Abstract
Precision medicine (PM), specifically genetic-based testing, is currently used in over 140,000 individual tests to inform the clinical management of disease. Though several databases (e.g., the NIH Genetic Testing Registry) demonstrate the availability of these sequencing-based tests, we do not currently understand the extent to which these tests are used. There exists a need to synthesize the body of real-world data (RWD) describing the use of sequencing-based tests to inform their appropriate use. To accomplish this, we performed a scoping review to examine what RWD sources have been used in studies of PM utilization between January 2015 and August 2021 to characterize the use of genome sequencing (GS), exome sequencing (ES), tumor sequencing (TS), next-generation sequencing-based panels (NGS), gene expression profiling (GEP), and pharmacogenomics (PGx) panels. We abstracted variables describing the use of these types of tests and performed a descriptive statistical analysis. We identified 440 articles in our search and included 72 articles in our study. Publications based on registry databases were the most common, followed by studies based on private insurer administrative claims. Slightly more than one-third (38%) used integrated datasets. Two thirds (67%) of the studies focused on the use of tests for oncological clinical applications. We summarize the RWD sources used in peer-reviewed literature on the use of PM. Our findings will help improve future study design by encouraging the use of centralized databases and registries to track the implementation and use of PM.
1. Introduction
Since the sequencing of the first human genome, there has been an explosion in the development of precision medicine (PM) tests. This, in conjunction with the rapid reduction in the cost of genomic sequencing over the 15 years from approximately USD 14 million in 2006 to USD 1000 in 2021 [1], as well as the rise in bioinformatic and digital data-driven approaches to health care delivery [2], puts our health care system in a position to utilize PM technologies with significantly greater ubiquity and effectiveness. Therefore, there is a need to understand how PM is being used.
The implementation of PM is increasing, and thus it is important to understand how it is used in routine clinical care [3]. Achieving this requires the use of real-world data (RWD): data related to patient health states and/or the delivery of care routinely collected from a variety of sources, e.g., electronic health records (EHR), claims data, registries, etc. However, it is well known that the use of RWD presents numerous challenges (e.g., data validity and quality relating to unstructured data, haphazard and unstandardized collection, methods of obtaining RWD, and issues such as data access, privacy, and the ability to combine data), and these challenges may be magnified in the context of PM [4,5,6].
We define PM as the use of genetic testing to target interventions, including the use of genomic tests for diagnosis, screening in asymptomatic patients, risk assessment, determining the prognosis of a diagnosed disease, and predicting treatment responses or adverse events [7]. For example, the interrogation of the single genes BRCA1 and BRCA2 for the risk assessment of hereditary breast and ovarian cancer (HBOC) has been available since 1996 [8], and the use of whole-exome and whole-genome sequencing in the diagnosis of suspected genetic diseases has developed much more recently. While these tests are being used, the extent of their utilization individually or as a whole, and their impact on clinical management worldwide, is unknown [3,9]. One reason for this is the lack of a central repository of these data.
At present, there is no centrally integrated source of utilizable data (e.g., tests performed with the intent of using the results in clinical management) in the real world. For example, some data can be found in the gray literature (e.g., white papers, health system reports, regulatory filings, company websites, news reports, and national/international consortia websites), and some data can be obtained from administrative/clinical resources (e.g., EHR, claims data, fee schedules, industry databases, and registries) [3]. However, very few of the data records are linked, highlighting a need to synthesize real-world data (RWD) describing the use of PM sequencing-based tests in order to obtain an overall utilization picture and to understand their impact on clinical management.
Our objective was to identify where PM use data have been described in peer-reviewed publications and how they may be linked in the real world to track utilization. To accomplish this, we performed a scoping review to examine what real-world data sources have been used in studies of PM utilization. We add to the literature by providing a systematic assessment of the real-world databases and registries that have been used in existing peer-reviewed studies. Our findings can be used to inform future study design and encourage the development and use of centralized databases and registries to track the real-world use of PM-based tests. These centralized databases or registries would inform future studies on appropriate test utilization.
2. Materials and Methods
We sought to identify peer-reviewed publications that used RWD sources to describe the use of PM using a scoping review. We searched PubMed for articles written in English and published between January 2015 and August 2021 (Supplementary Materials S1. Search Terms). We chose to limit our search to 5 years plus the current year, as this would identify the majority of articles on the topic (only nine articles would have been identified in the search prior to 2015, none of which met the inclusion criteria).
2.1. Inclusion/Exclusion
We included original research studies that examined PM, as defined above (e.g., whole-exome/genome sequencing (WES/WGS), tumor sequencing, next-generation sequencing (NGS) panels, gene expression profiling, pharmacogenetics, and hereditary cancer panels, including BCRA1/2), and its utilization (number and/or type of genetic tests performed) using real-world data (RWD; data related to patient health states and/or the delivery of care routinely collected from a variety of sources, e.g., electronic health records, claims data, registries, laboratories, or integrated datasets from these sources). We excluded non-genetic tests and single-gene test studies (while relevant, we wanted to focus on multi-gene tests, as those are more difficult to identify in databases such as claims databases due to coding issues), studies that did not fit our RWD definition (as defined above), and studies that were focused on economic evaluation versus test utilization.
