Abstract
Coronavirus disease 2019 (COVID-19) is now being investigated for its distinctive patterns in the course of disease development which can be indicated with miscellaneous immune responses in infected individuals. Besides this series of investigations on the pathophysiology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), significant fundamental immunological and physiological processes are indispensable to address clinical markers of COVID-19 disease and essential to identify or design effective therapeutics. Recent developments in the literature suggest that deficiency of type I interferon (IFN) in serum samples can be used to represent a severe progression of COVID-19 disease and can be used as the basis to develop combined immunotherapeutic strategies. Precise control over inflammatory response is a significant aspect of targeting viral infections. This account presents a brief review of the pathophysiological characteristics of the SARS-CoV-2 virus and the understanding of the immune status of infected patients. We further discuss the immune system’s interaction with the SARS-CoV-2 virus and their subsequent involvement of dysfunctional immune responses during the progression of the disease. Finally, we highlight some of the implications of the different approaches applicable in developing promising therapeutic interventions that redirect immunoregulation and viral infection.
1. Introduction
In consideration of public health emergency and global reach, on 11 March 2020, the World Health Organization (WHO) specified coronavirus disease 2019 (COVID-19) as a global pandemic outbreak of international public health concern [1]. A novel, highly transmissible enveloped RNA betacoronavirus unexpectedly emerged in December 2019 in Wuhan, China, and then was formally named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2. The most common clinical symptoms and manifestations of SARS-CoV-2 infection are pneumonia-like, including fever, hypoxia, dyspnea (labored breathing), headache, myalgia, cough, and in some cases, intestinal symptoms [2,3]. COVID-19 is now characterized as a mild to severe respiratory disease, and its clinical presentation to be influenced by comorbidities (patients with cardiovascular or renal disorders) and age factors [4]. A growing literature pointed out that the asymptomatic or mild infections are a significant fraction [5], at large half of spread events reported from pre-symptomatic and asymptomatic infections [6] and has immense implications of silent transmission. However, approximately 10–30% of SARS-CoV-2 infected patients who were hospitalized are associated with severe illness requiring intensive care to aid respiratory comfort [7,8].
The factors triggering severe disease progression in SARS-CoV-2 infected patients is yet to be wholly understood. Progression of severe immune status is not exclusively associated with the viral load but could be relevant to weak interferon response [9,10]. An excess inflammatory immune response in individuals infected by SARS-CoV-2 viruses is also known to ensure the foremost cause of severe disease development, organ failure, and mortality [11,12]. Besides this, it is interrelated with elevated levels of cytokines, mainly circulating ones [12], deep lymphopenia [13], and involved with extensive mononuclear cell infiltration in different organs, including lungs [14], spleen [15], heart [16], lymph nodes [17] and kidney [18], as observed in a post-mortem examination. Our previous report emphasized the significance of understanding the biological characteristics of the SARS-CoV-2 virus and its biomarkers applicable to developing new diagnostic kits and establishing point-of-care testing and surveillance measures [19].
The mortality concerns and morbidity-related complications realized in SARS-CoV-2 infection are associated with extensive inflammation [20]. Therefore, the need for a clear understanding of immunopathological factors underpinning the diverse responses observed in the infected patients is of paramount significance to identify appropriate therapeutic targets [21]. Several immunomodulatory agents are currently underpinning clinical trials at a rapid pace [22], and some of them are already being under routine use in the clinical practices in off-label use of drugs [23]. An in-depth understanding of each specific inflammatory pathway and cell-type shows promising results in choosing appropriate immunotherapeutic targets. Some of the recently developed strategies could be advantageous to avoid detrimental consequences in individual patients and/or during different stages of the SARS-CoV-2 infection. Presentation of specified therapeutic options, including intravenous immunoglobulin, steroid medications (methy/prednisolone, dexamethasone), personalized cytokine blockade (e.g., tocilizumab or anakinra), and JAK inhibition so far have been shown as potential alternatives in those individuals suffering from severe SARS-CoV-2 infection. In this account, we discuss potential inflammatory responses that have been identified so far in individuals with COVID-19 disease. In particular, we briefly highlight emerging pharmaceutical interventions, therapeutic modulations for pulmonary phase, inflammatory immunopathogenesis, immune-boosting strategies, the significance of tracking immune status, and specific aspects of macrophages and monocytes in the pathophysiology of SARS-CoV-2 infection.
2. Therapeutic Interventions
2.1. Pharmaceutical Interventions
The most important principle can be proactive efforts in preventing and controlling infectious diseases by reducing the source of contagion, interventions against most of the routes of spread, and protecting the susceptible old and juvenile populations. SARS-CoV-2 viruses mainly spread via respiratory aerosol droplets and rarely by surface contacts. Basic personnel and public protective measures are found to be helpful to restrict the transmission of SARS-CoV-2 viruses. At present, supportive therapies are mostly being adopted to treat symptomatic SARS-CoV-2 patients, including the treatments of other common colds, symptom aid, and complete protection. Supportive treatments are adopted to protect internal organs such as the lungs or brain, proactive prevention, emerging treatment options, pursuing individual complications, and ventilation support, if essential. Additional care should be provided to maintain the balance of electrolytes and water, thus comforting the strength of the internal organs.
Implementing vaccination programs is an effective measure to protect overall populations, but to date, the effective vaccines for SARS-CoV-2 viruses are now available in public service; some laboratories and private companies successfully completed clinical trials and distribution for vaccination. Several research institutions and private enterprises have reproposed several different technologies, including nanovaccine technology using mRNA, recombinant vaccine or inactivated vaccine, and DNA vaccine, to establish vaccine adjuncts against SARS-CoV-2 infections. For instance, a possible mRNA-based vaccine against SARS-CoV-2 viruses has been claimed by a Biotech Company called Moderna after tirelessly working in collaboration with pioneer institutes in health. AbMax Biotechnology Co., Ltd, Kechuang, China and several other private enterprises have also announced that they successfully develop vaccines against the SARS-CoV-2 viruses.
At present, few specific or compelling antiviral drugs are available to treat SARS-CoV-2 infected patients in the hospital. The discovery of new drugs, performing all steps in trials, and using them as antiviral drugs for SARS-CoV-2 viruses is a time-consuming process; instead, the most influential research strategy could be the use of old drugs. Gilead Sciences in the United States claimed that remdesivir a nucleoside analog prodrug is under development that is a broad-spectrum antiviral medication found efficient against Ebola virus [24]. Both in-vitro and in-vivo trials confirmed that even a low dose of remdesivir drug could have good inhibitory effects against the growth of both the SARS-CoV and MERS-CoV [25]. The remdesivir drug, as most potential drugs against the SARS-CoV-2 viruses, could be adopted after complete pharmacokinetic studies and completing safety protocols. A phase III clinical trial has been completed for a radcivir drug that was recently launched to treat SARS-CoV-2 infected patients by Jinyintan Hospital in the first week of February 2020, in Wuhan [26]. Its validation has been validated using a double-blind test, a clinically proven method. Recently, in vitro studies on chloroquine and radcivir and in combination have shown good inhibitory potential against the SARS-CoV-2 viruses [26].
As per the news reports, India-based enterprise Glenmark has secured government approval for the large-scale manufacturing and antiviral Favipiravir drug to treat SARS-CoV-2 infected patients [27]. Furthermore, molnupiravir parent drug (NHC) combination of drugs against SARS-CoV-2 viruses are recommended in a trial version of the treatment plan [28], including ritonavir/ralproveravir or ribavirin by intravenous injection in addition to the inhalation of alpha interferon [26]. Another example is the cheap and widely-available steroid drug called Dexamethasone, pushed by UK experts as a potential treatment of SARS-CoV-2 infection, with suggestions signifying great success in reducing deaths up to a third in patients showing severe symptoms, thus proving it to be a life-saving drug [29].
In May 2020, the Food and Drug Administration (FDA) of the United States was issued emergency use authorization, allowing distribution and use of remdesivir in the U.S [30], and suggested administering intravenously to treat the SARS-CoV-2 infected patients. Some reports have indicated that hydroxychloroquine and chloroquine help treat SARS-CoV-2 infected patients; however, the FDA is cautious about using both drugs [31,32]. Different patients need to be treated differently based on the symptom differentiation: the early symptom stage with stagnant lung, the middle symptom stage with pestilence, the severe symptom stage, and the final recovery stage [33]. At present, scientists from various interdisciplinary fields are working tirelessly to investigate a plethora of antiviral drugs to fight against the existing pandemic crisis posed by the COVID-19 disease (Table 1). To date, several preventive and treatment drug studies have been observed in the different clinical trial registration centers involving a variety of medicines. These include Darunavir, Corbistar, Ritonavir, Lopinavir, Camostat, Nafamostat, Umifenovir, Favipiravir, Famotidine, Ivermectin, Nitazoxanide, Tocilizumab, Corticosteroids, Sarilumab, Fluvoxamine, Bevacizumab, etc.

Table 1.
Mechanism of types of drugs and therapies useful in the treatment of SARS-CoV-2-infected patients [3,27,32].
2.2. Therapeutic Models for Pulmonary Phase
An intense understanding of COVID-19 disease pathogenesis is necessary to provide a scientific foundation in developing antiviral drugs and designing vaccines. As per the recent literature developments, the progression of pathogenesis is believed to manifest in three chronological phases, including pulmonary, pro-inflammatory, and prothrombic phases.
Here, we present SARS-CoV-2-host cell interactions, key pathogenic mechanisms, and potential clinical development in the pulmonary phase and key highlights on antithrombotic therapies. In pulmonary pathogenesis, excessive cytokine production in individuals infected with SARS-CoV-2 viruses causes enhanced membrane permeability and results in pulmonary dyspnea, edema, and hypoxemia [54]. Potential therapeutics in the treatment of SARS-CoV-2 infected patients with the pulmonary phase include viral entry inhibitors, protease inhibitors, RAS inhibitors, and replication inhibitors. In particular, RAS inhibitors can be classified into angiotensin-II receptor blockers (ARBs) and inhibitors of ACE, which are involved in alleviating the RAS over-activation exerted by ACE2 deficiency in SARS-CoV-2 infected patients. However, inhibitors that target viral entry, protease, and replication are also known for direct antiviral activities via targeting specific stages of the SARS-CoV-2 life cycles.
First, RAS inhibitors such as ARBs and ACE inhibitors are generally prescribed as hypertensive drugs. Therefore, its use as a SARS-CoV-2 therapeutic raises safety concerns owing to its ability to elevate ACE2 expression, particularly in hypertensive individuals [55]. Principally, ACE2 upregulation triggered by RAS inhibition leads to the more effective entry of SARS-CoV-2 viruses; thus, there is a severe threat for the users of RAS inhibitors of becoming more vulnerable to SARS-CoV-2 infection [56]. Regardless of such health concerns, most clinicians are against the quick rejection of RAS inhibitors, particularly in high-risk individuals, including those on the verge of myocardial infarction or who have a history of heart failure. Therefore, there is a possibility of developing clinical encounters and hostile health consequences [57]. Until further development in clinical trials, clinicians are advised to continue using RAS inhibitors to achieve stable conditions in high-risk SARS-CoV-2 infected patients [58].
At large, RAS inhibitors seem to be advantageous to SARS-CoV-2 infected patients since they can downregulate the RAS pathway when it gets overactivated after infection, as suggested described formerly [59]. This may benefit in reducing new infections of SARS-CoV-2 viruses simply via disruption or enzymatic digestion of coronavirus spike (S) glycoprotein by transmembrane serine protease-2 (TMPRSS2), wherein involves a membrane-fusion mechanism [60]. G-protein-coupled mas receptor (MasR) agonists and ACE2-mediated stimulation of signaling through ACE2 gene delivery together with the use of angiotensin 1 to 7 have been proposed as potential immunotherapeutic approaches to encounter RAS signaling pathways, enhanced ACE2 deficiency in SARS-CoV-2 infected patients [61]. It was also shown that the ACE inhibitors suppress the expression of TMPRSS2, a key co-receptor for the entry of SARS-CoV-2 viruses [62]. Thus, it can also be a promising pathway in developing therapeutics approaches for treating SARS-CoV-2 infected patients after blocking the target ACE2 host receptors or TMPRSS2 (Figure 1).
