Investigating Psychological Impact after Receiving Genetic Risk Results—A Survey of Participants in a Population Genomic Screening Program
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting
2.2. Participants
2.3. Procedures
2.4. Measures
2.5. Analyses
3. Results
3.1. Overall Sample Characteristics
3.2. FACToR Scale
3.2.1. Positive Subscale
3.2.2. Negative Subscale
3.2.3. Privacy Subscale
3.2.4. Uncertainty Subscale
3.3. PANAS
3.3.1. Positive Affect
3.3.2. Negative Affect
3.3.3. Discrete Emotions
Decision Regret
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abul-Husn, N.S.; Manickam, K.; Jones, L.K.; Wright, E.A.; Hartzel, D.N.; Gonzaga-Jauregui, C.; O’Dushlaine, C.; Leader, J.B.; Kirchner, H.L.; Lindbuchler, D.M.; et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science 2016, 354, aaf7000. [Google Scholar] [CrossRef] [PubMed]
- Grzymski, J.J.; Elhanan, G.; Rosado, J.A.M.; Smith, E.; Schlauch, K.A.; Read, R.; Rowan, C.; Slotnick, N.; Dabe, S.; Metcalf, W.J.; et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 2020, 26, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Manickam, K.; Buchanan, A.H.; Schwartz, M.L.B.; Hallquist, M.L.G.; Williams, J.L.; Rahm, A.K.; Rocha, H.; Savatt, J.M.; Evans, A.E.; Butry, L.M.; et al. Exome Sequencing–Based Screening for BRCA1/2 Expected Pathogenic Variants Among Adult Biobank Participants. JAMA Netw. Open 2018, 1, e182140. [Google Scholar] [CrossRef] [PubMed]
- David, S.P.; Dunnenberger, H.M.; Ali, R.; Matsil, A.; Lemke, A.A.; Singh, L.; Zimmer, A.; Hulick, P.J. Implementing Primary Care Mediated Population Genetic Screening Within an Integrated Health System. J. Am. Board Fam. Med. 2021, 34, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.K.; Strande, N.T.; Calvo, E.M.; Chen, J.; Rodriguez, G.; McCormick, C.Z.; Hallquist, M.L.G.; Savatt, J.M.; Rocha, H.; Williams, M.S.; et al. A RE-AIM Framework Analysis of DNA-Based Population Screening: Using Implementation Science to Translate Research Into Practice in a Healthcare System. Front. Genet. 2022, 13, 883073. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, A.H.; Kirchner, H.L.; Schwartz, M.L.B.; Kelly, M.A.; Schmidlen, T.; Jones, L.K.; Hallquist, M.L.G.; Rocha, H.; Betts, M.; Schwiter, R.; et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet. Med. 2020, 22, 1874–1882. [Google Scholar] [CrossRef]
- Buchanan, A.H.; Manickam, K.; Meyer, M.N.; Wagner, J.K.; Hallquist, M.L.G.; Williams, J.L.; Rahm, A.K.; Williams, M.S.; Chen, Z.-M.E.; Shah, C.K.; et al. Correction to: Early cancer diagnoses through BRCA1/2 screening of unselected adult biobank participants. Genet. Med. 2018, 20, 554–558. Genet. Med. 2021, 23, 2470. [Google Scholar] [CrossRef]
- Murray, M.F.; Giovanni, M.A.; Doyle, D.L.; Harrison, S.M.; Lyon, E.; Manickam, K.; Monaghan, K.G.; Rasmussen, S.A.; Scheuner, M.T.; Palomaki, G.E.; et al. DNA-based screening and population health: A points to consider statement for programs and sponsoring organizations from the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Linderman, M.D.; Nielsen, D.E.; Green, R.C. Personal Genome Sequencing in Ostensibly Healthy Individuals and the PeopleSeq Consortium. J. Pers. Med. 2016, 6, 14. [Google Scholar] [CrossRef]
- The All of Us Research Program Investigators The “All of Us” Research Program. N. Engl. J. Med. 2019, 381, 668–676. [CrossRef]
- Venner, E.; Muzny, D.; Smith, J.D.; Walker, K.; Neben, C.L.; Lockwood, C.M.; Empey, P.E.; Metcalf, G.A.; Kachulis, C.; Mian, S.; et al. Whole-genome sequencing as an investigational device for return of hereditary disease risk and pharmacogenomic results as part of the All of Us Research Program. Genome Med. 2022, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Foss, K.S.; O’Daniel, J.M.; Berg, J.S.; Powell, S.N.; Cadigan, R.J.; Kuczynski, K.J.; Milko, L.V.; Saylor, K.W.; Roberts, M.; Weck, K.; et al. The Rise of Population Genomic Screening: Characteristics of Current Programs and the Need for Evidence Regarding Optimal Implementation. J. Pers. Med. 2022, 12, 692. [Google Scholar] [CrossRef]