2.2. Data Abstraction
We developed a data abstraction spreadsheet (Supplementary Materials Table S2. Data Abstracted), and data were extracted independently by the two co-authors (A.K., M.P.D.). Discrepancies in coding, such as details regarding database names or test names or if datasets were integrated, were minimal and were resolved by discussion. We categorized studies using the following six categories (items in paratheses correspond to data abstracted):
- Demographic (PubMed ID, first author, year);
- Research question (research question);
- Name of RWD sources and whether or not the study used an integrated or merged dataset from two or more sources (clinical data source, admin claims (commercial) source, admin claims (public) source, registry source);
- Laboratory name that performed the PM testing, if available (lab name, other);
- Condition or disease, including if it was cancer or non-cancer condition (condition);
- Test name and/or genes, including if the test was BRCA, gene expression profiling/Oncotype Dx, multigene pane, tumor sequencing, WES/WGS, or other (test name and/or genes).
2.3. Analysis
Descriptive statistics were used to analyze data across variables.
3. Results
We identified 440 articles in our search and excluded 368, leaving a remaining 72 articles to be included in our study (Figure 1, Supplementary Materials S3. Studies Included, Supplementary Materials Table S4. Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) Checklist). We analyzed abstracted variables to determine the type of databases used, the laboratories used, if the condition/disease was cancer/non-cancer, and the type of test.
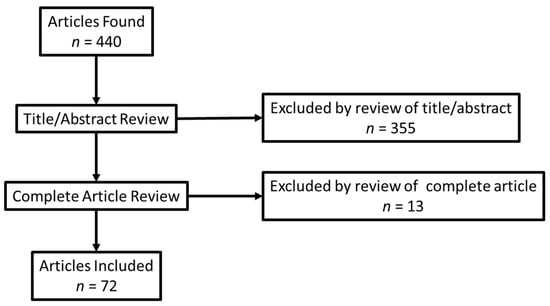
Figure 1.
Prisma diagram.
The two most common RWD types were administrative claims data (58%) and registry data (55%) (Table 1) [10]. Commonly used data sources were the SEER-Medicare (Surveillance, Epidemiology, and End Results Program-Medicare) linked database, the National Cancer Database, and Truven Health Analytics claims databases (now part of IBM Watson Health, Cambridge, MA, USA). Of importance is that over one-third (38%) of studies used an integrated data source (e.g., SEER-Medicare) or combined data from more than one source for their analyses. The use of such datasets was often in response to the challenges of using RWD from a single source. For example, several studies linked data from laboratories to registries and claims data, thereby enabling the analyses of test results that were otherwise missing. As one example, Roberts et al., 2019 linked data from Genomic Health’s clinical laboratory to SEER data to examine the utilization of a gene expression profiling test for breast cancer (Oncotype DX Breast Recurrence Score) [11].

Table 1.
Percentage of studies by characteristics (n = 72).
Two-thirds (67%) of studies focused on cancer, and slightly more than one-third (38%) focused on just two types of commonly used tests (22% of studies focused on gene expression profiling tests for breast cancer and 16% on BRCA1/2 tests for breast cancer risk) (Table 2). The remaining 33% of studies examined non-cancer conditions, such as metabolic deficiencies, prenatal abnormalities, various neurological conditions, familial hypercholesterolemia, and miscellaneous pharmacogenetic testing. Of the 72 included studies, 22 included the name of the test and laboratory that was utilized to perform the genetic tests (e.g., Genomic Health, GeneSight®, Ambry, GeneDx, Myriad, and Foundation Health) (Table 2).

Table 2.
Percentage of test types and examples (n = 88) *.
The studies we identified included a variety of research questions. These research questions addressed healthcare utilization (with various perspectives, including geographic, racial, and socioeconomic), the implementation of testing, costs (with various perspectives, including hospital and payer), diagnostic yield, the efficacy of treatment (informed by test results), clinical utility, a description of the referral patterns for test utilization, and appropriate test utilization.
4. Discussion
We identified 72 articles that described the use of PM in peer-reviewed RWD studies for a wide variety of tests, nearly 67% of which were cancer-based. Many of these studies were performed using proprietary or subscription-based databases or registries.
We found five categories of databases used in these studies. First, private administrative claims data were frequently used by insurance companies to measure utilization by “claim event.” In this case, the databases were limited to single insurance company claims and were often proprietary or costly to access. Furthermore, in order to accurately characterize the use of PM tests, individual claim events had to be able to be assigned to a specific test, and many PM tests were billed as unlisted or in non-descriptive ways (e.g., CPT81479) as a result. Second, public databases were identified in the form of Medicare claims. Similar to private claims, Medicare claims were also challenging in that they were limited to data collected to the population they served and were subject to billing coding issues. Third, clinical databases, such as medical systems EHR, were commonly used. The prevalence of these datasets could be descriptive of the use of PM at a specific hospital system; however, while such databases are informative, they are only representative of a localized diagnostic and treatment protocol in place at a particular hospital or clinic and may not be depictive of how PM is used across various other types of patient settings or populations. Fourth, laboratory databases, such as those maintained by test manufacturers, were limited to the specific tests offered by the laboratory and were largely proprietary in nature. Additionally, laboratories mainly only offered access to their individual test utilization for clinical studies in conjunction with a research partnership. Last, 55% of RWD used were registries that were created to track a specific test, condition, or population. These types of databases were helpful to study a specific test or condition and were informative, provided that the data included were needed for a particular study. Registries could be limiting if the necessary data were not included and if access was usually only available to particular researchers. Ultimately, each type of database, whether it be private/public claims, clinical, laboratory, or a registry dataset, held advantages and limitations that made each better suited for an understanding of PM utilization across various parts of the United States and the world.
