievals
2.4. Data Analysis
3. Results
3.1. Demographic and Clinical Characteristics of PLHIV
3.2. Genotypic Distributions of GSTs and HLA-B among Healthy Volunteers, ARVDILI and Non-ARVDILI Groups
3.3. Effects of Multiple Genes on ARVDILI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stern, J.O.; Robinson, P.A.; Love, J.; Lanes, S.; Imperiale, M.S.; Mayers, D.L. A Comprehensive Hepatic Safety Analysis of Nevirapine in Different Populations of HIV Infected Patients. JAIDS J. Acquir. Immune Defic. Syndr. 2003, 34, S21–S33. [Google Scholar] [CrossRef] [PubMed]
- Reiter, G.S. Hepatitis in an HIV-infected man. AIDS Clin. Care 1997, 9, 78–81. [Google Scholar] [PubMed]
- Parsons, M.; Campa, A.; Lai, S.; Li, Y.; Martinez, J.D.; Murillo, J.; Greer, P.; Martinez, S.S.; Baum, M.K. Effect of GSTM1-Polymorphism on Disease Progression and Oxidative Stress in HIV Infection: Modulation by HIV/HCV Co-Infection and Alcohol Consumption. J. AIDS Clin. Res. 2013, 4, 10002337. [Google Scholar] [CrossRef]
- Singh, H.O.; Lata, S.; Angadi, M.; Bapat, S.; Pawar, J.; Nema, V.; Ghate, M.V.; Sahay, S.; Gangakhedkar, R.R. Impact of GSTM1, GSTT1 and GSTP1 gene polymorphism and risk of ARV-associated hepatotoxicity in HIV-infected individuals and its modulation. Pharm. J. 2017, 17, 53–60. [Google Scholar] [CrossRef]
- Ebeshi, B.U.; Bolaji, O.O.; Masimirembwa, C.M. Glutathione-S-transferase (M1 and T1) polymorphisms in Nigerian populations. J. Med. Genet. Genom. 2011, 3, 56–60. [Google Scholar]
- Soto-Quintana, O.; Zúñiga-González, G.M.; Ramírez-Patiño, R.; Ramos-Silva, A.; Figuera, L.E.; Carrillo-Moreno, D.I.; Gutiérrez-Hurtado, I.A.; Puebla-Pérez, A.M.; Sánchez-Llamas, B.; Gallegos-Arreola, M.P. Association of the GSTM1 null polymorphism with breast cancer in a Mexican population. Genet. Mol. Res. 2015, 14, 13066–13075. [Google Scholar] [CrossRef]
- Chanhom, N.; Udomsinprasert, W.; Chaikledkaew, U.; Mahasirimongkol, S.; Wattanapokayakit, S.; Jittikoon, J. GSTM1 and GSTT1 genetic polymorphisms and their association with antituberculosis drug-induced liver injury. Biomed. Rep. 2020, 12, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Shokeer, A.; Mannervik, B. Residue 234 is a master switch of the alternative-substrate activity profile of human and rodent theta class glutathione transferase T1-1. Biochim. Biophys. Acta 2010, 1800, 466–473. [Google Scholar] [CrossRef]
- Tars, K.; Larsson, A.K.; Shokeer, A.; Olin, B.; Mannervik, B.; Kleywegt, G.J. Structural basis of the suppressed catalytic activity of wild-type human glutathione transferase T1-1 compared to its W234R mutant. J. Mol. Biol. 2006, 355, 96–105. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress during HIV Infection: Mechanisms and Consequences. Oxid. Med. Cell. Longev. 2016, 2016, 8910396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, A.M.; Nolan, D.; James, I.; Cameron, P.; Keller, J.; Moore, C.; Phillips, E.; Christiansen, F.T.; Mallal, S. Predisposition to nevirapine hypersensitivity associated with HLA-DRB1*0101 and abrogated by low CD4 T-cell counts. AIDS 2005, 19, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Chantarangsu, S.; Mushiroda, T.; Mahasirimongkol, S.; Kiertiburanakul, S.; Sungkanuparph, S.; Manosuthi, W.; Tantisiriwat, W.; Charoenyingwattana, A.; Sura, T.; Chantratita, W.; et al. HLA-B*3505 allele is a strong predictor for nevirapine-induced skin adverse drug reactions in HIV-infected Thai patients. Pharm. Genom. 2009, 19, 139–146. [Google Scholar] [CrossRef]