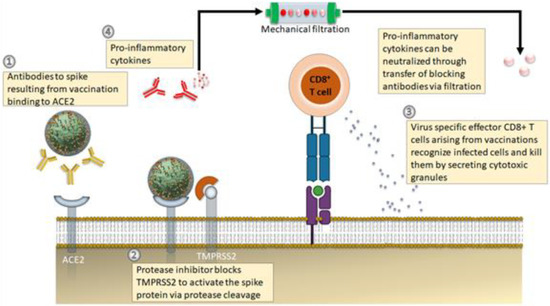
Figure 1.
Emerging therapeutic approaches against COVID-19 disease. (1) Antibody-based therapeutics against the S protein (either via adoptive transfer of vaccination) to avoid progression of severe infections. (2) Application of protease inhibitors against serine protease (TMPRSS2) prevents cleavage of S protein. (3) SRS-CoV-2 virus-specific memory CD8+ T cells from vaccination or earlier infection. (4) A new treatment approach targets the symptoms of cytokine storm, wherein the blood of infected patients is passed through customized filtration columns to capture pro-inflammatory cytokines before the pureblood returns to patients. Adapted and modified from [63].
Presently, some compounds identified that could target these molecules have been under clinical approval. For instance, applications of machine learning algorithms used to predict that JAK inhibitor called baricitinib, useful in the treatment of rheumatoid arthritis, could also be applied to inhibit endocytosis mediated by ACE2 [63]. That report also reveals clinical trial status for another potential JAK inhibitor called ruxolitinib, and it is expected to add a new treatment option. The delivery of high concentrations of ACE2 in soluble form is also considered a promising strategy to reduce the virus’s entry into host cells possibly. APEIRON is now under clinical trials with a recombinant form of ACE2 called APN01. In the case entry of SARS-CoV-2 viruses, ACE2 plays a key role, considering that neutralizing SARS-CoV-2 viruses via the administration of recombinant ACE2 protein is now being proposed as a potential therapeutic model [64]. Remdesivir and chloroquine functions at a phase post virus entry [65]. However, camostat mesylate [66], and Nafamostat mesylate [67] is an excellent inhibitor of TMPRSS2 and is under approval in several regions and countries. It is important to note that camostat mesylate is now being tested against isolated viruses from SARS-CoV-2 infected patients. It successfully prevents the SARS-CoV-2 viruses from entering into lung cells [68]. If this approach gets validation and completes clinical trials, promising repurposing of these antiviral drugs will possibly treat the SARS-CoV-2 infected patients.
At large, CD147 plays a crucial role in controlling the SARS-CoV-2 infection, including its variants and modulating their pathogenesis. The CD147 is now revealed as a universal entry receptor for the SARS-CoV-2 viruses, including its predominant variants and signaling pathway initiator, particularly cytokine storms. CD147 antibody effectively and specifically inhibit infection and block cellular entry of SARS-CoV-2 viruses and also helps in alleviating cytokine storm for almost all variants, including alpha, delta, beta, and gamma [69]. CD147 antibody named meplazumab effectively inhibits the SARS-CoV-2 virus and reduces cytokine storm by blocking the direct interactions between S glycoprotein and CD147 [70]. That report revealed that the meplazumab is appropriate in terms of tolerance and safety also recommended accelerating the recovery of severe SARS-CoV-2 infected patients. According to recent developments, the efficiency of humanized antibodies (e.g., meplazumab) have been examined against surface molecule CD147, clinical trials confirm its promising potential to treat SARS-CoV-2 infected patients [70]. Preliminary clinical trials advocate that surface molecule CD147 can also be a promising pathway to block the virus from entering the host cells, as S protein of the SARS-CoV-2 virus is also known to bind small surface molecules such as CD147 [71].
Antiviral effects of azithromycin, a macrolide antibiotic, is being presented since it reduces the viral load in hospitalized severe patients due to its ability to create interference in the course of ligand interactions with CD147 receptor [72]. Besides this, azithromycin also influences key immune parameters, including enhancing the expression of antiviral interferon and with antiviral and anti-inflammatory functions. Thus, it can be a promising therapeutic agent, but it needs validation [73].
Replication inhibitors are mainly applicable to suppressing the replication of the viral RNA genome via antagonizing the enzymatic activity such as RNA-dependent RNA polymerase (RDRP) of the SARS-CoV-2 virus. Therefore, they are mostly considered RDRP inhibitors, including favipiravir, remdesivir, and ribavirin. Favipiravir and remdesivir were specifically developed to treat influenza [74] and the Ebola virus [75], respectively. They are the nucleotide analogs helpful in hindering the functions of endogenous nucleotides in the course of viral RNA synthesis. Targets for remdesivir also include membrane protein (M protein) and RDRP inhibitors. It is the first drug approved clinically and repurposed as a potent antiviral agent for treating SARS-CoV-2 infected patients [76,77]. A well-recognized host protease inhibitor, including camostat mesylate and nafamostat, was useful in inhibiting the host TMPRSS-2 protease that restricts the entry of SARS-CoV-2 viruses into target host cells [78]. Consequently, host protease inhibitors can effectively block the virus entry; however, adverse events involve nafamostat mesylate and favipiravir in treating SARS-CoV-2 infected patients [79]. Lopinavir/ritonavir was first developed as an anti-HIV agent, now repurposed due to its inhibitory actions against protease of the SARS-CoV-2 virus [80,81].
Antithrombotic therapies are also identified as a significant approach since coagulation cascade dysregulates in SARS-CoV-2, anticoagulation can be explored in the treatment, essentially for patients with the event of venous thromboembolism (VTE) [82]. Indeed, a recent report stated approximately about 40% of SARS-CoV-2 patients hospitalized were found at high risk of VTE [83], and research data reported from extensive retrospective cohort studies for SARS-CoV-2 patients suggest that anticoagulation therapy can reduce mortality rate, especially among those patients with high severity [84]. Recent studies on the heparin-based treatment of SARS-CoV-2 have shown promising results [82,85]. This is reflected as one of the key agents to treat SARS-CoV-2 either as a therapeutic or prophylactic routine [86]. The applications of a selective antithrombin-dependent factor Xa inhibitor called fondaparinux in SARS-CoV-2 patients were reported [87]. Further studies on the application of direct oral anticoagulants in SARS-CoV-2 patients have to be studied [88,89].
Another approach to interrupt coagulation cascade via pharmacological approaches includes blockade of factor XII (FXII). It has been revealed to safeguard SARS-CoV-2 infected patients from occlusive thrombosis without any impairment in hemostasis [90,91]. Particularly, increased FXII activity was noticed in platelets isolated from the SARS-CoV-2 infected patients, which was also accounted for by the shortening of the activated thromboplastin time [92]. Likewise, serine protease inhibitors of plasmin, trypsin, and thrombin are also recognized as nafamostat mesylate, being used to treat pancreatitis and during dialysis [93], and its clinical trials are under investigation in the combination of heparin and nafamostat for SARS-CoV-2 (NCT04418128, NCT04352400).
The hypofibrinolytic state was detected in the acute respiratory distress syndrome (ARDS) and mainly being directed by the application of specific tissue-type plasminogen activators, which seems typically accountable in the transformation of plasminogen to plasmin, leads to the collapse of the cross-linked fibrin structures [94]. Indeed, infusion of tissue-type plasminogen activators has been revealed useful in severe SARS-CoV-2 infected patients [95], and current clinical trials on ARDS-related conditions are in progress with SARS-CoV-2 infected patients (NCT04357730).
Dipyridamole, an antiplatelet agent, is reported to have great therapeutic potential [96]. In addition to its antiplatelet use has also been shown multiple functions, including antiviral activity, mainly suppressing excessive inflammation and supporting mucosal healing, and preventing acute fibrosis in the lungs and other organs [97,98,99]. Treatment of SARS-CoV-2 infected patients using dipyridamole was beneficial due to its ability to prevent NETosis and promotes 3′,5′-cyclic adenosine monophosphate (cAMP) formation in neutrophils [100]. Liu et al. revealed that dipyridamole is also helpful in suppressing in vitro replication of the SARS-CoV-2 viruses and improving lung pathology in an animal model via type-I INF response [101]. Clinical trials for dipyridamole are currently under progression (NCT04391179). Alternative agent ticagrelor and antiplatelet agents also showed the ability to attenuate the formation of Neutrophil extracellular traps (NETs) [102]. Consequently, clinical trial-II on ticagrelor has been accomplished (NCT02735707, NCT04518735).
2.3. Anti-Inflammatory Therapeutics
The WHO advised against the clinical use of corticosteroids in treating SARS-CoV-2 infected patients, but some hospitals are still using them to treat the hyper-inflammatory symptoms [103]. However, many researchers are hopeful that corticosteroids could precisely inhibit specific pro-inflammatory pathways. One of the initial clinical reports showed elevated levels of interleukin (IL-6), which is associated mainly with the low oxygen saturation reported for SARS-CoV-2 infected patients [104], tocilizumab treatment in patients show symptoms of severe SARS-CoV-2 cases caused in better conditions, oxygen saturation, and lymphopenia conditions within a few days after treatment [105]. Similarly, in the German case study reported for 40 patients, higher levels of IL-6 are predictable for SARS-CoV-2 infected patients who will be at risk of respiratory failure. The detrimental effects of excessive levels of IL-6 are possible to be raised from both its potential to direct adverse effects on organs and arouse the immune system [106]. Thus, IL-6 receptor blocking antibodies may provide hope after challenging the hypothesis that directs anti-inflammatories pathways, thus providing great therapeutic relief [107]. According to the preliminary results of clinical trials performed for antibody therapies, it was stated to target IL-6 or IL-6R and manage appropriately. Clinical trials on mononuclear macrophages tocilizumab in fewer patients from China and France suggested that the treatment reduces the mortality and need for intensive care unit admission. Helpfully, a trial in the U.S. in a 154-patient, Tocilizumab ably reduced mortality rate in severely symptomatic patients was on ventilation [108]. The molecular mechanism of tocilizumab, binds to both the membrane and soluble forms of IL-6 receptors, thus ably suppresses the Janus kinase-signal transducer and activating transcription factors in signaling pathways [109], thus finally, the formation of inflammatory molecules [110,111]. However, IL-6-blockade may be challenging since it partly suppresses T cells’ activation. This therapy should consider disease pathophysiology without detrimental effects on organs.
3. Inflammatory Immunopathogenesis
Damage of lung cells after SARS-CoV-2 infections elicits local systemic and mucosal immune responses, recruits macrophages and monocytes to respond against disease progression, and also triggers cytokine storm and adaptive immune system (B and T cells). In most cases, such a response resolves SARS-CoV-2 infection. However, a dysfunction of the immune system sometimes follows, resulting in a severe condition that can distort the linings and walls of the lung’s air sacs and even cause systemic disease [112].
SARS-CoV-2 is also a cytopathic virus; it induces destruction and injury to virus-affected tissues and death of infected cells in the course of the replicative virus cycle [113]. As part of viral infection and its replication cycles in epithelial cells, it could cause severe pyroptosis and vascular leakage similar to patients with SARS-CoV [114]. This is a kind of programmed cell death manifest in highly inflammatory cells, and it can be seen in most cytopathic viruses. Pyroptosis is expected to trigger subsequent inflammatory responses. A key cytokine (IL-1β) is released during pyroptosis and elevated levels during the SARS-CoV-2 infection [2].
In most SARS-CoV-2 infected patients, recruited cells typically resolve the lung infection as immune systems respond well and infected patients recover without severe damage to lung health and other organs [63]. However, immune response dysfunction follows in several patients, which causes cytokine storm and mediates widespread inflammation in lung tissues. It can be seen in severe SARS-CoV-2 infections. Those who need intensive care or treatment in hospitals exhibit elevated levels of biomarkers in blood plasma, including interleukins (IL-2, -7, -10), interferon (IP-10), monocyte chemoattractant protein-1 (MCP1), granulocyte colony-stimulating factor (G-CSF), macrophage inflammatory protein 1α (MIP1α), and tumor necrosis factor-alpha (TNFα) [2] (see Figure 2). Also, infected individuals with severe pathology show a considerably higher count of inflammatory monocytes in peripheral blood samples than those with mild pathology [115]. These cells trigger the secretion of inflammatory cytokines that also intensify the risk of cytokine storms, including IP-10, MCP1, and MIP1α. The mechanisms associated with SARS-CoV-2 infection subvert the innate antiviral cytokine response, which is yet to be well understood. Recent research efforts show that numerous non-structural and structural proteins of viruses antagonize interferon response. Furthermore, as observed in transcriptomic analysis, dampening immune function and hyper-inflammation. Consequently, the mechanisms involved in reduced the cytokines are the most damaging. Thus, it could be assumed that TNF, IL-6, and IL-8 could be potential agents in boosting B- and T-cell response [116].