We provide two examples where the use of a database of PM utilization can have clinical, economic, and social implications. We found that nearly 38% of RWD studies on PM were focused on the use of BRCA1/2 testing or gene expression profiling (GEP). While these tests were important in cancer risk and treatment, they have been in use for many years with established PM utility, and utilization may no longer be tracked. For example, the Surveillance, Epidemiology, and End Results Program (SEER) database is a commonly used database for analyzing breast cancer cases, but the version containing Oncotype Dx data is no longer being updated and only contains data from 2004 to 2016 [12]. On the other hand, tests that are relatively new to PM, such as WES/WGS, were rarely described in the RWD databases (our study found only two such studies), which could have been useful in informing their clinical utility and application. Furthermore, there were many studies of one test type in specific settings that were only informative to that setting or location, and not generalizable across different populations and regions. To demonstrate these last two points, the use of rapid GS has been demonstrated by Dimmock et al., 2021 for diagnosis, change management, and saving money in critically ill infants at five California children’s hospitals, but these data are only found in study documentation and are limited to 184 infants within California’s Medicaid program [13].
Given our findings, there is a great need for a publicly available real-world dataset for PM. This ideal dataset should be cross-cutting in terms of source (e.g., EHR, public/private claims data, and individual registries), the types of conditions (e.g., cancer and non-cancer), and the specific tests used. A dataset, as described, would be beneficial to many stakeholders, such as insurance companies, clinical researchers, and test implementation specialists. The Association of Molecular Pathology Clinical Genomic Data Working Group describes the challenges, opportunities, and solutions surrounding the complexity, lack of standards, fluency, and functionality of genomic data generation, interoperability, and utilization within electronic health records [14]. Beckman and Lew describe the necessity of international data sharing and bioinformatic data consolidation in translating PM into clinically relevant, evidence-based medicine [15]. Additionally, the Professional Society for Health Economics and Outcomes Research (ISPOR) announced that the Real-World Evidence Transparency Initiative has launched the Real-World Evidence Registry [16]. While this registry is open to many types of medical interventions, it is a good launching platform that could inform registries for PM.
Limitations
Our study has limitations related to our ability to aggregate all appropriate studies in our scoping review. It was infeasible to perform a comprehensive assessment of all studies of PM utilization; specifically, identifying articles that described utilization was difficult due to the lack of “utilization” subject heading or MeSH term in PubMed. Furthermore, we limited our search to articles only found in PubMed, which may limit or show bias toward US publications. Finally, we did not abstract and analyze the sources of funding for each included study. For these reasons, our results are illustrative in nature.
5. Conclusions
In conclusion, RWD is essential to understanding the use of PM. Our findings identified studies describing the use of PM in specific settings/tests (e.g., specific EHR, Oncotype Dx) or using specific datasets (e.g., Truvan). As highlighted by others, there are many challenges to using existing databases [4,5,6]. There are no publicly available datasets that can be used to describe the overall use of PM. Future databases and registries are necessary to inform the overall utilization of PM.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm12040557/s1, S1: Search Terms; Table S2: Data Abstracted; S3. Studies Included; Table S4. Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) Checklist. References [11,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, M.P.D.; methodology, M.P.D.; validation, M.P.D. and A.K.; formal analysis, M.P.D. and A.K.; investigation, M.P.D.; resources, M.P.D.; data curation, M.P.D.; writing—original draft preparation, M.P.D.; writing—review and editing, M.P.D. and A.K.; visualization, M.P.D.; supervision, M.P.D.; project administration, M.P.D.; funding acquisition, M.P.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by an unrestricted consulting agreement with Illumina (no number) and grants from the National Human Genome Research Institute (R01 HG011792) and from the National Cancer Institute (R01 CA221870).
Institutional Review Board Statement
Not applicable; no human subjects/data/tissue were used in this study.
Informed Consent Statement
Not applicable; no human subjects/data/tissue were used in this study.
Data Availability Statement
Data are available upon request from corresponding author.
Acknowledgments
Authors wish to thank Kathryn Phillips and Patricia Deverka, University of California, San Francisco and Center for Translational and Policy Research on Precision Medicine (TRANSPERS), for their thoughtful critique of the draft manuscript.
Conflicts of Interest
M.P.D. receives consulting income from Illumina to support the research conducted for this publication. A.K. declares no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: https://www.genome.gov/sequencingcostsdata (accessed on 18 February 2022).
