An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
2.2. Oligos and crRNA Preparation
2.3. Verification of LbuCas13a Activity
2.4. PCR-Cas13a Assay
2.5. Sensitivity and Specificity of the PCR-Cas13a Fluorescence Detection
2.6. Q-PCR Assay
2.7. Statistical Analysis
3. Results
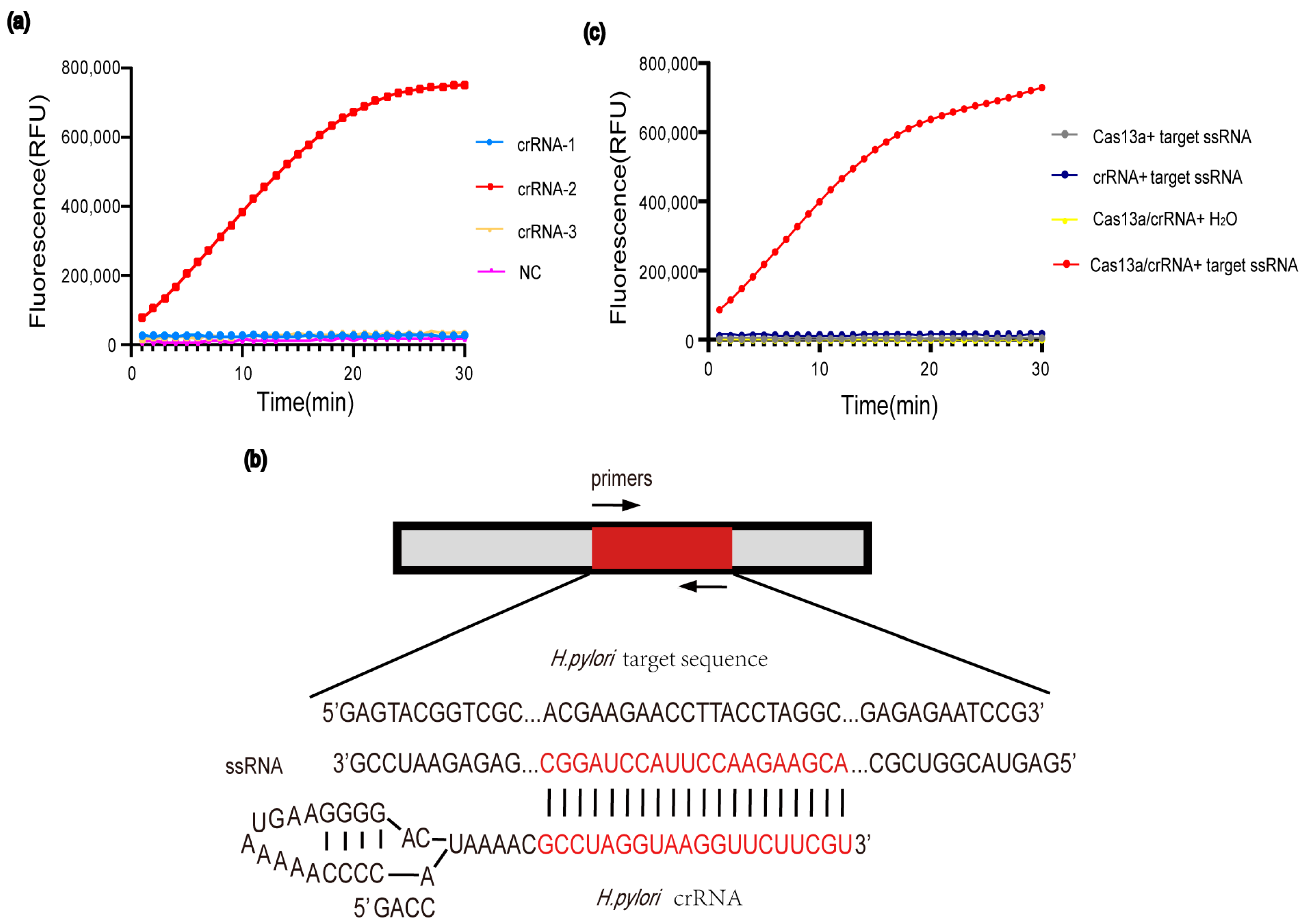
3.1. Optimization of the CRISPR-Cas13a Reaction
3.2. Establishment of the PCR-Cas13a Detection System
3.3. Evaluation of Sensitivity and Specificity of PCR-Cas13a on H. pylori
3.4. PCR-Cas13a Diagnostic Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dooley, C.; Cohen, H.; Fitzgibbons, P.; Bauer, M.; Appleman, M.; Perez-Perez, G.; Blaser, M.J. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N. Engl. J. Med. 1989, 321, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Li, Z.; Li, R.; Tan, P.; Zhang, R.; Li, J. mNGS in clinical microbiology laboratories: On the road to maturity. Crit. Rev. Microbiol. 2019, 45, 668–685. [Google Scholar] [CrossRef] [PubMed]
- Qasem, A.; Shaw, A.M.; Elkamel, E.; Naser, S.A. Coronavirus Disease 2019 (COVID-19) Diagnostic Tools: A Focus on Detection Technologies and Limitations. Curr. Issues Mol. Biol. 2021, 43, 728–748. [Google Scholar] [CrossRef]
- Maiti, B.; Anupama, K.P.; Rai, P.; Karunasagar, I.; Karunasagar, I. Isothermal amplification-based assays for rapid and sensitive detection of severe acute respiratory syndrome coronavirus 2: Opportunities and recent developments. Rev. Med. Virol. 2022, 32, e2274. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Diaz de Leon Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.; Abudayyeh, O.; Lee, J.; Essletzbichler, P.; Dy, A.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.; Freije, C.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.; Gootenberg, J.; Konermann, S.; Joung, J.; Slaymaker, I.; Cox, D.; Shmakov, S.; Makarova, K.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Zhou, X.; Huang, R.; Xing, D. High-Fidelity and Rapid Quantification of miRNA Combining crRNA Programmability and CRISPR/Cas13a trans-Cleavage Activity. Anal. Chem. 2019, 91, 5278–5285. [Google Scholar] [CrossRef]
- Sha, Y.; Huang, R.; Huang, M.; Yue, H.; Shan, Y.; Hu, J.; Xing, D. Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity. Chem. Commun. 2021, 57, 247–250. [Google Scholar] [CrossRef]
- Tian, T.; Shu, B.; Jiang, Y.; Ye, M.; Liu, L.; Guo, Z.; Han, Z.; Wang, Z.; Zhou, X. An Ultralocalized Cas13a Assay Enables Universal and Nucleic Acid Amplification-Free Single-Molecule RNA Diagnostics. ACS Nano 2021, 15, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Tian, T.; Sun, J.; Hu, M.; Wang, X.; Xiong, E.; Cheng, M.; Bao, Y.; Lin, W.; Jiang, J.; et al. Universal and Naked-Eye Gene Detection Platform Based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas12a/13a System. Anal. Chem. 2020, 92, 4029–4037. [Google Scholar] [CrossRef]