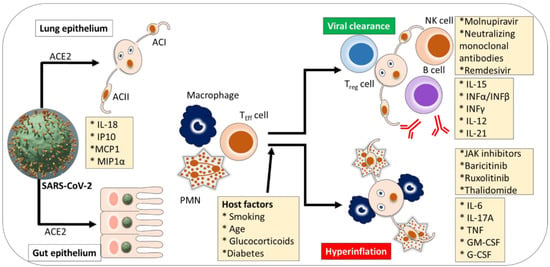
Figure 2.
Cytokine pathogenesis of SARS-CoV-2 infection. Intensive care or appropriate treatment is recommended for hospitalized severe SARS-CoV-2 patients who exhibit elevated levels of biomarkers in blood plasma. Alveolar cell (AC), angiotensin-converting enzyme 2 (ACE2), atopic dermatitis (AD), ancestry clusters (AC1-4) granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF or GCSF), interleukin (IL), interferon (INF), monocyte chemoattractant protein 1 (MCP1), macrophage inflammatory proteins (MIP1α), natural killer (NK), polymorphonuclear granulocyte (PMN), T-effector cell (TEFF cell), tumor necrosis factor (TNF), regulatory T cell (Treg cell). Adapted and modified from [116].
Recent reports suggest that the SARS-CoV-2 infections drive a diverse range of immune cascades, thus raising the risk of immunosuppressant agents tested in clinical trials that might vary from patient to patient and sometimes can be detrimental to some patients [117]. A diminished immune system when SARS-CoV-2 infected patients struggle off viruses can be an adverse pathophysiological state. Those infected patients had acute respiratory distress syndrome in previous times, apparently instigated by fugitive immune responses, thus resulting in cytokine storms [118,119]; clinicians are confused about the overuse of anti-inflammatory agents that helps to keep immune response in progress and avoid any collateral damages [120]. Furthermore, the upsurge in cytokines released by immune systems causes hyperinflammation in SARS-CoV-2 infection, may result in sepsis and cytokine storm, and lead to morbidity. In such cases, controlled inflammation can help avoid multi-organ failure, particularly the respiratory, hepatic, cardiac, and urinary systems. Some of the SARS-CoV-2 infected patients develop severe symptoms or organ failure eventually encounter mortality [121,122].
Both B and T cell responses were observed in blood samples for about one week from the onset of severe symptoms of the SARS-CoV-2 infection [63]. B cells response in individuals with SARS-CoV-2 disease concurrently with helper cells (T follicular) response, about one week of severe symptoms onset. Furthermore, two types of T cells include CD8+ T and CD4+ T cells play an essential role in death virus-infected cells, wherein CD4+ T cells are primarily accountable for controlling the production of cytokines and driving the recruitment of immune cells. If the patients with low-dose showed indication of early inflammation, the SARS-CoV-2 virus could survive even after the initial boost of immune activity. T cells are identified to battle against the SARS-CoV-2 infections; there is a chance of causing dysfunctional and metabolically exhaustion; thus, the exhausted T cells need to be substituted with fresh ones [123]. Severity progresses in SARS-CoV-2 infected patients with the simultaneous increase in the level of inflammatory cytokine may determine the exhaustion and depletion of T cell abundance.
4. Immune-Boosting Strategies
There is ever-growing worldwide interest in immune-boosting agents, nutrients, and yoga, which is on the rise. The changes in the immune response vary with infectious diseases, and such changes also rely on the duration of infection. Other viruses, for example, human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV), take several months to produce symptoms. After that, infections turn into chronic [124]. However, infections such as the influenza virus show apparent symptoms within two days of the infection, and most people recover from such virus within one week of the period [125]. However, SARS-CoV-2 infections either show no symptoms or some symptoms within four to six days of infection and, for some patients, the disease can continue for about fifteen days [126]. Subsequently, SARS-CoV-2 infection is not a chronic disease such as HIV or HBV, but also not a truly acute infection such as the H1N1 virus [127]. Various strategies could be applied to treat SARS-CoV-2 infected patients, in particular IL-7, which could promote T-cell proliferation, prevent T-cell death, and help reverse lymphopenia in severe patients [128,129].
The chronicity of the SARS-CoV-2 infection needs to be explained, especially both immune-boosting and -suppressing approaches could be potential approaches to address at different time courses [130]. Until then, the ambiguity over whether to boost or suppress the immune response led to clinical trial results contrasting to choosing appropriate treatment strategies.
After decades of R&D efforts, most of the clinical trials with anti-inflammatories resulted in failure to avoid organ damage and overcome cytokine storms. The sepsis field is now turned to address immune-activating strategies [131]. These R&D efforts are yet to deliver convincing evidence of their effectiveness. However, anti-PD1/PDL1 and IL-7 therapeutics have shown signs of usefulness and efficacy during clinical trials [132,133]. Thus, similar strategies would be effective in treating SARS-CoV-2 infected patients. Bekele et al. further revealed that IL-7 could aid to protect T cells from death, promote the proliferation of T cells, and support reversing lymphopenia in SARS-CoV-2 infected patients suffering from sepsis [133].
4.1. Interferon Mediated Interventions
IFN, a critical inflammatory cytokine agent that can be detected in SARS-CoV-2 infected patients, is controlled mainly by histone markers. It aids in preventing viral infections [134]. In recent studies, transcriptomic results suggested that its application may be inappropriate to control interferon response in SARS-CoV-2 infected patients [135]. That report suggests both in-vitro and in-vivo studies further reveal it to aggravate low transcripts downstream levels of both type-I and type-III interferons compared with the high level of cytokines (IL-6). Such monitoring studies proved that interferons could be considered a first-line defense against the SARS-CoV-2 infection. Thus, early improvement of interferon responses, both type-I and type-III, could be potentially adjunct patients while fighting against SARS-CoV-2 infections, allowing clearance of the virus particles before hyper-inflammation and intervention turn out to be significant challenge [63].
Furthermore, activation of IFN can be precisely modulated using epigenetic regulators, including H3K27me3, H3K4me3, and H3K9me2 [136,137]. Besides this, SARS-CoV viruses have an IFN-stimulated gene 15 (ISG-15) effector role, are essentially linked with the histone marker of ISG-15 genes on the promoters, and deviate among a range of viruses [138,139]. IFNs are also useful in controlling certain cancers (Borden 2019) and hepatitis C [140]. IFN-α combined with anti-viral drug ribavirin was identified as the backbone for HCV treatment up to 2014. SARS-CoV-2 viruses were identified to interact characteristically with the ubiquitin-like ISG-15, a critical innate-immune controller of host cells. Additionally, preferential cleavage of ISG-15 via protease of the virus (PLpro) may weaken the signaling pathways of type-I IFN, which is a vital component in most of the antiviral responses, and IFN responsive factor-3 (IRF3) [141]. The PLpro is essential for the formation of appropriate function replicase complex and promoting viral spread. Several key proteins of the SARS-CoV-2 virus, including N protein, structural proteins, and some other accessory proteins such as ORF8 and ORF6, were recently established as potential inhibitors of the type-I IFN pathway [142]. However, recent clinical studies have demonstrated an absence of noticeable type-I IFN among most of SARS-CoV-2 infected patients [143].
CD4+ T cells that are reactive to M protein are identified as multifunctional accompanied by increased levels of IL-2, TNF-α, and IFN-γ, consequently S protein and ultimately CD4+ T cells reactive to N proteins [144]. Furthermore, CD8+ T cells were recently studied in SARS-CoV-2 for IFN-γ production. However, levels of CD8+ T cells remain lower than that of CD4+ T cells [145]. Another important clinical study suggested that in response to S or N proteins, the level of IFN-γ remains higher in SARS-CoV-2 infected patients with mild infections than severe ones [146].
A recently published report showed that triple combination of ribavirin, ritonavir/lopinavir, and IFNβ-1b was found to be effective and safe in patients with mild symptoms compared to ritonavir/lopinavir in concern to control symptoms, promote viral shedding, and shorten the hospital stay [147]. However, a clinical trial performed with 127-person showed evidence supporting the importance of interferons in decreasing the course of SARS-CoV-2 infection by simply using repurposed drugs ritonavir, ribavirin, and lopinavir in combination with IFNβ [147]. Therefore, a type I interferon (IFNβ) is a solitary immune-boosting approach that has to be further investigated in the large-scale trial in combination with nominated anti-viral drugs [148]. In addition, a type III interferon (IFNλ) in patients has to be tested with those mild or moderate symptoms [149]. Since IFNλ receptors are expressed on epithelial cells, some normal cells thus need to be tested for any complications related to cytokine production and organ-damaging, as reported previously [150]. Furthermore, previous studies showed no advantage from IFN-α/β in severe patients with SARS and MERS viruses [151,152,153]. In contrast, IFNs show noticeable adverse symptoms, including flu-like, headaches, gastrointestinal issues, and allergic reactions. Further studies are required to validate the data and evaluate the clinical uses and potential toxicity concerns of IFNs.
4.2. Restoring Distressed T-Cells
Other strategies have reflected advantages to achieve immune-boosting and restoring exhausted T-cells. Often T cells cannot keep up with the vitality demanded during prolonged battles against infection since, by time course, they get a decline in their function through a series of processes as reported for lymphocytic choriomeningitis virus [154]. The report has shown that patients with chronic HIV, HBV, or HCV infections can cause exhaustion of T-cells, though T-cells could keep the infection on a smoldering level. However, they seem powerless to eradicate infection [155]. An important marker for T cell fatigue receptor is that the programmed death (PD1) gets upregulated in infected cells and dampens the normal functioning of T cells. However, antibodies targeting PD1 and its ligands programmed death-ligand (PDL1), cytotoxic T-lymphocyte-associated protein-4, and some other inhibitory surface receptors can transform SARS-CoV-2 treatment performed simply by reenergizing the immune response [156]. Recently it was evaluated whether activation of human macrophage, astrocytes, microglia, brain endothelial cells, and neurons using safe level concentrations of ethanol exposure ably alters expression of PD-1/PD-L1 [157]. However, PD1-targeting antibodies showed good efficacy, demonstrating that the PD1 inhibition is applicable in human diseases, wherein PD1-blocking antibodies assist to increases the active T-cell population. Thus, it helps in chronic infection such as HIV [158]. Trials examining applications of anti-PD1 treatments in SARS-CoV-2 are yet to be completed [159]. Another approach emphasizes the significance of providing an energy boost to T-cells using antioxidants and restoring immune balance in SARS-CoV-2 infected patients, hoping that antioxidant compounds such as N-acetylcysteine could restore T-cell function [160]. Those clinical trials have to examine whether repurposed strategies for immune-boosting are effective, safe, or address hyper-inflamed states in patients.
5. Tracking Immune Status
Severe SARS-CoV-2 infected patients have to be timely treated on the basis of their immune status. However, there is a need to define how this could be achieved [161]. According to the current WHO guidelines, which are often recommended to arrange trials based on exclusion and inclusion criteria, infected individuals transition from mild to severe or need ventilator support [162]. However, the immune status reflects the level of healthcare needs for the particular patient instead of the underlying immune status or biological processes. Therefore, considering several biomarkers could recognize the patients who are mostly expected to respond positively to a specified immunomodulating agent [163]. Most biomarkers are based on a single protein being developed to stratify sepsis individuals [164]. However, it is challenging to discern or validate as SARS-CoV-2 virus biomarkers. There is a need for specific functional assays to identify or count the active T cells in the blood samples [165]. Integrating several key biomarkers into a single signature can be a promising approach. Metabolomic and proteomic characterization of serum samples was reported by a research group led by Tiannan Guo [166], wherein blood samples from 46 SARS-CoV-2 infected patients and 53 uninfected individuals were analyzed using mass spectrometry. That report suggests that proteomic and metabolomic fingerprints can be prepared and validated in other cohort studies, including main 22 proteins and seven other metabolites are proposed to evaluate the severity of the disease. Another report similarly suggested a broad approach-based signature, wherein samples were collected from early hospitalized cases and used to identify potential proteins about 27 potential biomarkers for their increasing or decreasing levels. In Table 2, an important pro-inflammatory signaling pathway is also discussed, which expresses differentially both downstream and upstream of IL-6, is in agreement with WHO severity grade provided for the SARS-CoV-2 infection [167]. Understanding the immune response at a molecular and functional level for SARS-CoV-2 infections is vital, which is not either acute or chronic infection. Such studies could also be indispensable to be prepared against future pandemic viruses [168] and also help in developing promising vaccines against SARS-CoV-2 viruses [169].