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine. Omics 2018, 22, 630–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, K.A.; Douglas, M.P.; Marshall, D.A. Expanding Use of Clinical Genome Sequencing and the Need for More Data on Implementation. JAMA 2020, 324, 2029–2030. [Google Scholar] [CrossRef] [PubMed]
- Deverka, P.A.; Douglas, M.P.; Phillips, K.A. Use of Real-World Evidence in US Payer Coverage Decision-Making for Next-Generation Sequencing-Based Tests: Challenges, Opportunities, and Potential Solutions. Value Health 2020, 23, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Rudrapatna, V.A.; Butte, A.J. Opportunities and challenges in using real-world data for health care. J. Clin. Investig. 2020, 130, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Crown, W.H. Real-World Evidence, Causal Inference, and Machine Learning. Value Health 2019, 22, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Teutsch, S.M.; Bradley, L.A.; Palomaki, G.E.; Haddow, J.E.; Piper, M.; Calonge, N.; Dotson, W.D.; Douglas, M.P.; Berg, A.O. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: Methods of the EGAPP Working Group. Genet. Med. 2009, 11, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolata, G. Breaking Ranks, Lab Offers Test to Assess Risk of Breast Cancer. New York Times, 1 April 1996. Available online: https://www.nytimes.com/1996/04/01/us/breaking-ranks-lab-offers-test-to-assess-risk-of-breast-cancer.html (accessed on 11 December 2021).
- Phillips, K.A.; Douglas, M.P.; Wordsworth, S.; Buchanan, J.; Marshall, D.A. Availability and funding of clinical genomic sequencing globally. BMJ Glob. Health 2021, 6, e004415. [Google Scholar] [CrossRef] [PubMed]
- Garrison, L.P., Jr.; Neumann, P.J.; Erickson, P.; Marshall, D.; Mullins, C.D. Using real-world data for coverage and payment decisions: The ISPOR Real-World Data Task Force report. Value Health 2007, 10, 326–335. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.C.; Kurian, A.W.; Petkov, V.I. Uptake of the 21-Gene Assay among Women with Node-Positive, Hormone Receptor-Positive Breast Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Oncotype DX Database (2004–2016) with or without SES/Rurality: National Cancer Institute Surveillance, Epidemiology, and End Results Program. 2022. Available online: https://seer.cancer.gov/seerstat/databases/oncotype-dx/index.html (accessed on 18 February 2022).
- Dimmock, D.; Caylor, S.; Waldman, B.; Benson, W.; Ashburner, C.; Carmichael, J.L.; Carroll, J.; Cham, E.; Chowdhury, S.; Cleary, J.; et al. Project Baby Bear: Rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care. Am. J. Hum. Genet. 2021, 108, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.B.; Abruzzo, L.V.; Hirschhorn, J.W.; Jones, D.; Jordan, D.C.; Nassiri, M.; Ogino, S.; Patel, N.R.; Suciu, C.G.; Temple-Smolkin, R.L.; et al. Electronic Health Records and Genomics: Perspectives from the Association for Molecular Pathology Electronic Health Record (EHR) Interoperability for Clinical Genomics Data Working Group. J. Mol. Diagn. 2022, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.S.; Lew, D. Reconciling evidence-based medicine and precision medicine in the era of big data: Challenges and opportunities. Genome Med. 2016, 8, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- RWE Registry Developed From the Real-World Evidence Transparency Initiative: The Professional Society for Health Economics and Outcomes Research (ISPOR). 2021. Available online: https://www.ispor.org/heor-resources/news/2021/10/26/new-real-world-evidence-registry-launches (accessed on 11 December 2021).
- Abul-Husn, N.S.; Manickam, K.; Jones, L.K.; Wright, E.A.; Hartzel, D.N.; Gonzaga-Jauregui, C.; O’Dushlaine, C.; Leader, J.B.; Kirchner, H.L.; Lindbuchler, D.M.; et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science 2016, 354, aaf7000. [Google Scholar] [CrossRef] [PubMed]
- Altman, A.M.; Marmor, S.; Tuttle, T.M.; Hui, J.Y.C. 21-Gene Recurrence Score Testing in HER2-positive Patients. Clin. Breast Cancer 2019, 19, 126–130. [Google Scholar] [CrossRef]
- Anderson, H.D.; Crooks, K.R.; Kao, D.P.; Aquilante, C.L. The landscape of pharmacogenetic testing in a US managed care population. Genet. Med. 2020, 22, 1247–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, J.; Toscano, M.; Kotchko, N.; Friedman, S.; Schwartz, M.D.; Virgo, K.S.; Lynch, K.; Andrews, J.E.; Loi, C.X.A.; Bauer, J.E.; et al. Utilization and Outcomes of BRCAGenetic Testing and Counseling in a National Commercially Insured Population. JAMA Oncol. 2015, 1, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardakjian, T.M.; Helbig, I.; Quinn, C.; Elman, L.B.; McCluskey, L.F.; Scherer, S.S.; Gonzalez-Alegre, P. Genetic test utilization and diagnostic yield in adult patients with neurological disorders. Neurogenetics 2018, 19, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Benitez, J.; Cool, C.L.; Scotti, D.J. Use of combinatorial pharmacogenomic guidance in treating psychiatric disorders. Pers. Med. 2018, 15, 481–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhutiani, N.; Vuong, B.; Egger, M.E.; Eldredge-Hindy, H.; McMasters, K.M.; Ajkay, N. Evaluating patterns of utilization of gene signature panels and impact on treatment patterns in patients with ductal carcinoma in situ of the breast. Surgery 2019, 166, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Blaes, A.H.; Jewett, P.I.; McKay, K.; Riley, D.; Jatoi, I.; Trentham-Dietz, A.; Chrischilles, E.; Klemp, J.R. Factors associated with genetic testing in a cohort of breast cancer survivors. Breast J. 2019, 25, 1241–1244. [Google Scholar] [CrossRef]
- Blagec, K.; Kuch, W.; Samwald, M. The Importance of Gene-Drug-Drug-Interactions in Pharmacogenomics Decision Support: An Analysis Based on Austrian Claims Data. In Health Informatics Meets eHealth; Schreier, G., Ammenwerth, E., Hörbst, A., Hayn, D., Eds.; Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2017; Volume 236, pp. 121–127. Available online: https://ebooks.iospress.nl/publication/46468 (accessed on 18 February 2022).