- Zhou, T.; Huang, M.; Lin, J.; Huang, R.; Xing, D. High-Fidelity CRISPR/Cas13a-Cleavage-Triggered Rolling Circle Amplified DNAzyme for Visual Profiling of MicroRNA. Anal Chem. 2021, 93, 2038–2044. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Qiu, Z.; Jiang, Y.; Zhu, D.; Zhou, X. Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 2022, 196, 113701. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, X.; Shan, Y.; Yue, H.; Huang, R.; Hu, J.; Xing, D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, T.; Okuda, M.; Hashiguchi, K.; Imamura, I.; Nakayama, A.; Matsuo, M. Evaluation of a Novel Stool Antigen Rapid Test Kit for Detection of Helicobacter pylori Infection. J. Clin. Microbiol. 2019, 57, e01825-18. [Google Scholar] [CrossRef]
- Kobayashi, D.; Eishi, Y.; Ohkusa, T.; Ishige, I.; Suzuki, T.; Minami, J.; Yamada, T.; Takizawa, T.; Koike, M. Gastric mucosal density of Helicobacter pylori estimated by real-time PCR compared with results of urea breath test and histological grading. J. Med. Microbiol. 2002, 51, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.; Warren, J. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984, 323, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Hooi, J.; Lai, W.; Ng, W.; Suen, M.; Underwood, F.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.; Wong, V.; Wu, J.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Yeh, J.; Kuntz, K.; Ezzati, M.; Goldie, S. Exploring the cost-effectiveness of Helicobacter pylori screening to prevent gastric cancer in China in anticipation of clinical trial results. Int. J. Cancer 2009, 124, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ndip, R.; MacKay, W.; Farthing, M.; Weaver, L. Culturing Helicobacter pylori from clinical specimens: Review of microbiologic methods. J. Pediatric Gastroenterol. Nutr. 2003, 36, 616–622. [Google Scholar] [CrossRef]
- MacOni, G.; Vago, L.; Galletta, G.; Imbesi, V.; Sangaletti, O.; Parente, F.; Cucino, C.; Bonetto, S.; Bianchi Porro, G. therapeutics. Is routine histological evaluation an accurate test for Helicobacter pylori infection? Aliment. Pharmacol. Ther. 1999, 13, 327–331. [Google Scholar] [CrossRef]
- Tseng, C.A.; Wang, W.M.; Wu, D.C. Comparison of the clinical feasibility of three rapid urease tests in the diagnosis of Helicobacter pylori infection. Dig. Dis. Sci. 2005, 50, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, B.; Stoller, M. Nonsurgical management of infection-related renal calculi. Urol. Clin. N. Am. 1999, 26, 765–778, viii. [Google Scholar] [CrossRef] [PubMed]
- Herbrink, P.; van Doorn, L.J. Serological methods for diagnosis of Helicobacter pylori infection and monitoring of eradication therapy. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Hirschl, A.; Rotter, M. Serological tests for monitoring Helicobacter pylori eradication treatment. J. Gastroenterol. 1996, 31, 33–36. [Google Scholar] [PubMed]