Table 2.
List of potential biomarkers of SARS-CoV-2 infection.
Confronting the challenge brought by the COVID-19 pandemic crisis to public health, we still lack the ways to explain the differences among the immune response and symptoms of infected patients. The fundamental key is a comprehensive understanding and significance of SARS-CoV-2 infected patients for their diverse immune response or immune indicators to inflammatory changes from initial onset to the end. There is scope to provide new approaches to overcome the existing pandemic by performing dynamic analysis of lymphocyte subsets, leukocyte classification, and cytokines. It was suggested that the total counts of lymphocytes, leucocytes, and eosinophils in SARS-CoV-2 infected patients decline. However, there was a significant difference for monocytes and neutrophils compared to non-infected individuals [203]. That report further reveals that the counts of leukocytes and neutrophils increase, eosinophils and lymphocytes continue to decline, and monocytes count remains stable (particularly in severe SARS-CoV-2 infected patients, which is partly consistent with another report [1]. The increase of monocytes in SARS-CoV-2 infected patients compared to healthy individuals is caused by inflammation-triggered monocyte infiltration and activation [204]. There is an observation on the increase in levels of leukocytes and neutrophils, mainly in severe patients and those with secondary infections. It can be explained reasonably as downregulation of ACE2 occurs on the onset of the COVID-19 disease and causes infiltration of neutrophils, which further cause tissue damage and then manifests into venous thrombosis [205]. Furthermore, a recent report showed a significant decrease in eosinophil count compared to the healthy control. Therefore, involvement of eosinophils is expected in immune response and viral clearance [206].
In the recent literature, we noted an interesting and perhaps controversial concern about the existing pandemic and still considerable uncertainty about the success of the current therapeutic interventions on the immune response [100]. This would interfere with the longitudinal detection approaches available so far to examine the immune status of SARS-CoV-2 infected patients. Firstly, virus-targeting anti-viral drugs were commonly applied for SARS-CoV-2 infected patients, including lopinavir or ritonavir and ribavirin and α-IFN and chloroquine as host-targeting drugs, were reported in a previous report [1]. These drugs were initially used as protease inhibitors for several other viruses and also investigated for SARS and MERS infections. However, chloroquine was found to be an outstanding candidate since it acts as an immune modulator and suppresses the SARS-CoV-2 infection [207]. Several anti-viral agents are underpinning clinical trials and are yet to be confirmed for their efficiency against SARS-CoV-2 viruses and their effect on the immune response [208]. Recently it was proposed that α-IFN regulates monocyte-derived macrophages, NK cells, and T cells in SARS-CoV-2 infected patients. However, the immune response triggered by α-IFN is still unclear [209]. The recent report on the effects of chloroquine suggests promising results in treating SARS-CoV-2 infected patients; efficacy and impact on immunity must be in the final stage of clinical trials [210,211,212]. So far, we are on the verge of rapid progress in evaluating the dynamic changes between the human host and SARS-CoV-2 viruses. It is under current treatment through monitoring immune response and a well-thought-out guide to choosing the right drugs optimal medication time. Second, an appropriate selection of therapeutic interventions is quite challenging to apply firmly, followed by a new variant of SARS-CoV-2 virus for both mild and severe patients. However, it would be reasonable to successfully treat SARS-CoV-2 infected patients based on the approved evaluation criteria and treatment principles. Therefore, this report suggests validating the effects of different treatment options, preparing the database for several severity groups among SARS-CoV-2 infected patients, and matching the results using a continuous monitoring approach for immune indexes. Furthermore, the biomarkers monitored by clinical teams could more accurately reflect the clinical significance and immune status of SARS-CoV-2 infected individuals.
6. Future Perspectives
Researchers have made enormous efforts to improve our understanding of the pathogenesis of the SARS-CoV-2 infections and find therapeutics to relieve the existing pandemic crisis. After the beginning pandemic, most research was focused on viral proteins. The basis for this was previous developments on SARS and MERS, since they share identical genomic features and some of the same viral proteins. Therefore, repurposing previous drugs was the ideal choice, as it can save the time required for drug discovery, and its safety and efficacy needed to be clinically validated. However, some satisfactory results from the clinical trials for repurposing drugs were reported. This review highlights recent advancements in emergent interventional potential therapeutic targets and clinical strategies based on the target host and virus.
Due to structural proteins of the SARS-CoV-2 virus, S protein seems to be the most promising target to develop direct antiviral agents, and current developments on crystal structure play a significant role in blocking the replication cycle of the SARS-CoV-2 virus. The common structural proteins among coronaviruses may assist in determining the results of the antiviral strategies. Some of the nonstructural proteins of the target virus are vital in both the replication cycle of the target virus and understanding virus-host interactions, which can be promising indirect targets for developing antiviral therapeutics. Therefore, precise knowledge about structures of target proteins and the clinical pathogenesis of this infection progression may benefit from revealing potential therapeutic agents needed to encounter SARS-CoV-2 infections. However, with an insignificant accomplishment for potential drugs targets/emerging therapeutics in battling SARS-CoV-2 infections, we suggest that most repurposed or newly developed drugs had only minor success in their preclinical trials. Although few targeted antiviral drugs were fortunately entered in the clinical trials, some of them also failed in the final phase of the clinical trials (including some of the vaccines), which may be reconsidered for further investigation by researchers. There is still a huge information gap among new targets, potential drug discovery, and clinical trials. Several new strategies should be deliberated to address the existing gap in the near future and to anticipate breakthrough research achievements combating the COVID-19 pandemic by focusing on R&D efforts for most of the potential drug targets in the current drug development programs, strengthening collaboration among various disciplines, and monitoring mutation event in long-term is a must.
In consideration of host immune response, both B and T cell immune responses to SARS-CoV-2 infections is remained poorly understood. Some of the recent studies suggest that immune response, particularly an aggressive one, leads to immunopathology, whereas other reports indicated that the mechanism of T cell exhaustion or dysfunction is also marked [213,214,215]. Examinations showed high levels of viral particles in the respiratory tract and some other tissues [119], representing an incompetent immune response. Nevertheless, asymptomatic or mild patients who do recover from infections had evidence of SARS-CoV-2-specific T cell memory [216]. Antibodies specific to SARS-CoV-2 are found in recovering patients, and individuals are under treatment with plasma therapy [217,218].
Conversely, SARS-CoV-2 infected patients under intensive care units have virus-specific antibodies, raising questions concerning why those patients cannot manage the disease. At large, some of the previous studies have stated observations on few patients or small cohorts’ studies, and thus further comprehensive studies with deep immune profiling of hospitalized SARS-CoV-2 infected patients are needed. Such revelations would address the critical questions, including common immune dysfunction profiles in severely ill patients. Such developments will also aid in preparing a framework of testing key therapeutic agents to inhibit, enhance, or otherwise engineer the immune response in SARS-CoV-2 infected patients.
A significant implication is that the potential to combine immune features with the severity of disease at the sampling time along in the course of disease severity changes over a progression. Application of correlative examination allows one to observe relationships among characteristics of the diverse immunotypes, comorbidity factors, and clinical aspects of the SARS-CoV-2 virus. After integrating several immune features using comprehensive clinical data, temporal changes, and severity scores, one can develop a computational model that integrates and connects the phenotype of patient immune response with the severity of the disease. This approach may allow us to connect dots to integrate immune signatures data to clinically measurable disease features. In the future, comprehensive data sets on circulating inflammatory mediators and immune cell types for most patients will improve understanding of immune responses. However, such findings may incite the idea of modifying clinical treatments or immune-based future clinical trials to individuals whose immunotype shows potential benefits. Respiratory viral diseases cause clinical pathology due to an immune response that is either too weak with virus-induced pathology or too strong with immunopathology [135]. Recent developments in the scientific literature suggest that the immune response of severe SARS-CoV-2 infected patients may fall across the range of immune response patterns that exist as diverse immunotypes associated with clinical features, severity, and temporal changes in immune response and pathogenesis. This account highlights the significance of immune response and developing an integrated framework to link clinical pathology with the disease severity.
7. Conclusions
Herein, we present the significance of understanding the severe symptoms in SARS-CoV-2 infected patients and the need for potential anti-inflammatory measures, which is of great interest since hyper inflammation enhances the risk and severity of the disease. Strategic investigations are also needed to track immune responses and explain how some infected people recover naturally and others did not. Such knowledge would also lead us to develop effective vaccines and therapeutics. Based on recent developments in the literature, we believe that deficiency of type-I IFN is a sign of severe SARS-CoV-2 infected patients. Thus, we infer that the severity in SARS-CoV-2 infected patients can be comforted after considering fulfilling IFN deficiency via administration of IFN and addressing excessive inflammation via appropriate anti-inflammatory therapies that target TNF-α or IL-6. However, this hypothesis needs further testing and elucidating regarding how SARS-CoV-2 viruses cause cascade pathways in the immune system. Interdisciplinary efforts from expertise in microbiology, medicine, public health, pharmacology, and information technology and the contribution of environmental scientists are immediately needed to battle against the prevailing crisis. Continuous monitoring of immune responses onset of SARS-CoV-2 infection is still challenging considering both diverse patterns and complexity. Thus, comprehensive revelations on an immune response would be a practical approach to challenge the current pandemic crisis, the emergence of SARS-CoV-2 variants, and future pandemic pathogens.