- Gatanaga, H.; Yazaki, H.; Tanuma, J.; Honda, M.; Genka, I.; Teruya, K.; Tachikawa, N.; Kikuchi, Y.; Oka, S. HLA-Cw8 primarily associated with hypersensitivity to nevirapine. AIDS 2007, 21, 264–265. [Google Scholar] [CrossRef]
- Littera, R.; Carcassi, C.; Masala, A.; Piano, P.; Serra, P.; Ortu, F.; Corso, N.; Casula, B.; La Nasa, G.; Contu, L.; et al. HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients. AIDS 2006, 20, 1621. [Google Scholar] [CrossRef]
- Yuan, J.; Guo, S.; Hall, D.; Cammett, A.M.; Jayadev, S.; Distel, M.; Storfer, S.; Huang, Z.; Mootsikapun, P.; Ruxrungtham, K.; et al. Toxicogenomics of nevirapine-associated cutaneous and hepatic adverse events among populations of African, Asian, and European descent. AIDS 2011, 25, 1271–1280. [Google Scholar] [CrossRef]
- Esmaeilzadeh, H.; Farjadian, S.; Alyasin, S.; Nemati, H.; Nabavizadeh, H.; Esmaeilzadeh, E. Epidemiology of Severe Cutaneous Adverse Drug Reaction and Its HLA Association among Pediatrics. Iran. J. Pharm. Res. 2019, 18, 506–522. [Google Scholar]
- Phanuphak, P.; Leechawengwongs, M.; Siraprapasiri, T.; Chantratita, W.; Techasathit, W.; Teeraratkul, A.; Chokephaibulkit, K. Thailand National Guidelines on HIV/AIDs Diagnosis and Treatment, 1st ed.; Division of AIDS and STIs, Department of Disease Control: Nonthaburi, Thailand, 2010.
- Hoofnagle, J.H.; Navarro, V.J. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012.
- Kiran, B.; Karkucak, M.; Ozan, H.; Yakut, T.; Ozerkan, K.; Sag, S.; Ture, M. GST (GSTM1, GSTT1, and GSTP1) polymorphisms in the genetic susceptibility of Turkish patients to cervical cancer. J. Gynecol. Oncol. 2010, 21, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Demidov, G.; Ossowski, S. ClinCNV: A Tool for Large-Scale CNV and CNA Detection. 2019; Unpublished work. [Google Scholar]
- UNAIDS Data 2020. Available online: https://www.unaids.org/en/resources/documents/2020/unaids-data (accessed on 6 January 2021).
- Nishijima, T.; Inaba, Y.; Kawasaki, Y.; Tsukada, K.; Teruya, K.; Kikuchi, Y.; Gatanaga, H.; Oka, S. Mortality and causes of death in people living with HIV in the era of combination antiretroviral therapy compared with the general population in Japan. AIDS 2020, 34, 913. [Google Scholar] [CrossRef]
- Kovari, H.; Sabin, C.A.; Ledergerber, B.; Ryom, L.; Worm, S.W.; Smith, C.; Phillips, A.; Reiss, P.; Fontas, E.; Petoumenos, K.; et al. Antiretroviral drug-related liver mortality among HIV-positive persons in the absence of hepatitis B or C virus coinfection: The data collection on adverse events of anti-HIV drugs study. Clin. Infect. Dis. 2013, 56, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Kowalska, J.D.; Friis-Møller, N.; Kirk, O.; Bannister, W.; Mocroft, A.; Sabin, C.; Reiss, P.; Gill, J.; Lewden, C.; Phillips, A.; et al. The Coding Causes of Death in HIV (CoDe) Project: Initial results and evaluation of methodology. Epidemiology 2011, 22, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Kasthurinaidu, S.P.; Ramasamy, T.; Ayyavoo, J.; Dave, D.K.; Adroja, D.A. GST M1-T1 null allele frequency patterns in geographically assorted human populations: A phylogenetic approach. PLoS ONE 2015, 10, e0118660. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.W.F.; Cavallari, L.H. Chapter 1—Principles of Pharmacogenomics: Pharmacokinetic, Pharmacodynamic, and Clinical Implications. In Pharmacogenomics; Lam, Y.-W.F., Cavallari, L.H., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 1–44. [Google Scholar]
- Ntais, C.; Polycarpou, A.; Ioannidis, J.P. Association of GSTM1, GSTT1, and GSTP1 gene polymorphisms with the risk of prostate cancer: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2005, 14, 176–181. [Google Scholar]
- Stehbens, W.E. Oxidative stress in viral hepatitis and AIDS. Exp. Mol. Pathol. 2004, 77, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Caswell, J.L.; Mannargudi, B.; Shenton, J.M.; Uetrecht, J.P. Study of the sequence of events involved in nevirapine-induced skin rash in Brown Norway rats. Chem. Res. Toxicol. 2006, 19, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Lian, L.-Y.; Maggs, J.L.; Chaponda, M.; Pirmohamed, M.; Williams, D.P.; Park, B.K. Quantifying the metabolic activation of nevirapine in patients by integrated applications of NMR and mass spectrometries. Drug. Metab. Dispos. 2010, 38, 122–132. [Google Scholar] [CrossRef] [Green Version]
- Claes, P.; Wintzen, M.; Allard, S.; Simons, P.; De Coninck, A.; Lacor, P. Nevirapine-induced toxic epidermal necrolysis and toxic hepatitis treated successfully with a combination of intravenous immunoglobulins and N-acetylcysteine. Eur. J. Intern. Med. 2004, 15, 255–258. [Google Scholar] [CrossRef]
| Variables | Before ART Treatment | p-Value | At 14 Days after Treatment | p-Value | At 42 Days after Treatment | p-Value | |||
|---|---|---|---|---|---|---|---|---|---|
| HIV+ without Hepatotoxicity | HIV+ with Hepatotoxicity | HIV+ without Hepatotoxicity | HIV+ with Hepatotoxicity | HIV+ without Hepatotoxicity | HIV+ with Hepatotoxicity | ||||
| N (%) | 312 (100.0) | 50 (100.0) | |||||||
| Age (years) | 36.32 ± 10.29 | 39.26 ± 11.29 | 0.080 | ||||||
| Sex (M/F) | 180/132 | 25/25 | 0.357 | ||||||
| BMI (kg/m2) | 22.58 ± 21.52 | 20.52 ± 3.58 | 0.098 | ||||||
| Smoking (%) | 70 (23.0) | 9 (18.4) | 0.581 | ||||||
| Alcohol (%) | 98 (34.4) | 16 (35.6) | 0.868 | ||||||
| CD4+ (cells/mm3) | 144.23 ± 105.11 | 167.49 ± 114.14 | 0.224 | N/A | N/A | N/A | 269.12 ± 151.24 | 303.97 ± 144.38 | 0.129 |
| Viral load (copies/mm3) | 129,027.37 ± 261,163.86 | 105,268.46 ± 170,374.59 | 0.332 | N/A | N/A | N/A | 16,760.24 ± 163,799.31 | 138.63 ± 445.04 | 0.386 |
| AST (IU/L) | 31.74 ± 15.47 | 32.76 ± 13.42 | 0.299 | 28.09 ± 13.58 | 61.72 ± 78.32 | <0.001 | 28.22 ± 14.84 | 69.02 ± 108.36 | <0.001 |
| ALT (IU/L) | 33.48 ± 18.86 | 36.66 ± 18.33 | 0.235 | 32.82 ± 17.67 | 74.80 ± 102.83 | <0.001 | 35.75 ± 29.40 | 89.19 ± 144.50 | <0.001 |
| Genotypic Distribution of GSTs in Non-ARVDILI and ARVDILI Patients | ||||
|---|---|---|---|---|