- Byfield, S.D.; Wei, H.; DuCharme, M.; Lancaster, J.M. Economic impact of multigene panel testing for hereditary breast and ovarian cancer. J. Comp. Eff. Res. 2021, 10, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Caplan, E.O.; Wong, W.B.; Ferries, E.; Hulinsky, R.; Brown, V.T.; Bordenave, K.; Suehs, B.T. Novel Approach Using Administrative Claims to Evaluate Trends in Oncology Multigene Panel Testing for Patients Enrolled in Medicare Advantage Health Plans. JCO Precis. Oncol. 2021, 5, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Kolor, K.; Grosse, S.D.; Rodriguez, J.L.; Lynch, J.A.; Green, R.F.; Dotson, W.D.; Bowen, M.S.; Khoury, M.J. Trends in utilization and costs of BRCA testing among women aged 18–64 years in the United States, 2003–2014. Genet. Med. 2018, 20, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Childers, K.K.; Maggard-Gibbons, M.; Macinko, J.; Childers, C.P. National Distribution of Cancer Genetic Testing in the United States. JAMA Oncol. 2018, 4, 876. [Google Scholar] [CrossRef] [PubMed]
- Chitty, L.S.; Wright, D.; Hill, M.; Verhoef, T.I.; Daley, R.; Lewis, C.; Mason, S.; McKay, F.; Jenkins, L.; Howarth, A.; et al. Uptake, outcomes, and costs of implementing non-invasive prenatal testing for Down’s syndrome into NHS maternity care: Prospective cohort study in eight diverse maternity units. BMJ 2016, 354, i3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cress, R.D.; Chen, Y.S.; Morris, C.R.; Chew, H.; Kizer, K.W. Underutilization of gene expression profiling for early-stage breast cancer in California. Cancer Causes Control 2016, 27, 721–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalal, A.A.; Guerin, A.; Mutebi, A.; Culver, K.W. Economic analysis of BRAF gene mutation testing in real world practice using claims data: Costs of single gene versus panel tests in patients with lung cancer. J. Med. Econ. 2018, 21, 649–655. [Google Scholar] [CrossRef] [PubMed]
- DeFrancesco, M.S.; Waldman, R.N.; Pearlstone, M.M.; Karanik, D.; Bernhisel, R.; Logan, J.; Alico, L.; Adkins, R.T. Hereditary Cancer Risk Assessment and Genetic Testing in the Community-Practice Setting. Obstet. Gynecol. 2018, 132, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.; Jena, A.B. Do celebrity endorsements matter? Observational study of BRCA gene testing and mastectomy rates after Angelina Jolie’s New York Times editorial. BMJ 2016, 355, i6357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinan, M.A.; Mi, X.; Reed, S.D.; Hirsch, B.R.; Lyman, G.H.; Curtis, L.H. Initial Trends in the Use of the 21-Gene Recurrence Score Assay for Patients With Breast Cancer in the Medicare Population, 2005–2009. JAMA Oncol. 2015, 1, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Dinan, M.A.; Mi, X.; Reed, S.D.; Lyman, G.H.; Curtis, L.H. Association Between Use of the 21-Gene Recurrence Score Assay and Receipt of Chemotherapy Among Medicare Beneficiaries With Early-Stage Breast Cancer, 2005–2009. JAMA Oncol. 2015, 1, 1098–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinan, M.A.; Wilson, L.; Reed, S.D. Chemotherapy Costs and 21-Gene Recurrence Score Genomic Testing Among Medicare Beneficiaries With Early-Stage Breast Cancer, 2005 to 2011. J. Natl. Compr. Cancer Netw. 2019, 17, 245–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinan, M.A.; Wilson, L.E.; Reed, S.D.; Griggs, J.J.; Norton, E.C. Association of 21-Gene Assay (OncotypeDX) Testing and Receipt of Chemotherapy in the Medicare Breast Cancer Patient Population Following Initial Adoption. Clin. Breast Cancer 2020, 20, 487–494.e1. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, E.; Raymond, S.; Chun, J.; Fong, A.; Patel, N.; Guth, A.; Schnabel, F. Genomic testing in early stage invasive male breast cancer: An NCDB analysis from 2008 to 2014. Breast J. 2019, 25, 425–433. [Google Scholar] [CrossRef]