Author Contributions
Conceptualization, G.S.G. and D.-Y.K.; Methodology, A.A.K.; Software, S.K.S.; Validation A.H.B.; Formal Analysis and Resources A.S. and D.-Y.K.; Data Curation, A.A.K.; Writing-Original Draft Preparation, G.D.S., R.G.S. and G.S.G.; Writing-Review & Editing, G.D.S., G.S.G. and H.-S.S.; Visualization, M.K. and G.D.S.; Supervision, D.-Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
This work was supported by the Dongguk University Research Fund of 2021. The author extends the appreciation to the King Saud University, Riyadh, Saudi Arabia, for the kind support.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Guan, J.; Wei, X.; Qin, S.; Liu, X.; Jiang, Y.; Chen, Y.; Chen, Y.; Lu, H.; Qian, J.; Wang, Z.; et al. Continuous tracking of COVID-19 patients’ immune status. Int. Immunopharmacol. 2020, 89, 107034. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Mehta, O.P.; Bhandari, P.; Raut, A.; Kacimi, S.E.O.; Huy, N.T. Coronavirus Disease (COVID-19): Comprehensive Review of Clinical Presentation. Front. Public Health 2020, 8, 582932. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Moghadas, S.M.; Fitzpatrick, M.C.; Sah, P.; Pandey, A.; Shoukat, A.; Singer, B.H.; Galvani, A.P. The implications of silent transmission for the control of COVID-19 outbreaks. Proc. Natl. Acad. Sci. USA 2020, 117, 17513–17515. [Google Scholar] [CrossRef]
- Daher, A.; Balfanz, P.; Aetou, M.; Hartmann, B.; Müller-Wieland, D.; Müller, T.; Marx, N.; Dreher, M.; Cornelissen, C.G. Clinical course of COVID-19 patients needing supplemental oxygen outside the intensive care unit. Sci. Rep. 2021, 11, 2256. [Google Scholar] [CrossRef] [PubMed]
- Llitjos, J.-F.; Bredin, S.; Lascarrou, J.-B.; Soumagne, T.; Cojocaru, M.; Leclerc, M.; Lepetit, A.; Gouhier, A.; Charpentier, J.; Piton, G.; et al. Increased susceptibility to intensive care unit-acquired pneumonia in severe COVID-19 patients: A multicentre retrospective cohort study. Ann. Intensive Care 2021, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-Y.; Shinde, S.K.; Lone, S.; Palem, R.R.; Ghodake, G.S. COVID-19 Pandemic: Public Health Risk Assessment and Risk Mitigation Strategies. J. Pers. Med. 2021, 11, 1243. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Olbei, M.; Hautefort, I.; Modos, D.; Treveil, A.; Poletti, M.; Gul, L.; Shannon-Lowe, C.D.; Korcsmaros, T. SARS-CoV-2 Causes a Different Cytokine Response Compared to Other Cytokine Storm-Causing Respiratory Viruses in Severely Ill Patients. Front. Immunol. 2021, 12, 629193. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, A.; Jafarzadeh, S.; Nozari, P.; Mokhtari, P.; Nemati, M. Lymphopenia an important immunological abnormality in patients with COVID-19: Possible mechanisms. Scand. J. Immunol. 2021, 93, e12967. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Maccio, U.; Zinkernagel, A.S.; Shambat, S.M.; Zeng, X.; Cathomas, G.; Ruschitzka, F.; Schuepbach, R.A.; Moch, H.; Varga, Z. SARS-CoV-2 leads to a small vessel endotheliitis in the heart. EBioMedicine 2021, 63, 103182. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.L.; Dmytrenko, O.; Greenberg, L.; Bredemeyer, A.L.; Ma, P.; Liu, J.; Penna, V.; Winkler, E.S.; Sviben, S.; Brooks, E.; et al. SARS-CoV-2 Infects Human Engineered Heart Tissues and Models COVID-19 Myocarditis. JACC Basic Transl. Sci. 2021, 6, 331–345. [Google Scholar] [CrossRef]
- Taoufik, Y.; de Goër de Herve, M.-G.; Corgnac, S.; Durrbach, A.; Mami-Chouaib, F. When Immunity Kills: The Lessons of SARS-CoV-2 Outbreak. Front. Immunol. 2021, 12, 3833. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; Liu, Y.; Liu, Y.; et al. Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. medRxiv 2020. [Google Scholar] [CrossRef]
- Ghodake, G.S.; Shinde, S.K.; Kadam, A.A.; Saratale, R.G.; Saratale, G.D.; Syed, A.; Elgorban, A.M.; Marraiki, N.; Kim, D.-Y. Biological characteristics and biomarkers of novel SARS-CoV-2 facilitated rapid development and implementation of diagnostic tools and surveillance measures. Biosens. Bioelectron. 2021, 177, 112969. [Google Scholar] [CrossRef]
- Gelaye, B.; Foster, S.; Bhasin, M.; Tawakol, A.; Fricchione, G. SARS-CoV-2 morbidity and mortality in racial/ethnic minority populations: A window into the stress related inflammatory basis of health disparities? Brain Behav. Immun. Health 2020, 9, 100158. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Nie, M.; Pang, H.; Wang, B.; Hu, J.; Meng, X.; Li, K.; Ran, X.; Long, Q.; Deng, H.; et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021, 12, 1618. [Google Scholar] [CrossRef] [PubMed]
- Panovska-Stavridis, I.; Ridova, N.; Stojanoska, T.; Demiri, I.; Stevanovic, M.; Stojanovska, S.; Ristevska, T.; Dimkovski, A.; Filipce, V.; Dimovski, A.; et al. Insight in the Current Progress in the Largest Clinical Trials for COVID-19 Drug Management (As of January 2021). Prilozi 2021, 42, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Dhupkar, P.; Mukherjee, S. Ethical dimensions in randomized trials and off-label use of investigational drugs for COVID-19 treatment. Clin. Ethics 2022, 17, 95–104. [Google Scholar] [CrossRef]
- Mulangu, S.; Dodd, L.E.; Davey, R.T.; Tshiani Mbaya, O.; Proschan, M.; Mukadi, D.; Lusakibanza Manzo, M.; Nzolo, D.; Tshomba Oloma, A.; Ibanda, A.; et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, X. COVID-19: A new challenge for human beings. Cell. Mol. Immunol. 2020, 17, 555–557. [Google Scholar] [CrossRef]
- Arora, G.; Shrivastava, R.; Kumar, P.; Bandichhor, R.; Krishnamurthy, D.; Sharma, R.K.; Matharu, A.S.; Pandey, J.; Rizwan, M. Recent advances made in the synthesis of small drug molecules for clinical applications: An insight. Curr. Res. Green Sustain. Chem. 2021, 4, 100097. [Google Scholar] [CrossRef]
- Prince, T.; Donovan-Banfield, I.a.; Goldswain, H.; Penrice-Randal, R.; Turtle, L.; Fletcher, T.; Khoo, S.; Hiscox, J.A. Antiviral activity of molnupiravir precursor NHC against SARS-CoV-2 Variants of Concern (VOCs) and its therapeutic window in a human lung cell model. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2020, 384, 693–704. [CrossRef]
- Ison, M.G.; Wolfe, C.; Boucher, H.W. Emergency Use Authorization of Remdesivir: The Need for a Transparent Distribution Process. JAMA 2020, 323, 2365–2366. [Google Scholar] [CrossRef] [PubMed]
- Gendrot, M.; Javelle, E.; Clerc, A.; Savini, H.; Pradines, B. Chloroquine as a prophylactic agent against COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105980. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, A.; Shaikh, A.; Singh, R.; Misra, A. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr. 2020, 14, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Cai, S.; Li, Y.; Li, Y.; Fan, Y.; Li, L.; Lei, C.; Tang, X.; Hu, F.; Li, F.; et al. Prognostic Factors for COVID-19 Pneumonia Progression to Severe Symptoms Based on Earlier Clinical Features: A Retrospective Analysis. Front. Med. 2020, 7, 643. [Google Scholar] [CrossRef]
- Horby, P.W.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Emberson, J.; Palfreeman, A.; Raw, J.; Elmahi, E.; Prudon, B.; et al. Lopinavir–ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef]
- Fischer, W.; Eron, J.J.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv 2021. [Google Scholar] [CrossRef]
- Robinson, P.C.; Morand, E. Divergent effects of acute versus chronic glucocorticoids in COVID-19. Lancet Rheumatol. 2021, 3, e168–e170. [Google Scholar] [CrossRef]
- Zhuravel, S.V.; Khmelnitskiy, O.K.; Burlaka, O.O.; Gritsan, A.I.; Goloshchekin, B.M.; Kim, S.; Hong, K.Y. Nafamostat in hospitalized patients with moderate to severe COVID-19 pneumonia: A randomised Phase II clinical trial. eClinicalMedicine 2021, 41, 101169. [Google Scholar] [CrossRef]
- Axfors, C.; Schmitt, A.M.; Janiaud, P.; van’t Hooft, J.; Abd-Elsalam, S.; Abdo, E.F.; Abella, B.S.; Akram, J.; Amaravadi, R.K.; Angus, D.C.; et al. Mortality outcomes with hydroxychloroquine and chloroquine in COVID-19 from an international collaborative meta-analysis of randomized trials. Nat. Commun. 2021, 12, 2349. [Google Scholar] [CrossRef]
- Khalil, A.; Kamar, A.; Nemer, G. Thalidomide-Revisited: Are COVID-19 Patients Going to Be the Latest Victims of Yet Another Theoretical Drug-Repurposing? Front. Immunol. 2020, 11, 1248. [Google Scholar] [CrossRef]
- Loffredo, M.; Lucero, H.; Chen, D.-Y.; O’Connell, A.; Bergqvist, S.; Munawar, A.; Bandara, A.; De Graef, S.; Weeks, S.D.; Douam, F.; et al. The in-vitro effect of famotidine on SARS-CoV-2 proteases and virus replication. Sci. Rep. 2021, 11, 5433. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, H.; Li, Y.; Jian, X.; Hou, X.; Zhong, N.; Fei, J.; Su, D.; Bian, Z.; Zhang, Y.; et al. Efficacy and Safety of Leflunomide for Refractory COVID-19: A Pilot Study. Front. Pharmacol. 2021, 12, 1217. [Google Scholar] [CrossRef] [PubMed]
- Durán-Méndez, A.; Aguilar-Arroyo, A.D.; Vivanco-Gómez, E.; Nieto-Ortega, E.; Pérez-Ortega, D.; Jiménez-Pérez, C.; Hernández-Skewes, K.Y.; Montiel-Bravo, G.; Roque-Reyes, O.J.; Romero-Lechuga, F.; et al. Tocilizumab reduces COVID-19 mortality and pathology in a dose and timing-dependent fashion: A multi-centric study. Sci. Rep. 2021, 11, 19728. [Google Scholar] [CrossRef] [PubMed]
- Kyriazopoulou, E.; Poulakou, G.; Milionis, H.; Metallidis, S.; Adamis, G.; Tsiakos, K.; Fragkou, A.; Rapti, A.; Damoulari, C.; Fantoni, M.; et al. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: A double-blind, randomized controlled phase 3 trial. Nat. Med. 2021, 27, 1752–1760. [Google Scholar] [CrossRef]
- Pang, J.; Xu, F.; Aondio, G.; Li, Y.; Fumagalli, A.; Lu, M.; Valmadre, G.; Wei, J.; Bian, Y.; Canesi, M.; et al. Efficacy and tolerability of bevacizumab in patients with severe COVID-19. Nat. Commun. 2021, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Vankadari, N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 2020, 56, 105998. [Google Scholar] [CrossRef] [PubMed]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; de Cassia Pellegrini, R.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- Nojomi, M.; Yassin, Z.; Keyvani, H.; Makiani, M.J.; Roham, M.; Laali, A.; Dehghan, N.; Navaei, M.; Ranjbar, M. Effect of Arbidol (Umifenovir) on COVID-19: A randomized controlled trial. BMC Infect. Dis. 2020, 20, 954. [Google Scholar] [CrossRef]
- Blum, V.F.; Cimerman, S.; Hunter, J.R.; Tierno, P.; Lacerda, A.; Soeiro, A.; Cardoso, F.; Bellei, N.C.; Maricato, J.; Mantovani, N.; et al. Nitazoxanide superiority to placebo to treat moderate COVID-19—A Pilot prove of concept randomized double-blind clinical trial. eClinicalMedicine 2021, 37, 100981. [Google Scholar] [CrossRef]
- Saleh, M.; Vaezi, A.A.; Aliannejad, R.; Sohrabpour, A.A.; Kiaei, S.Z.F.; Shadnoush, M.; Siavashi, V.; Aghaghazvini, L.; Khoundabi, B.; Abdoli, S.; et al. Cell therapy in patients with COVID-19 using Wharton’s jelly mesenchymal stem cells: A phase 1 clinical trial. Stem Cell Res. Ther. 2021, 12, 410. [Google Scholar] [CrossRef]