- Williams, M.S. Population Screening in Health Systems. Annu. Rev. Genom. Hum. Genet. 2022, 23, 549–567. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.G.; Judge, D.P.; Levin, E.; Sterba, K.; Hunt, K.; Ramos, P.S.; Melvin, C.; Wager, K.; Catchpole, K.; Clinton, C.; et al. A pragmatic implementation research study for In Our DNA SC: A protocol to identify multi-level factors that support the implementation of a population-wide genomic screening initiative in diverse populations. Implement. Sci. Commun. 2022, 3, 48. [Google Scholar] [CrossRef] [PubMed]
- Lacaze, P.A.; Tiller, J.; Winship, I.; Brotchie, A.; McNeil, J.; Zalcberg, J.; Thomas, D.; Milne, R.; James, P.; Delatycki, M.; et al. Population DNA screening for medically actionable disease risk in adults. Med. J. Aust. 2022, 216, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Leitsalu, L.; Palover, M.; Sikka, T.T.; Reigo, A.; Kals, M.; Pärn, K.; Nikopensius, T.; Esko, T.; Metspalu, A.; Padrik, P.; et al. Genotype-first approach to the detection of hereditary breast and ovarian cancer risk, and effects of risk disclosure to biobank participants. Eur. J. Hum. Genet. 2020, 29, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Haverfield, E.V.; Esplin, E.D.; Aguilar, S.J.; Hatchell, K.E.; Ormond, K.E.; Hanson-Kahn, A.; Atwal, P.S.; Macklin-Mantia, S.; Hines, S.; Sak, C.W.M.; et al. Correction to: Physician-directed genetic screening to evaluate personal risk for medically actionable disorders: A large multi-center cohort study. BMC Med. 2021, 19, 288. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Tier 1 Genomics Applications and Their Importance to Public Health. 2014. Available online: https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm (accessed on 22 September 2022).
- Guzauskas, G.F.; Garbett, S.; Zhou, Z.; Spencer, S.J.; Smith, H.S.; Hao, J.; Hassen, D.; Snyder, S.R.; Graves, J.A.; Peterson, J.F.; et al. Cost-effectiveness of Population-Wide Genomic Screening for Hereditary Breast and Ovarian Cancer in the United States. JAMA Netw. Open 2020, 3, e2022874. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.J.; Feero, W.G.; Chambers, D.A.; Brody, L.C.; Aziz, N.; Green, R.C.; Janssens, A.C.J.W.; Murray, M.F.; Rodriguez, L.L.; Rutter, J.L.; et al. Correction: A collaborative translational research framework for evaluating and implementing the appropriate use of human genome sequencing to improve health. PLOS Med. 2018, 15, e1002650. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.F.; Evans, J.P.; Khoury, M.J. DNA-Based Population Screening: Potential Suitability and Important Knowledge Gaps. JAMA 2020, 323, 307–308. [Google Scholar] [CrossRef]
- Peterson, J.F.; Roden, D.M.; Orlando, L.A.; Ramirez, A.H.; Mensah, G.A.; Williams, M.S. Building evidence and measuring clinical outcomes for genomic medicine. Lancet 2019, 394, 604–610. [Google Scholar] [CrossRef]
- Mackley, M.P.; Fletcher, B.; Parker, M.; Watkins, H.; Ormondroyd, E. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: A systematic review of quantitative and qualitative studies. Anesthesia Analg. 2017, 19, 283–293. [Google Scholar] [CrossRef]
- de Wert, G.; Hentze, S.; Dondorp, W.; Clarke, A.; Dequeker, E.M.C.; Cordier, C.; Deans, Z.; van El, C.G.; Fellmann, F.; Hastings, R.; et al. Opportunistic genomic screening. Recommendations of the European Society of Human Genetics. Eur. J. Hum. Genet. 2021, 29, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Tolwinski, K.; Atondo, J.; Davis, F.D.; Goehringer, J.; Jones, L.K.; Pisieczko, C.J.; Sturm, A.C.; Williams, J.L.; Williams, M.S.; et al. Understanding the Patient Experience of Receiving Clinically Actionable Genetic Results from the MyCode Community Health Initiative, a Population-Based Genomic Screening Initiative. J. Pers. Med. 2022, 12, 1511. [Google Scholar] [CrossRef] [PubMed]
- Halverson, C.M.E.; Jones, S.H.; Novak, L.; Simpson, C.; Edwards, D.R.V.; Zhao, S.K.; Clayton, E.W. What Results Should Be Returned from Opportunistic Screening in Translational Research? J. Pers. Med. 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Lemke, A.A.; Amendola, L.M.; Thompson, J.; Dunnenberger, H.M.; Kuchta, K.; Wang, C.; Dilzell-Yu, K.; Hulick, P.J. Patient-Reported Outcomes and Experiences with Population Genetic Testing Offered Through a Primary Care Network. Genet. Test. Mol. Biomark. 2021, 25, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, E.A.; Hesse, C. Investigating Uncertainty and Emotions in Conversations About Family Health History: A Test of the Theory of Motivated Information Management. J. Health Commun. 2014, 19, 939–954. [Google Scholar] [CrossRef]