- El Rouby, N.; Alrwisan, A.; Langaee, T.; Lipori, G.; Angiolillo, D.J.; Franchi, F.; Riva, A.; Elsey, A.; Johnson, J.A.; Cavallari, L.H.; et al. Clinical Utility of Pharmacogene Panel-Based Testing in Patients Undergoing Percutaneous Coronary Intervention. Clin. Transl. Sci. 2019, 13, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Fohner, A.E.; Ranatunga, D.K.; Thai, K.K.; Lawson, B.L.; Risch, N.; Oni-Orisan, A.; Jelalian, A.T.; Rettie, A.E.; Liu, V.X.; Schaefer, C.A. Assessing the clinical impact of CYP2C9 pharmacogenetic variation on phenytoin prescribing practice and patient response in an integrated health system. Pharmacogenet. Genom. 2019, 29, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Franc, B.L.; Copeland, T.P.; Thombley, R.; Park, M.; Marafino, B.; Dean, M.; Boscardin, W.J.; Rugo, H.S.; Dudley, R.A. Geographic and Patient Characteristics Associated With Election of Prophylactic Mastectomy in Young Breast Cancer Patients With Early Disease. Am. J. Clin. Oncol. 2018, 41, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Geddes, G.C.; Basel, D.; Frommelt, P.; Kinney, A.; Earing, M. Genetic Testing Protocol Reduces Costs and Increases Rate of Genetic Diagnosis in Infants with Congenital Heart Disease. Pediatr. Cardiol. 2017, 38, 1465–1470. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Bs, M.S.; Fuchs, E.L.; Berenson, A.B.; Kuo, Y. BRCA testing in unaffected young women in the United States, 2006–2017. Cancer 2020, 126, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Scholl, M.; Fuchs, E.L.; Wong, R.; Kuo, Y.-F.; Berenson, A.B. Trends in Positive BRCA Test Results Among Older Women in the United States, 2008–2018. JAMA Netw. Open 2020, 3, e2024358. [Google Scholar] [CrossRef] [PubMed]
- Hefti, E.; Jacobs, D.M.; Rana, K.; Blanco, J.G. Analysis of outpatient HER2 testing in New York state using the statewide planning and research cooperative system. Pharmacogenomics 2018, 19, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Kamath, P.; Schlumbrecht, M.; Miao, F.; Driscoll, D.; Oldak, S.; Slomovitz, B.; Koru-Sengul, T.; George, S. Identifying disparities in germline and somatic testing for ovarian cancer. Gynecol. Oncol. 2019, 153, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Hutchinson, B.; Poulton, A.; Halliday, J. Population-based impact of noninvasive prenatal screening on screening and diagnostic testing for fetal aneuploidy. Genet. Med. 2017, 19, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Hull, L.E.; Lynch, J.A.; Berse, B.B.; DuVall, S.L.; Chun, D.S.; Venne, V.L.; Efimova, O.V.; Icardi, M.S.; Kelley, M.J. Clinical Impact of 21-Gene Recurrence Score Test Within the Veterans Health Administration: Utilization and Receipt of Guideline-Concordant Care. Clin. Breast Cancer 2017, 18, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.J.; Bondarenko, I.; Ward, K.C.; Hamilton, A.S.; Morrow, M.; Kurian, A.W.; Hofer, T.P. Association of Attending Surgeon With Variation in the Receipt of Genetic Testing After Diagnosis of Breast Cancer. JAMA Surg. 2018, 153, 909. [Google Scholar] [CrossRef] [PubMed]
- Nzale, S.K.; Weeks, W.B.; Ouafik, L.; Rouquette, I.; Beau-Faller, M.; Lemoine, A.; Bringuier, P.-P.; Soriano, A.-G.L.C.; Barlesi, F.; Ventelou, B. Inequity in access to personalized medicine in France: Evidences from analysis of geo variations in the access to molecular profiling among advanced non-small-cell lung cancer patients: Results from the IFCT Biomarkers France Study. PLoS ONE 2020, 15, e0234387. [Google Scholar] [CrossRef]
- Kolor, K.; Chen, Z.; Grosse, S.D.; Rodriguez, J.L.; Green, R.F.; Dotson, W.D.; Bowen, M.S.; Lynch, J.A.; Khoury, M.J. BRCA Genetic Testing and Receipt of Preventive Interventions among Women Aged 18–64 Years with Employer-Sponsored Health Insurance in Nonmetropolitan and Metropolitan Areas-United States, 2009–2014, 2017. Centers for Diseases Control and Prevention Website. Available online: https://www.cdc.gov/mmwr/volumes/66/ss/ss6615a1.htm (accessed on 18 February 2022).