- Zanirati, G.; Provenzi, L.; Libermann, L.L.; Bizotto, S.C.; Ghilardi, I.M.; Marinowic, D.R.; Shetty, A.K.; Da Costa, J.C. Stem cell-based therapy for COVID-19 and ARDS: A systematic review. NPJ Regen. Med. 2021, 6, 73. [Google Scholar] [CrossRef]
- Bégin, P.; Callum, J.; Jamula, E.; Cook, R.; Heddle, N.M.; Tinmouth, A.; Zeller, M.P.; Beaudoin-Bussières, G.; Amorim, L.; Bazin, R.; et al. Convalescent plasma for hospitalized patients with COVID-19: An open-label, randomized controlled trial. Nat. Med. 2021, 27, 2012–2024. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Yamin, R.; Jones, A.T.; Hoffmann, H.-H.; Schäfer, A.; Kao, K.S.; Francis, R.L.; Sheahan, T.P.; Baric, R.S.; Rice, C.M.; Ravetch, J.V.; et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature 2021, 599, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Mir, T.; Almas, T.; Kaur, J.; Faisaluddin, M.; Song, D.; Ullah, W.; Mamtani, S.; Rauf, H.; Yadav, S.; Latchana, S.; et al. Coronavirus disease 2019 (COVID-19): Multisystem review of pathophysiology. Ann. Med. Surg. 2021, 69, 102745. [Google Scholar] [CrossRef] [PubMed]
- Aleksova, A.; Ferro, F.; Gagno, G.; Cappelletto, C.; Santon, D.; Rossi, M.; Ippolito, G.; Zumla, A.; Beltrami, A.P.; Sinagra, G. COVID-19 and renin-angiotensin system inhibition: Role of angiotensin converting enzyme 2 (ACE2)—Is there any scientific evidence for controversy? J. Intern. Med. 2020, 288, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Edin, M.L.; Zeldin, D.C.; Li, C.; Wang, D.W.; Chen, C. Good or bad: Application of RAAS inhibitors in COVID-19 patients with cardiovascular comorbidities. Pharmacol. Ther. 2020, 215, 107628. [Google Scholar] [CrossRef] [PubMed]
- Rico-Mesa, J.S.; White, A.; Anderson, A.S. Outcomes in Patients with COVID-19 Infection Taking ACEI/ARB. Curr. Cardiol. Rep. 2020, 22, 31. [Google Scholar] [CrossRef]
- Vaduganathan, M.; Vardeny, O.; Michel, T.; McMurray, J.J.V.; Pfeffer, M.A.; Solomon, S.D. Renin–Angiotensin–Aldosterone System Inhibitors in Patients with COVID-19. N. Engl. J. Med. 2020, 382, 1653–1659. [Google Scholar] [CrossRef]
- Dean, A.Q.; Bozza, W.P.; Twomey, J.D.; Luo, S.; Nalli, A.; Zhang, B. The fight against COVID-19: Striking a balance in the renin–angiotensin system. Drug Discov. Today 2021, 26, 2214–2220. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020, 76, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Hoshizaki, M.; Minato, T.; Nirasawa, S.; Asaka, M.N.; Niiyama, M.; Imai, M.; Uda, A.; Chan, J.F.-W.; Takahashi, S.; et al. ACE2-like carboxypeptidase B38-CAP protects from SARS-CoV-2-induced lung injury. Nat. Commun. 2021, 12, 6791. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choi, W.J. Overview of COVID-19 inflammatory pathogenesis from the therapeutic perspective. Arch. Pharmacal. Res. 2021, 44, 99–116. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Yamamoto, M.; Matsuyama, S.; Li, X.; Takeda, M.; Kawaguchi, Y.; Inoue, J.-i.; Matsuda, Z. Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrob. Agents Chemother. 2016, 60, 6532–6539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Geng, J.; Chen, L.; Yuan, Y.; Wang, K.; Wang, Y.; Qin, C.; Wu, G.; Chen, R.; Zhang, Z.; Wei, D.; et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021, 6, 347. [Google Scholar] [CrossRef]
- Bian, H.; Zheng, Z.-H.; Wei, D.; Wen, A.; Zhang, Z.; Lian, J.-Q.; Kang, W.-Z.; Hao, C.-Q.; Wang, J.; Xie, R.-H.; et al. Safety and efficacy of meplazumab in healthy volunteers and COVID-19 patients: A randomized phase 1 and an exploratory phase 2 trial. Signal Transduct. Target. Ther. 2021, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Gubernatorova, E.O.; Gorshkova, E.A.; Polinova, A.I.; Drutskaya, M.S. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020, 53, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Pillat, M.M. CD147 as a Target for COVID-19 Treatment: Suggested Effects of Azithromycin and Stem Cell Engagement. Stem Cell Rev. Rep. 2020, 16, 434–440. [Google Scholar] [CrossRef]
- Schijns, V.; Lavelle, E.C. Prevention and treatment of COVID-19 disease by controlled modulation of innate immunity. Eur. J. Immunol. 2020, 50, 932–938. [Google Scholar] [CrossRef]
- Reina, J.; Reina, N. Favipiravir, a new concept of antiviral drug against influenza viruses. Rev. Esp. Quimioter. Publ. Of. Soc. Esp. Quimioter. 2017, 30, 79–83. [Google Scholar]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Buckland, M.S.; Galloway, J.B.; Fhogartaigh, C.N.; Meredith, L.; Provine, N.M.; Bloor, S.; Ogbe, A.; Zelek, W.M.; Smielewska, A.; Yakovleva, A.; et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: A case report. Nat. Commun. 2020, 11, 6385. [Google Scholar] [CrossRef]
- Khan, F.I.; Kang, T.; Ali, H.; Lai, D. Remdesivir Strongly Binds to RNA-Dependent RNA Polymerase, Membrane Protein, and Main Protease of SARS-CoV-2: Indication From Molecular Modeling and Simulations. Front. Pharmacol. 2021, 12, 1616. [Google Scholar] [CrossRef]
- Doi, K.; Ikeda, M.; Hayase, N.; Moriya, K.; Morimura, N.; Maehara, H.; Tagami, S.; Fukushima, K.; Misawa, N.; Inoue, Y.; et al. Nafamostat mesylate treatment in combination with favipiravir for patients critically ill with COVID-19: A case series. Crit. Care 2020, 24, 392. [Google Scholar] [CrossRef]
- Hifumi, T.; Isokawa, S.; Otani, N.; Ishimatsu, S. Adverse events associated with nafamostat mesylate and favipiravir treatment in COVID-19 patients. Crit. Care 2020, 24, 497. [Google Scholar] [CrossRef]
- Osborne, V.; Davies, M.; Lane, S.; Evans, A.; Denyer, J.; Dhanda, S.; Roy, D.; Shakir, S. Lopinavir-Ritonavir in the Treatment of COVID-19: A Dynamic Systematic Benefit-Risk Assessment. Drug Saf. 2020, 43, 809–821. [Google Scholar] [CrossRef]
- Stower, H. Lopinavir–ritonavir in severe COVID-19. Nat. Med. 2020, 26, 465. [Google Scholar] [CrossRef] [PubMed]
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Nigoghossian, C.D.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2020, 75, 2950–2973. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, R.; Liu, C.; Liang, W.; Guan, W.; Tang, R.; Tang, C.; Zhang, N.; Zhong, N.; Li, S. Attention should be paid to venous thromboembolism prophylaxis in the management of COVID-19. Lancet Haematol. 2020, 7, e362–e363. [Google Scholar] [CrossRef]
- Paranjpe, I.; Fuster, V.; Lala, A.; Russak, A.J.; Glicksberg, B.S.; Levin, M.A.; Charney, A.W.; Narula, J.; Fayad, Z.A.; Bagiella, E.; et al. Association of Treatment Dose Anticoagulation With In-Hospital Survival Among Hospitalized Patients With COVID-19. J. Am. Coll. Cardiol. 2020, 76, 122–124. [Google Scholar] [CrossRef]
- Barnes, G.D.; Burnett, A.; Allen, A.; Blumenstein, M.; Clark, N.P.; Cuker, A.; Dager, W.E.; Deitelzweig, S.B.; Ellsworth, S.; Garcia, D.; et al. Thromboembolism and anticoagulant therapy during the COVID-19 pandemic: Interim clinical guidance from the anticoagulation forum. J. Thromb. Thrombolysis 2020, 50, 72–81. [Google Scholar] [CrossRef]
- Paar, V.; Wernly, B.; Zhou, Z.; Motloch, L.J.; Hoppe, U.C.; Egle, A.; Lichtenauer, M. Anti-coagulation for COVID-19 treatment: Both anti-thrombotic and anti-inflammatory? J. Thromb. Thrombolysis 2021, 51, 226–231. [Google Scholar] [CrossRef]
- Russo, V.; Cardillo, G.; Viggiano, G.V.; Mangiacapra, S.; Cavalli, A.; Fontanella, A.; Agrusta, F.; Bellizzi, A.; Amitrano, M.; Iannuzzo, M.; et al. Fondaparinux Use in Patients With COVID-19: A Preliminary Multicenter Real-World Experience. J. Cardiovasc. Pharmacol. 2020, 76, 369–371. [Google Scholar] [CrossRef]
- Flam, B.; Wintzell, V.; Ludvigsson, J.F.; Mårtensson, J.; Pasternak, B. Direct oral anticoagulant use and risk of severe COVID-19. J. Intern. Med. 2021, 289, 411–419. [Google Scholar] [CrossRef]
- Rossi, R.; Coppi, F.; Talarico, M.; Boriani, G. Protective role of chronic treatment with direct oral anticoagulants in elderly patients affected by interstitial pneumonia in COVID-19 era. Eur. J. Intern. Med. 2020, 77, 158–160. [Google Scholar] [CrossRef]
- Meini, S.; Zanichelli, A.; Sbrojavacca, R.; Iuri, F.; Roberts, A.T.; Suffritti, C.; Tascini, C. Understanding the Pathophysiology of COVID-19: Could the Contact System Be the Key? Front. Immunol. 2020, 11, 2014. [Google Scholar] [CrossRef]
- Mizurini, D.M.; Hottz, E.D.; Bozza, P.T.; Monteiro, R.Q. Fundamentals in COVID-19-Associated Thrombosis: Molecular and Cellular Aspects. Front. Cardiovasc. Med. 2021, 8, 785738. [Google Scholar] [CrossRef] [PubMed]
- Taus, F.; Salvagno, G.; Canè, S.; Fava, C.; Mazzaferri, F.; Carrara, E.; Petrova, V.; Barouni, R.M.; Dima, F.; Dalbeni, A.; et al. Platelets Promote Thromboinflammation in SARS-CoV-2 Pneumonia. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2975–2989. [Google Scholar] [CrossRef] [PubMed]
- Asakura, H.; Ogawa, H. Potential of heparin and nafamostat combination therapy for COVID-19. J. Thromb. Haemost. 2020, 18, 1521–1522. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, A.; Montecucco, F.; Dallegri, F. Update on the effects of treatment with recombinant tissue-type plasminogen activator (rt-PA) in acute ischemic stroke. Expert Opin. Biol. Ther. 2016, 16, 1323–1340. [Google Scholar] [CrossRef]
- Wang, J.; Hajizadeh, N.; Moore, E.E.; McIntyre, R.C.; Moore, P.K.; Veress, L.A.; Yaffe, M.B.; Moore, H.B.; Barrett, C.D. Tissue plasminogen activator (tPA) treatment for COVID-19 associated acute respiratory distress syndrome (ARDS): A case series. J. Thromb. Haemost. 2020, 18, 1752–1755. [Google Scholar] [CrossRef] [PubMed]
- Kanthi, Y.; Knight, J.S.; Zuo, Y.; Pinsky, D.J. New (re)purpose for an old drug: Purinergic modulation may extinguish the COVID-19 thromboinflammatory firestorm. JCI Insight 2020, 5, e140971. [Google Scholar] [CrossRef]
- Fata-Hartley, C.L.; Palmenberg, A.C. Dipyridamole Reversibly Inhibits Mengovirus RNA Replication. J. Virol. 2005, 79, 11062–11070. [Google Scholar] [CrossRef]
- Huang, B.; Chen, Z.; Geng, L.; Wang, J.; Liang, H.; Cao, Y.; Chen, H.; Huang, W.; Su, M.; Wang, H.; et al. Mucosal Profiling of Pediatric-Onset Colitis and IBD Reveals Common Pathogenics and Therapeutic Pathways. Cell 2019, 179, 1160–1176.e1124. [Google Scholar] [CrossRef]
- Insel, P.A.; Murray, F.; Yokoyama, U.; Romano, S.; Yun, H.; Brown, L.; Snead, A.; Lu, D.; Aroonsakool, N. cAMP and Epac in the regulation of tissue fibrosis. Br. J. Pharmacol. 2012, 166, 447–456. [Google Scholar] [CrossRef]