- Ramis, I.B.; de Moraes, E.P.; Fernandes, M.S.; Mendoza-Sassi, R.; Rodrigues, O.; Juliano, C.R.; Scaini, C.J.; da Silva, P.E. Evaluation of diagnostic methods for the detection of Helicobacter pylori in gastric biopsy specimens of dyspeptic patients. Braz. J. Microbiol. 2012, 43, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Chen, B.; Pan, H.; Wang, D.; Liu, M.; Yang, Y.; Zou, M.; Yang, J.; Xiao, K.; Zhao, R.; et al. Detection of Microbial 16S rRNA Gene in the Serum of Patients With Gastric Cancer. Front. Oncol. 2019, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Kisa, O.; Albay, A.; Mas, M.R.; Celasun, B.; Doganci, L. The evaluation of diagnostic methods for the detection of Helicobacter pylori in gastric biopsy specimens. Diagn. Microbiol. Infect. Dis. 2002, 43, 251–255. [Google Scholar] [CrossRef]
- Pohl, D.; Keller, P.M.; Bordier, V.; Wagner, K. Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J. Gastroenterol. 2019, 25, 4629–4660. [Google Scholar] [CrossRef]
- Redondo, J.; Keller, P.; Zbinden, R.; Wagner, K. A novel RT-PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn. Microbiol. Infect. Dis. 2018, 90, 1–6. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Hirschl, A.; Dragosics, B.; Hufnagl, P.; Puz, S.; Kovách, Z.; Rotter, M.; Makristathis, A. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J. Clin. Microbiol. 2004, 42, 4512–4518. [Google Scholar] [CrossRef]
- Lottspeich, C.; Schwarzer, A.; Panthel, K.; Koletzko, S.; Rüssmann, H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J. Clin. Microbiol. 2007, 45, 1718–1722. [Google Scholar] [CrossRef] [PubMed]
- Vécsei, A.; Innerhofer, A.; Binder, C.; Gizci, H.; Hammer, K.; Bruckdorfer, A.; Riedl, S.; Gadner, H.; Hirschl, A.; Makristathis, A. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin. Gastroenterol. Hepatol. 2010, 8, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Scaletsky, I.; Aranda, K.; Garcia, G.; Gonçalves, M.; Cardoso, S.; Iriya, K.; Silva, N. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter 2011, 16, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. Engl. 2019, 58, 17399–17405. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, S.X.; Wang, F.; Zeng, M.S. Room Temperature Detection of Plasma Epstein-Barr Virus DNA with CRISPR-Cas13. Clin. Chem. 2019, 65, 591–592. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.; Alcantar, M.; Lape, I.; Greensmith, R.; Huske, A.; Valeri, J.; Marty, F.; Klämbt, V.; Azzi, J.; Akalin, E.; et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 2020, 4, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.W.; Zhou, X.; Xu, T.; Yang, M.; Chen, Y.; He, G.Q.; Pan, N.; Cai, Y.; Li, Y.; Wang, X.; et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019, 8, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Qiu, E.; Jin, S.; Xiao, Z.; Chen, Q.; Wang, Q.; Liu, H.; Xie, C.; Chen, C.; Li, Z.; Han, S. CRISPR-based detection of Helicobacter pylori in stool samples. Helicobacter 2021, 26, e12828. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, L.; Dong, Y.; Peng, L.; Liu, G.; Man, S.; Ma, L. CRISPR-Cas13a based bacterial detection platform: Sensing pathogen Staphylococcus aureus in food samples. Anal. Chim. Acta. 2020, 1127, 225–233. [Google Scholar] [CrossRef]