- Larson, K.L.; Huang, B.; Chen, Q.; Tucker, T.; Schuh, M.; Arnold, S.M.; Kolesar, J.M. EGFR testing and erlotinib use in non-small cell lung cancer patients in Kentucky. PLoS ONE 2020, 15, e0237790. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fan, J.; Vitali, F.; Berghout, J.; Aberasturi, D.; Li, J.; Wilson, L.; Chiu, W.; Pumarejo, M.; Han, J.; et al. Novel disease syndromes unveiled by integrative multiscale network analysis of diseases sharing molecular effectors and comorbidities. BMC Med. Genom. 2018, 11, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liede, A.; Cai, M.; Crouter, T.F.; Niepel, D.; Callaghan, F.; Evans, D.G. Risk-reducing mastectomy rates in the US: A closer examination of the Angelina Jolie effect. Breast Cancer Res. Treat. 2018, 171, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.A.; Berse, B.; Coomer, N.; Kautter, J. 21-Gene recurrence score testing among Medicare beneficiaries with breast cancer in 2010–2013. Genet. Med. 2017, 19, 1134–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.A.; Berse, B.; Dotson, W.D.; Khoury, M.J.; Coomer, N.; Kautter, J. Utilization of genetic tests: Analysis of gene-specific billing in Medicare claims data. Genet. Med. 2017, 19, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, S.J.; Lin, C.C.; Todd, P.K.; Burke, J.F.; Callaghan, B.C. Genetic testing utilization for patients with neurologic disease and the limitations of claims data. Neurol. Genet. 2020, 6, e405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathias, P.C.; Hendrix, N.; Wang, W.-J.; Keyloun, K.; Khelifi, M.; Tarczy-Hornoch, P.; Devine, B. Characterizing Pharmacogenomic-Guided Medication Use With a Clinical Data Repository. Clin. Pharmacol. Ther. 2017, 102, 340–348. [Google Scholar] [CrossRef] [PubMed]
- McCuaig, J.M.; Care, M.; Ferguson, S.E.; Kim, R.H.; Stockley, T.L.; Metcalfe, K.A. Year 1: Experiences of a tertiary cancer centre following implementation of reflex BRCA1 and BRCA2 tumor testing for all high-grade serous ovarian cancers in a universal healthcare system. Gynecol. Oncol. 2020, 158, 747–753. [Google Scholar] [CrossRef]
- Mittmann, N.; Earle, C.C.; Cheng, S.Y.; Julian, J.A.; Rahman, F.; Seung, S.J.; Levine, M.N. Population-Based Study to Determine the Health System Costs of Using the 21-Gene Assay. J. Clin. Oncol. 2018, 36, 238–243. [Google Scholar] [CrossRef]
- Muller, C.; Lee, S.M.; Barge, W.; Siddique, S.M.; Berera, S.; Wideroff, G.; Tondon, R.; Chang, J.; Peterson, M.; Stoll, J.; et al. Low Referral Rate for Genetic Testing in Racially and Ethnically Diverse Patients Despite Universal Colorectal Cancer Screening. Clin. Gastroenterol. Hepatol. 2018, 16, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.C.; Isaacs, C.; Chao, C.; Tsai, H.-T.; Liu, C.; Ekezue, B.F.; Selvam, N.; Kessler, L.G.; Schwartz, M.D.; Lobo, T.; et al. Adoption of Gene Expression Profiling for Breast Cancer in US Oncology Practice for Women Younger Than 65 Years. J. Natl. Compr. Cancer Netw. 2015, 13, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- O’neill, S.C.; Isaacs, C.; Lynce, F.; Graham, D.M.A.; Chao, C.; Sheppard, V.B.; Zhou, Y.; Liu, C.; Selvam, N.; Schwartz, M.D.; et al. Endocrine therapy initiation, discontinuation and adherence and breast imaging among 21-gene recurrence score assay-eligible women under age 65. Breast Cancer Res. 2017, 19, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orucevic, A.; Heidel, R.E.; Bell, J.L. Utilization and impact of 21-gene recurrence score assay for breast cancer in clinical practice across the United States: Lessons learned from the 2010 to 2012 National Cancer Data Base analysis. Breast Cancer Res. Treat. 2016, 157, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, L.E.; Baum, C.F.; Horvath, K.; Raja, S.; Cohen, J.; Hawkins, S.S. BRCA1/2 Testing in Massachusetts Among Women With Private Insurance or Medicaid, 2011–2015. Med. Care 2020, 58, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, L.L.; Sheehan, D.F.; Tramontano, A.C.; Kong, C.Y. Disparities and Trends in Genetic Testing and Erlotinib Treatment among Metastatic Non–Small Cell Lung Cancer Patients. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 926–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkhurst, E.; Calonico, E.; Abboy, S. Utilization of Genetic Testing for RET Mutations in Patients with Medullary Thyroid Carcinoma: A Single-Center Experience. J. Genet. Couns. 2018, 27, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Pavey, A.R.; Bodian, D.L.; Vilboux, T.; Khromykh, A.; Hauser, N.S.; Huddleston, K.; Klein, E.; Black, A.; Kane, M.S.; Iyer, R.K.; et al. Utilization of genomic sequencing for population screening of immunodeficiencies in the newborn. Genet. Med. 2017, 19, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Perlis, R.H.; Mehta, R.; Edwards, A.M.; Tiwari, A.; Imbens, G.W. Pharmacogenetic testing among patients with mood and anxiety disorders is associated with decreased utilization and cost: A propensity-score matched study. Depress. Anxiety 2018, 35, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Petelin, L.; James, P.A.; Trainer, A.H. Changing landscape of hereditary breast and ovarian cancer germline genetic testing in Australia. Intern. Med. J. 2018, 48, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, H.P.; Hacker, N.F.; Andrews, L. Changing patterns of referrals and outcomes of genetic participation in gynaecological-oncology multidisciplinary care. Aust. N. Z. J. Obstet. Gynaecol. 2016, 56, 633–638. [Google Scholar] [CrossRef]