- Anderson, R.M.; Vegvari, C.; Hollingsworth, T.D.; Pi, L.; Maddren, R.; Ng, C.W.; Baggaley, R.F. The SARS-CoV-2 pandemic: Remaining uncertainties in our understanding of the epidemiology and transmission dynamics of the virus, and challenges to be overcome. Interface Focus 2021, 11, 20210008. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Liu, S.; Sun, J.; Chen, Z.; Jiang, M.; Zhang, Q.; Wei, Y.; Wang, X.; Huang, Y.-Y.; et al. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B 2020, 10, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Mitsios, A.; Chrysanthopoulou, A.; Arampatzioglou, A.; Angelidou, I.; Vidali, V.; Ritis, K.; Skendros, P.; Stakos, D. Ticagrelor Exerts Immune-Modulatory Effect by Attenuating Neutrophil Extracellular Traps. Int. J. Mol. Sci. 2020, 21, 3625. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Majumdar, S.; Singh, R.; Misra, A. Role of corticosteroid in the management of COVID-19: A systemic review and a Clinician’s perspective. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 971–978. [Google Scholar] [CrossRef]
- Magro, G. SARS-CoV-2 and COVID-19: Is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine X 2020, 2, 100029. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef] [PubMed]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136.e124. [Google Scholar] [CrossRef]
- Liu, B.; Li, M.; Zhou, Z.; Guan, X.; Xiang, Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020, 111, 102452. [Google Scholar] [CrossRef]
- Somers, E.C.; Eschenauer, G.A.; Troost, J.P.; Golob, J.L.; Gandhi, T.N.; Wang, L.; Zhou, N.; Petty, L.A.; Baang, J.H.; Dillman, N.O.; et al. Tocilizumab for Treatment of Mechanically Ventilated Patients With COVID-19. Clin. Infect. Dis. 2020, 73, e445–e454. [Google Scholar] [CrossRef]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Rawling, M.; Savory, E.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef]
- Riegler, L.L.; Jones, G.P.; Lee, D.W. Current approaches in the grading and management of cytokine release syndrome after chimeric antigen receptor T-cell therapy. Clin. Risk Manag. 2019, 15, 323–335. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. ImmunoTherapy Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Synowiec, A.; Szczepański, A.; Barreto-Duran, E.; Lie, L.K.; Pyrc, K. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): A Systemic Infection. Clin. Microbiol. Rev. 2021, 34, e00133-00120. [Google Scholar] [CrossRef]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Tran, B.N.; Cheng, Y.-S.; Xu, M.; Pradhan, M.; Henderson, M.; Zhu, W.; et al. The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infect. Dis. 2021, 7, 1389–1408. [Google Scholar] [CrossRef]
- Chen, I.-Y.; Moriyama, M.; Chang, M.-F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Sticherling, M.; Neurath, M.F. COVID-19: Risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol. 2020, 20, 271–272. [Google Scholar] [CrossRef] [PubMed]
- Venerito, V.; Lopalco, G.; Iannone, F. COVID-19, rheumatic diseases and immunosuppressive drugs: An appeal for medication adherence. Rheumatol. Int. 2020, 40, 827–828. [Google Scholar] [CrossRef]
- Nile, S.H.; Nile, A.; Qiu, J.; Li, L.; Jia, X.; Kai, G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020, 53, 66–70. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Zhang, F.; Wang, Q.; Li, T.; Liu, Z.; Wang, J.; Qin, Y.; Zhang, X.; Yan, X.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Dariya, B.; Nagaraju, G.P. Understanding novel COVID-19: Its impact on organ failure and risk assessment for diabetic and cancer patients. Cytokine Growth Factor Rev. 2020, 53, 43–52. [Google Scholar] [CrossRef]
- Ronco, C.; Reis, T.; Husain-Syed, F. Management of acute kidney injury in patients with COVID-19. Lancet Respir. Med. 2020, 8, 738–742. [Google Scholar] [CrossRef]
- Moon, C. Fighting COVID-19 exhausts T cells. Nat. Rev. Immunol. 2020, 20, 277. [Google Scholar] [CrossRef] [PubMed]
- Côté, P.; Baril, J.-G.; Hébert, M.-N.; Klein, M.; Lalonde, R.; Poliquin, M.; Rouleau, D.; Therrien, R.; Vézina, S.; Willems, B.; et al. Management and Treatment of Hepatitis C Virus in Patients with HIV and Hepatitis C Virus Coinfection: A Practical Guide for Health Care Professionals. Can. J. Infect. Dis. Med. Microbiol. 2007, 18, 631054. [Google Scholar] [CrossRef][Green Version]
- Taubenberger, J.K.; Morens, D.M. The Pathology of Influenza Virus Infections. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 499–522. [Google Scholar] [CrossRef]
- Kim, Y.-I.; Kim, S.-G.; Kim, S.-M.; Kim, E.-H.; Park, S.-J.; Yu, K.-M.; Chang, J.-H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709.e702. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Philip, A.T.; Stoppacher, R. Pathologic Findings in Novel Influenza A (H1N1) Virus (“Swine Flu”) Infection: Contrasting Clinical Manifestations and Lung Pathology in Two Fatal Cases. Am. J. Clin. Pathol. 2010, 133, 380–387. [Google Scholar] [CrossRef][Green Version]
- Pascal, L.; Hivert, B.; Trauet, J.; Deberranger, E.; Dessaint, J.-P.; Yakoub-Agha, I.; Labalette, M. A Low Effective Dose of Interleukin-7 Is Sufficient to Maintain Cord Blood T Cells Alive without Potentiating Allo-Immune Responses. Biol. Blood Marrow Transplant. 2015, 21, 625–631. [Google Scholar] [CrossRef][Green Version]
- Zhong, J.; Tang, J.; Ye, C.; Dong, L. The immunology of COVID-19: Is immune modulation an option for treatment? Lancet Rheumatol. 2020, 2, e428–e436. [Google Scholar] [CrossRef]
- Rovere Querini, P.; De Lorenzo, R.; Conte, C.; Brioni, E.; Lanzani, C.; Yacoub, M.R.; Chionna, R.; Martinenghi, S.; Vitali, G.; Tresoldi, M.; et al. Post-COVID-19 follow-up clinic: Depicting chronicity of a new disease. Acta Biomed. Atenei Parm. 2020, 91, 22–28. [Google Scholar] [CrossRef]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Yin, Q.; Wu, Y.; Zhang, W. Potential protective role of the anti-PD-1 blockade against SARS-CoV-2 infection. Biomed. Pharmacother. 2021, 142, 111957. [Google Scholar] [CrossRef] [PubMed]
- Bekele, Y.; Sui, Y.; Berzofsky, J.A. IL-7 in SARS-CoV-2 Infection and as a Potential Vaccine Adjuvant. Front. Immunol. 2021, 12, 3796. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Herbein, G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin. Epigenetics 2020, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e1039. [Google Scholar] [CrossRef]
- Aevermann, B.D.; Pickett, B.E.; Kumar, S.; Klem, E.B.; Agnihothram, S.; Askovich, P.S.; Bankhead, A.; Bolles, M.; Carter, V.; Chang, J.; et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci. Data 2014, 1, 140033. [Google Scholar] [CrossRef]
- Menachery, V.D.; Eisfeld, A.J.; Schäfer, A.; Josset, L.; Sims, A.C.; Proll, S.; Fan, S.; Li, C.; Neumann, G.; Tilton, S.C.; et al. Pathogenic Influenza Viruses and Coronaviruses Utilize Similar and Contrasting Approaches To Control Interferon-Stimulated Gene Responses. mBio 2014, 5, e01174-01114. [Google Scholar] [CrossRef]
- Akira, S. Pathogen recognition by innate immunity and its signaling. Proc. Jpn. Acad. Ser. B 2009, 85, 143–156. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Bruening, J.; Weigel, B.; Gerold, G. The Role of Type III Interferons in Hepatitis C Virus Infection and Therapy. J. Immunol. Res. 2017, 2017, 7232361. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Liu, X.; Cao, H.; Li, J.; Wang, B.; Zhang, P.; Dong Zhang, X.; Liu, Z.; Yuan, H.; Zhan, Z. Autophagy induced by DAMPs facilitates the inflammation response in lungs undergoing ischemia-reperfusion injury through promoting TRAF6 ubiquitination. Cell Death Differ. 2017, 24, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Schöllhorn, A.; Schuhmacher, J.; Besedovsky, L.; Fendel, R.; Jensen, A.T.R.; Stevanović, S.; Lange, T.; Rammensee, H.-G.; Born, J.; Gouttefangeas, C.; et al. Integrin Activation Enables Sensitive Detection of Functional CD4+ and CD8+ T Cells: Application to Characterize SARS-CoV-2 Immunity. Front. Immunol. 2021, 12, 522. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Maggi, L.; Capone, M.; Spinicci, M.; Salvati, L.; Colao, M.G.; Vanni, A.; Kiros, S.T.; Mencarini, J.; Zammarchi, L.; et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol. 2020, 50, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, M.; Spehner, L.; Vettoretti, L.; Bouard, A.; Eberst, G.; Pili Floury, S.; Capellier, G.; Lepiller, Q.; Orillard, E.; Mansi, L.; et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J. Infect. 2021, 82, 282–327. [Google Scholar] [CrossRef] [PubMed]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020, 179, 104811. [Google Scholar] [CrossRef]
- Choi, H.; Shin, E.-C. Roles of Type I and III Interferons in COVID-19. Yonsei Med. J. 2021, 62, 381–390. [Google Scholar] [CrossRef]
- Ramos, I.; Fernandez-Sesma, A. Modulating the Innate Immune Response to Influenza A Virus: Potential Therapeutic Use of Anti-Inflammatory Drugs. Front. Immunol. 2015, 6, 361. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Momattin, H.; Dib, J.; Memish, Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: An observational study. Int. J. Infect. Dis. 2014, 20, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Arabi, Y.M.; Shalhoub, S.; Mandourah, Y.; Al-Hameed, F.; Al-Omari, A.; Al Qasim, E.; Jose, J.; Alraddadi, B.; Almotairi, A.; Al Khatib, K.; et al. Ribavirin and Interferon Therapy for Critically Ill Patients With Middle East Respiratory Syndrome: A Multicenter Observational Study. Clin. Infect. Dis. 2019, 70, 1837–1844. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Chan, K.S.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.-H.; Barbi, J.; Pan, F. Hypoxia-inducible factors in T lymphocyte differentiation and function. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C580–C589. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Blum, H.E.; Thimme, R. T-cell responses in hepatitis B and C virus infection: Similarities and differences. Emerg. Microbes Infect. 2013, 2, e15. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Horn, L.A.; Haile, S.T. The Programmed Death-1 Immune-Suppressive Pathway: Barrier to Antitumor Immunity. J. Immunol. 2014, 193, 3835–3841. [Google Scholar] [CrossRef]
- Mishra, V.; Agas, A.; Schuetz, H.; Kalluru, J.; Haorah, J. Alcohol induces programmed death receptor-1 and programmed death-ligand-1 differentially in neuroimmune cells. Alcohol 2020, 86, 65–74. [Google Scholar] [CrossRef]
- Porichis, F.; Kaufmann, D.E. Role of PD-1 in HIV Pathogenesis and as Target for Therapy. Curr. HIV/AIDS Rep. 2012, 9, 81–90. [Google Scholar] [CrossRef]
- Bersanelli, M. Controversies about COVID-19 and anticancer treatment with immune checkpoint inhibitors. Immunotherapy 2020, 12, 269–273. [Google Scholar] [CrossRef]
- Poe, F.L.; Corn, J. N-Acetylcysteine: A potential therapeutic agent for SARS-CoV-2. Med. Hypotheses 2020, 143, 109862. [Google Scholar] [CrossRef]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Fogel, D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 2018, 11, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Gnjatic, S.; Bronte, V.; Brunet, L.R.; Butler, M.O.; Disis, M.L.; Galon, J.; Hakansson, L.G.; Hanks, B.A.; Karanikas, V.; Khleif, S.N.; et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J. ImmunoTherapy Cancer 2017, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Lei, H. A single transcript for the prognosis of disease severity in COVID-19 patients. Sci. Rep. 2021, 11, 12174. [Google Scholar] [CrossRef] [PubMed]