- Chandrasekaran, S.S.; Agrawal, S.; Fanton, A.; Jangid, A.R.; Charrez, B.; Escajeda, A.M.; Son, S.; McIntosh, R.; Tran, H.; Bhuiya, A.; et al. Rapid detection of SARS-CoV-2 RNA in saliva via Cas13. Nat. Biomed. Eng. 2022, 6, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Munawar, M.A. Critical insight into recombinase polymerase amplification technology. Expert Rev. Mol. Diagn. 2022, 22, 725–737. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Wang, J.P.; Osato, M.; Lachman, L.B. Real-time quantitative PCR for detection of Helicobacter pylori. J. Clin. Microbiol. 2002, 40, 3720–3728. [Google Scholar] [CrossRef] [PubMed]
- Tram, K.; Kanda, P.; Salena, B.J.; Huan, S.; Li, Y. Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem. Int. Ed. Engl. 2014, 53, 12799–12802. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Kellner, M.J.; Zhang, F. Nucleic Acid Detection of Plant Genes Using CRISPR-Cas13. CRISPR J. 2019, 2, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, e1905311. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence (5′-3′) |
|---|---|
| PCR-Cas13a | |
| T7-F1 | TAATACGACTCACTATAGGGGAGTACGGTCGCAAGATTA |
| R1 | CGGATTCTCTCAATGTCAAG |
| crRNA-1 | GACCACCCCAAAAAUGAAGGGGACUAAAACUCUCAAUGUCAAGCCUAGGUAAGG |
| crRNA-2 | GACCACCCCAAAAAUGAAGGGGACUAAAACGCCUAGGUAAGGUUCUUCGU |
| crRNA-3 | GACCACCCCAAAAAUGAAGGGGACUAAAACCAAGCCUAGGUAAGGUUCUUCGUG |
| RNA reporter | FAM-UUUUUU-BHQ1 |
| q-PCR [25] | |
| F2 | CTCATTGCGAAGGCGACCT |
| R2 | TCTAATCCTGTTTGCTCCCCA |
| Probe | FAM-ATTACTGACGCTGATTGCGCGAAAGC-TAMRA |
| Clinical Diagnosis | Total | Performance Characteristics | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity | Specificity | PPV * | NPV * | k | |||
| PCR-Cas13a | Positive | 48 | 0 | 48 | 98.0% | 100% | 0.967 | ||
| Negative | 1 | 22 | 23 | 100% | 95.7% | ||||
| Total | 49 | 22 | 71 | ||||||
| q-PCR | Positive | 46 | 0 | 46 | 93.9% | 100% | 0.905 | ||
| Negative | 3 | 22 | 25 | 100% | 88.0% | ||||
| Total | 49 | 22 | 71 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, L.; Liu, X.; Wu, K.; Zhu, X.; Ma, L.; Su, J. An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori. J. Pers. Med. 2022, 12, 2082. https://doi.org/10.3390/jpm12122082
Wang Y, Liu L, Liu X, Wu K, Zhu X, Ma L, Su J. An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori. Journal of Personalized Medicine. 2022; 12(12):2082. https://doi.org/10.3390/jpm12122082
Chicago/Turabian StyleWang, Yaxuan, Liyang Liu, Xiaochuan Liu, Kai Wu, Xiaoyan Zhu, Liyan Ma, and Jianrong Su. 2022. "An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori" Journal of Personalized Medicine 12, no. 12: 2082. https://doi.org/10.3390/jpm12122082
APA StyleWang, Y., Liu, L., Liu, X., Wu, K., Zhu, X., Ma, L., & Su, J. (2022). An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori. Journal of Personalized Medicine, 12(12), 2082. https://doi.org/10.3390/jpm12122082