| GST | HIV (n = 362) | Healthy (n = 85) | OR (95% CI) | p-Value |
| GSTM1 present | 151 (41.7%) | 40 (47.1%) | 1 | ref. |
| GSTM1 null | 211 (58.3%) | 45 (52.9%) | 1.257 (0.781–2.022) | 0.346 |
| GSTT1 present | 237 (65.5%) | 57 (67.1%) | 1 | ref. |
| GSTT1 null | 125 (34.5%) | 28 (32.9%) | 1.097 (0.662–1.817) | 0.720 |
| GSTM1 or GSTT1 present | 292 (80.7%) | 71 (83.5%) | 1 | ref. |
| GSTM1 and GSTT1 null | 70 (19.3%) | 14 (16.5%) | 1.268 (0.668–2.406) | 0.467 |
| GST | HIV+ with Hepatotoxicity | HIV+ without Hepatotoxicity | Odd Ratio (95% CI) | p-Value |
|---|---|---|---|---|
| N (%) | 50 (100%) | 312 (100%) | ||
| GST | ||||
| GSTM1 present | 21 (42.0%) | 130 (41.7%) | 1 | Ref. |
| GSTM1 null | 29 (58.0%) | 182 (58.3%) | 0.99 (0.54–1.81) | 0.965 |
| GSTT1 present | 39 (78.0%) | 198 (63.5%) | 1 | Ref. |
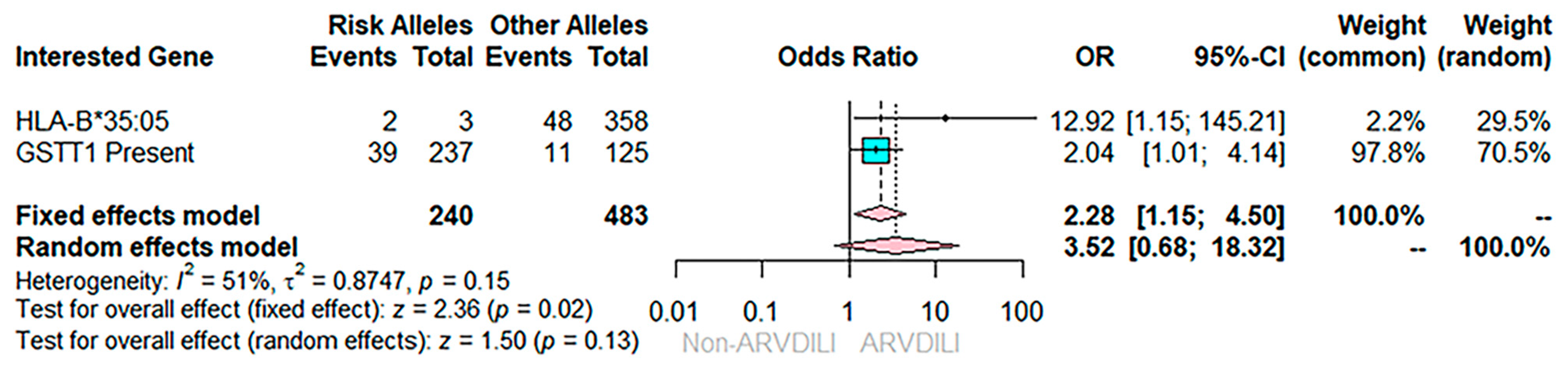
| GSTT1 null | 11 (22.0%) | 114 (36.5%) | 0.49 (0.24–0.99) | 0.045 |
| GSTM1 or GSTT1 present | 44 (88.0%) | 248 (79.5%) | 1 | Ref. |
| GSTM1 and GSTT1 null | 6 (12.0%) | 64 (20.5%) | 0.53 (0.22–1.29) | 0.157 |
| HLA-B | ||||
| *35:01 | 0 (0.0) | 6 (1.9) | 0.47 (0.03–8.39) | 1.000 |
| *35:03 | 0 (0.0) | 4 (1.3) | 0.68 (0.04–12.80) | 1.000 |
| *35:05 | 2 (4.0) | 1 (0.3) | 12.92 (1.15–145.21) | 0.052 |
| *35:60 | 0 (0.0) | 1 (0.3) | 2.06 (0.08–51.17) | 1.000 |
| *58:01 | 5 (10.0) | 45 (90.0) | 1.415 (0.59–3.40) | 0.427 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chanhom, N.; Jittikoon, J.; Wattanapokayakit, S.; Mahasirimongkol, S.; Charoenyingwattana, A.; Udomsinprasert, W.; Chaikledkaew, U.; Suvichapanich, S.; Mushiroda, T.; Kiertiburanakul, S.; et al. The Association of HLA-B*35 and GSTT1 Genotypes and Hepatotoxicity in Thai People Living with HIV. J. Pers. Med. 2022, 12, 940. https://doi.org/10.3390/jpm12060940
Chanhom N, Jittikoon J, Wattanapokayakit S, Mahasirimongkol S, Charoenyingwattana A, Udomsinprasert W, Chaikledkaew U, Suvichapanich S, Mushiroda T, Kiertiburanakul S, et al. The Association of HLA-B*35 and GSTT1 Genotypes and Hepatotoxicity in Thai People Living with HIV. Journal of Personalized Medicine. 2022; 12(6):940. https://doi.org/10.3390/jpm12060940
Chicago/Turabian StyleChanhom, Noppadol, Jiraphun Jittikoon, Sukanya Wattanapokayakit, Surakameth Mahasirimongkol, Angkana Charoenyingwattana, Wanvisa Udomsinprasert, Usa Chaikledkaew, Supharat Suvichapanich, Taisei Mushiroda, Sasisopin Kiertiburanakul, and et al. 2022. "The Association of HLA-B*35 and GSTT1 Genotypes and Hepatotoxicity in Thai People Living with HIV" Journal of Personalized Medicine 12, no. 6: 940. https://doi.org/10.3390/jpm12060940
APA StyleChanhom, N., Jittikoon, J., Wattanapokayakit, S., Mahasirimongkol, S., Charoenyingwattana, A., Udomsinprasert, W., Chaikledkaew, U., Suvichapanich, S., Mushiroda, T., Kiertiburanakul, S., Rojanawiwat, A., Wangsomboonsiri, W., Manosuthi, W., Kantipong, P., Apisarnthanarak, A., Sangsirinakakul, W., Wongprasit, P., Chaiwarith, R., Tantisiriwat, W., ... Chantratita, W. (2022). The Association of HLA-B*35 and GSTT1 Genotypes and Hepatotoxicity in Thai People Living with HIV. Journal of Personalized Medicine, 12(6), 940. https://doi.org/10.3390/jpm12060940