- Ganji, A.; Farahani, I.; Khansarinejad, B.; Ghazavi, A.; Mosayebi, G. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cells Mol. Dis. 2020, 83, 102437. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.e14. [Google Scholar] [CrossRef]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Tong, X.; Cheng, A.; Yuan, X.; Zhong, X.; Wang, H.; Zhou, W.; Xu, X.; Li, Y. Characteristics of peripheral white blood cells in COVID-19 patients revealed by a retrospective cohort study. BMC Infect. Dis. 2021, 21, 1236. [Google Scholar] [CrossRef]
- Lu, C.; Liu, Y.; Chen, B.; Yang, H.; Hu, H.; Liu, Y.; Zhao, Y. Prognostic value of lymphocyte count in severe COVID-19 patients with corticosteroid treatment. Signal Transduct Target 2021, 6, 106. [Google Scholar] [CrossRef]
- deKay, J.T.; Emery, I.F.; Rud, J.; Eldridge, A.; Lord, C.; Gagnon, D.J.; May, T.L.; Herrera, V.L.M.; Ruiz-Opazo, N.; Riker, R.R.; et al. DEspRhigh neutrophils are associated with critical illness in COVID-19. Sci. Rep. 2021, 11, 22463. [Google Scholar] [CrossRef] [PubMed]
- Dávila-Collado, R.; Jarquín-Durán, O.; Solís-Vallejo, A.; Nguyen, M.A.; Espinoza, J.L. Elevated Monocyte to Lymphocyte Ratio and Increased Mortality among Patients with Chronic Kidney Disease Hospitalized for COVID-19. J. Pers. Med. 2021, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Prozan, L.; Shusterman, E.; Ablin, J.; Mitelpunkt, A.; Weiss-Meilik, A.; Adler, A.; Choshen, G.; Kehat, O. Prognostic value of neutrophil-to-lymphocyte ratio in COVID-19 compared with Influenza and respiratory syncytial virus infection. Sci. Rep. 2021, 11, 21519. [Google Scholar] [CrossRef]
- Gumus, H.; Demir, A.; Yükkaldıran, A. Is mean platelet volume a predictive marker for the diagnosis of COVID-19 in children? Int. J. Clin. Pract. 2021, 75, e13892. [Google Scholar] [CrossRef] [PubMed]
- Alsuwaidi, L.; Al Heialy, S.; Shaikh, N.; Al Najjar, F.; Seliem, R.; Han, A.; Hachim, M. Monocyte distribution width as a novel sepsis indicator in COVID-19 patients. BMC Infect. Dis. 2022, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Yaghmaei, B.; Sharifzadeh Ekbatani, M.; Pourakbari, B.; Navaeian, A.; Parvaneh, N.; Haghi Ashtiani, M.T.; Mamishi, S. Effects of Coronavirus Disease 2019 (COVID-19) on Peripheral Blood Lymphocytes and Their Subsets in Children: Imbalanced CD4+/CD8+ T Cell Ratio and Disease Severity. Front. Pediatrics 2021, 9, 267. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rong, L.; Cui, R.; Feng, J.; Jin, Y.; Chen, X.; Xu, R. Dynamic changes in serum IL-6, IL-8, and IL-10 predict the outcome of ICU patients with severe COVID-19. Ann. Palliat. Med. 2021, 10, 3706–3714. [Google Scholar] [CrossRef]
- Hu, R.; Han, C.; Pei, S.; Yin, M.; Chen, X. Procalcitonin levels in COVID-19 patients. Int. J. Antimicrob. Agents 2020, 56, 106051. [Google Scholar] [CrossRef]
- Zinellu, A.; Paliogiannis, P.; Carru, C.; Mangoni, A.A. Serum amyloid A concentrations, COVID-19 severity and mortality: An updated systematic review and meta-analysis. Int. J. Infect. Dis. 2021, 105, 668–674. [Google Scholar] [CrossRef]
- Mardani, R.; Namavar, M.; Ghorbi, E.; Shoja, Z.; Zali, F.; Kaghazian, H.; Aghasadeghi, M.R.; Sadeghi, S.A.; Sabeti, S.; Darazam, I.A.; et al. Association between serum inflammatory parameters and the disease severity in COVID-19 patients. J. Clin. Lab. Anal. 2022, 36, e24162. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.-Y.; Yin, C.-H.; Yao, Y.-M. Update Advances on C-Reactive Protein in COVID-19 and Other Viral Infections. Front. Immunol. 2021, 12, 3153. [Google Scholar] [CrossRef] [PubMed]
- Tural Onur, S.; Altın, S.; Sokucu, S.N.; Fikri, B.; Barça, T.; Bolat, E.; Toptaş, M. Could ferritin level be an indicator of COVID-19 disease mortality? J. Med. Virol. 2021, 93, 1672–1677. [Google Scholar] [CrossRef]
- Marfia, G.; Navone, S.; Guarnaccia, L.; Campanella, R.; Mondoni, M.; Locatelli, M.; Barassi, A.; Fontana, L.; Palumbo, F.; Garzia, E.; et al. Decreased serum level of sphingosine-1-phosphate: A novel predictor of clinical severity in COVID-19. EMBO Mol. Med. 2021, 13, e13424. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.-C.; You, C.-Y.; Lu, S.-W.; Fu, Y.-Q. Characteristics of coagulation alteration in patients with COVID-19. Ann. Hematol. 2021, 100, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, X.; Fan, Q.; Liu, H.; Liu, X.; Liu, Z.; Zhang, Z. D-dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J. Thromb. Haemost. JTH 2020, 18, 1324–1329. [Google Scholar] [CrossRef]
- Long, H.; Nie, L.; Xiang, X.; Li, H.; Zhang, X.; Fu, X.; Ren, H.; Liu, W.; Wang, Q.; Wu, Q. D-Dimer and Prothrombin Time Are the Significant Indicators of Severe COVID-19 and Poor Prognosis. BioMed Res. Int. 2020, 2020, 6159720. [Google Scholar] [CrossRef]
- Han, H.; Yang, L.; Liu, R.; Liu, F.; Wu, K.L.; Li, J.; Liu, X.H.; Zhu, C.L. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin. Chem. Lab. Med. 2020, 58, 1116–1120. [Google Scholar] [CrossRef]
- Shimura, T.; Kurano, M.; Kanno, Y.; Ikeda, M.; Okamoto, K.; Jubishi, D.; Harada, S.; Okugawa, S.; Moriya, K.; Yatomi, Y. Clot waveform of APTT has abnormal patterns in subjects with COVID-19. Sci. Rep. 2021, 11, 5190. [Google Scholar] [CrossRef]
- Araya, S.; Mamo, M.A.; Tsegay, Y.G.; Atlaw, A.; Aytenew, A.; Hordofa, A.; Negeso, A.E.; Wordofa, M.; Niguse, T.; Cheru, M.; et al. Blood coagulation parameter abnormalities in hospitalized patients with confirmed COVID-19 in Ethiopia. PLoS ONE 2021, 16, e0252939. [Google Scholar] [CrossRef]
- Liaquat, H.; Shupp, B.; Rollins, S.; Schneider, Y.; Matin, A. Comparison of the impact of chronic corticosteroid therapy on critical care outcomes of COVID-19 patients with and without history of chronic liver disease. Sci. Rep. 2021, 11, 19245. [Google Scholar] [CrossRef] [PubMed]
- Szarpak, L.; Ruetzler, K.; Safiejko, K.; Hampel, M.; Pruc, M.; Kanczuga-Koda, L.; Filipiak, K.J.; Jaguszewski, M.J. Lactate dehydrogenase level as a COVID-19 severity marker. Am. J. Emerg. Med. 2021, 45, 638–639. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Wei, Y.; Lyu, X.; Zhao, B.; Feng, Y.; Li, T.; Cao, H.; Yang, X.; Zhou, X.; Wang, W.; et al. High aspartate aminotransferase to alanine aminotransferase ratio on admission as risk factor for poor prognosis in COVID-19 patients. Sci. Rep. 2020, 10, 16496. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, J.; Long, W.; Zeng, W.; Gao, R.; Zeng, G.; Chen, D.; Wang, S.; Li, Q.; Hu, D.; et al. Bilirubin Levels as Potential Indicators of Disease Severity in Coronavirus Disease Patients: A Retrospective Cohort Study. Front. Med. 2020, 7, 799. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, C.; Yang, Q.; Yuan, X.; Yang, F.; Li, P.; Chen, G.; Liang, W.; Yang, Y. The clinical implication of gamma-glutamyl transpeptidase in COVID-19. Liver Res. 2021, 5, 209–216. [Google Scholar] [CrossRef]
- Takeshita, Y.; Terada, J.; Hirasawa, Y.; Kinoshita, T.; Tajima, H.; Koshikawa, K.; Kinouchi, T.; Isaka, Y.; Shionoya, Y.; Fujikawa, A.; et al. Elevated TAT in COVID-19 Patients with Normal D-Dimer as a Predictor of Severe Respiratory Failure: A Retrospective Analysis of 797 Patients. J. Clin. Med. 2022, 11, 134. [Google Scholar] [CrossRef]
- Ponti, G.; Ruini, C.; Tomasi, A. Homocysteine as a potential predictor of cardiovascular risk in patients with COVID-19. Med. Hypotheses 2020, 143, 109859. [Google Scholar] [CrossRef]
- Pranata, R.; Huang, I.; Lukito, A.A.; Raharjo, S.B. Elevated N-terminal pro-brain natriuretic peptide is associated with increased mortality in patients with COVID-19: Systematic review and meta-analysis. Postgrad. Med. J. 2020, 96, 387–391. [Google Scholar] [CrossRef]
- Kim, C.W.; Aronow, W.S. COVID-19, cardiovascular diseases and cardiac troponins. Future Cardiol. 2022, 18, 135–142. [Google Scholar] [CrossRef]
- Akbar, M.R.; Pranata, R.; Wibowo, A.; Lim, M.A.; Sihite, T.A.; Martha, J.W. The prognostic value of elevated creatine kinase to predict poor outcome in patients with COVID-19—A systematic review and meta-analysis. Diabetes Metab. Syndr. 2021, 15, 529–534. [Google Scholar] [CrossRef]
- Zhu, F.; Li, W.; Lin, Q.; Xu, M.; Du, J.; Li, H. Myoglobin and troponin as prognostic factors in patients with COVID-19 pneumonia. Med. Clin. 2021, 157, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Sozio, E.; Tascini, C.; Fabris, M.; D’Aurizio, F.; De Carlo, C.; Graziano, E.; Bassi, F.; Sbrana, F.; Ripoli, A.; Pagotto, A.; et al. MR-proADM as prognostic factor of outcome in COVID-19 patients. Sci. Rep. 2021, 11, 5121. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Cai, X.; Wang, H.; He, G.; Lin, Y.; Lu, B.; Chen, C.; Pan, Y.; Hu, X. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta 2020, 507, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Tomar, B.; Anders, H.J.; Desai, J.; Mulay, S.R. Neutrophils and Neutrophil Extracellular Traps Drive Necroinflammation in COVID-19. Cells 2020, 9, 1383. [Google Scholar] [CrossRef]
- Lindsley, A.W.; Schwartz, J.T.; Rothenberg, M.E. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J. Allergy Clin. Immunol. 2020, 146, 122–129. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Tu, Y.-F.; Chien, C.-S.; Yarmishyn, A.A.; Lin, Y.-Y.; Luo, Y.-H.; Lin, Y.-T.; Lai, W.-Y.; Yang, D.-M.; Chou, S.-J.; Yang, Y.-P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef]
- Acharya, D.; Liu, G.; Gack, M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. [Google Scholar] [CrossRef]
- Hernandez, A.V.; Roman, Y.M.; Pasupuleti, V.; Barboza, J.J.; White, C.M. Update Alert: Hydroxychloroquine or Chloroquine for the Treatment or Prophylaxis of COVID-19. Ann. Intern. Med. 2020, 173, W78–W79. [Google Scholar] [CrossRef]
- Kupferschmidt, K. Big studies dim hopes for hydroxychloroquine. Science 2020, 368, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Saitz, R.; Schwitzer, G. Communicating Science in the Time of a Pandemic. JAMA 2020, 324, 443–444. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Braun, J.; Loyal, L.; Frentsch, M.; Wendisch, D.; Georg, P.; Kurth, F.; Hippenstiel, S.; Dingeldey, M.; Kruse, B.; Fauchere, F.; et al. Presence of SARS-CoV-2-reactive T cells in COVID-19 patients and healthy donors. medRxiv 2020. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Shen, C.; Wang, Z.; Zhao, F.; Yang, Y.; Li, J.; Yuan, J.; Wang, F.; Li, D.; Yang, M.; Xing, L.; et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 2020, 323, 1582–1589. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).