- Potosky, A.L.; O’Neill, S.C.; Isaacs, C.; Tsai, H.-T.; Chao, C.; Liu, C.; Ekezue, B.F.; Selvam, N.; Kessler, L.G.; Zhou, Y.; et al. Population-based study of the effect of gene expression profiling on adjuvant chemotherapy use in breast cancer patients under the age of 65 years. Cancer 2015, 121, 4062–4070. [Google Scholar] [CrossRef] [Green Version]
- Poulton, A.; Lewis, S.; Hui, L.; Halliday, J.L. Prenatal and preimplantation genetic diagnosis for single gene disorders: A population-based study from 1977 to 2016. Prenat. Diagn. 2018, 38, 904–910. [Google Scholar] [CrossRef]
- Ray, G.T.; Mandelblatt, J.; Habel, L.A.; Ramsey, S.; Kushi, L.H.; Li, Y.; Lieu, T.A. Breast cancer multigene testing trends and impact on chemotherapy use. Am. J. Manag. Care 2016, 22, e153. [Google Scholar]
- Ritter, A.; Bedoukian, E.; Berger, J.H.; Copenheaver, D.; Gray, C.; Krantz, I.; Izumi, K.; Juusola, J.; Leonard, J.; Lin, K.; et al. Clinical utility of exome sequencing in infantile heart failure. Genet. Med. 2019, 22, 423–426. [Google Scholar] [CrossRef]
- Roberts, M.C.; Dusetzina, S.B. Use and Costs for Tumor Gene Expression Profiling Panels in the Management of Breast Cancer From 2006 to 2012: Implications for Genomic Test Adoption Among Private Payers. J. Oncol. Pract. 2015, 11, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Dusetzina, S. The effect of a celebrity health disclosure on demand for health care: Trends in BRCA testing and subsequent health services use. J. Community Genet. 2017, 8, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Weinberger, M.; Dusetzina, S.; Dinan, M.A.; Reeder-Hayes, K.E.; Carey, L.A.; Troester, M.A.; Wheeler, S.B. Racial Variation in the Uptake of Oncotype DX Testing for Early-Stage Breast Cancer. J. Clin. Oncol. 2016, 34, 130–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoen, C.; Santolaya-Forgas, J.; Genc, M.; Ashkinadze, E. Differential utilization of expanded genetic screening tests in patients of reproductive ages from private and academic practices. J. Périnat. Med. 2015, 43, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.; Handorf, E.A.; Meyer, J.E.; Hall, M.J.; Esnaola, N.F. Mismatch Repair Deficiency Testing in Patients With Colorectal Cancer and Nonadherence to Testing Guidelines in Young Adults. JAMA Oncol. 2018, 4, e173580. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.; Hanna, J.W.; Visaria, J.; Gu, T.; McCoy, M.; Kloos, R.T. Impact of a gene expression classifier on the long-term management of patients with cytologically indeterminate thyroid nodules. Curr. Med. Res. Opin. 2016, 32, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Sivapiragasam, A.; Kumar, P.A.; Sokol, E.S.; Albacker, L.A.; Killian, J.K.; Ramkissoon, S.H.; Huang, R.S.P.; Severson, E.A.; Brown, C.A.; Danziger, N.; et al. Predictive Biomarkers for Immune Checkpoint Inhibitors in Metastatic Breast Cancer. Cancer Med. 2021, 10, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Stein, Q.P.; Vockley, C.W.; Edick, M.J.; Zhai, S.; Hiner, S.J.; Loman, R.S.; Davis-Keppen, L.; Zuck, T.A.; Cameron, C.A.; Berry, S.A.; et al. An Exploration of Genetic Test Utilization, Genetic Counseling, and Consanguinity within the Inborn Errors of Metabolism Collaborative (IBEMC). J. Genet. Couns. 2017, 26, 1238–1243. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.-A.; Brown, L.C.; Yu, K.; Li, J.; Dechairo, B.M. Canadian Medication Cost Savings Associated with Combinatorial Pharmacogenomic Guidance for Psychiatric Medications. Clin. Outcomes Res. 2019, ume 11, 779–787. [Google Scholar] [CrossRef] [Green Version]
- Tiller, G.E.; Kershberg, H.B.; Goff, J.; Coffeen, C.; Liao, W.; Sehnert, A.J. Women’s views and the impact of noninvasive prenatal testing on procedures in a managed care setting. Prenat. Diagn. 2015, 35, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Vaeth, S.; Christensen, R.; Dunø, M.; Lildballe, D.L.; Thorsen, K.; Vissing, J.; Svenstrup, K.; Hertz, J.M.; Andersen, H.; Jensen, U.B. Genetic analysis of Charcot-Marie-Tooth disease in Denmark and the implementation of a next generation sequencing platform. Eur. J. Med Genet. 2018, 62, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).