- Dean, M.; Fisher, C.L. Uncertainty and previvors’ cancer risk management: Understanding the decision-making process. J. Appl. Commun. Res. 2019, 47, 460–483. [Google Scholar] [CrossRef]
- Carey, D.J.; Fetterolf, S.N.; Davis, F.D.; Faucett, W.A.; Kirchner, H.L.; Mirshahi, U.; Murray, M.F.; Smelser, D.T.; Gerhard, G.S.; Ledbetter, D.H. The Geisinger MyCode community health initiative: An electronic health record–linked biobank for precision medicine research. Genet. Med. 2016, 18, 906–913. [Google Scholar] [CrossRef]
- Schwartz, M.L.; McCormick, C.Z.; Lazzeri, A.L.; Lindbuchler, D.M.; Hallquist, M.L.; Manickam, K.; Buchanan, A.H.; Rahm, A.K.; Giovanni, M.A.; Frisbie, L.; et al. A Model for Genome-First Care: Returning Secondary Genomic Findings to Participants and Their Healthcare Providers in a Large Research Cohort. Am. J. Hum. Genet. 2018, 103, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Faucett, W.A.; Davis, F.D. How Geisinger made the case for an institutional duty to return genomic results to biobank participants. Appl. Transl. Genom. 2016, 8, 33–35. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bennette, C.S.; Amendola, L.M.; Hart, M.R.; Heagerty, P.; Comstock, B.; Tarczy-Hornoch, P.; Fullerton, S.M.; Regier, D.A.; Burke, W.; et al. The Feelings About genomiC Testing Results (FACToR) Questionnaire: Development and Preliminary Validation. J. Genet. Couns. 2018, 28, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.; Clark, L.A.; Tellegen, A. Development and validation of brief measures of positive and negative affect: The PANAS scales. J. Pers. Soc. Psychol. 1988, 54, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Brehaut, J.C.; O’Connor, A.M.; Wood, T.; Hack, T.; Siminoff, L.; Gordon, E.; Feldman-Stewart, D. Validation of a Decision Regret Scale. Med. Decis. Mak. 2003, 23, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Zawatsky, C.L.B.; Shah, N.; Machini, K.; Perez, E.; Christensen, K.D.; Zouk, H.; Steeves, M.; Koch, C.; Uveges, M.; Shea, J.; et al. Returning actionable genomic results in a research biobank: Analytic validity, clinical implementation, and resource utilization. Am. J. Hum. Genet. 2021, 108, 2224–2237. [Google Scholar] [CrossRef]
- Beil, A.; Hornsby, W.; Uhlmann, W.R.; Aatre, R.; Arscott, P.; Wolford, B.; Eagle, K.A.; Yang, B.; McNamara, J.; Willer, C.; et al. Disclosure of clinically actionable genetic variants to thoracic aortic dissection biobank participants. BMC Med. Genom. 2021, 14, 66. [Google Scholar] [CrossRef]
- Zoltick, E.S.; Linderman, M.D.; McGinniss, M.A.; Ramos, E.; Ball, M.; Church, G.M.; Leonard, D.G.B.; Pereira, S.; McGuire, A.L.; Caskey, C.T.; et al. Predispositional genome sequencing in healthy adults: Design, participant characteristics, and early outcomes of the PeopleSeq Consortium. Genome Med. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Sapp, J.C.; Facio, F.M.; Cooper, D.; Lewis, K.L.; Modlin, E.; van der Wees, P.; Biesecker, L.G. A systematic literature review of disclosure practices and reported outcomes for medically actionable genomic secondary findings. Genet. Med. 2021, 23, 2260–2269. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, R.S. Cognition and motivation in emotion. Am. Psychol. 1991, 46, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Davidson, L.G. Previvors’ Uncertainty Management Strategies for Hereditary Breast and Ovarian Cancer. Health Commun. 2016, 33, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Seymour, K.C.; Addington-Hall, J.; Lucassen, A.M.; Foster, C.L. What Facilitates or Impedes Family Communication Following Genetic Testing for Cancer Risk? A Systematic Review and Meta-Synthesis of Primary Qualitative Research. J. Genet. Couns. 2010, 19, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Forrest, K.; Simpson, S.; Wilson, B.; Van Teijlingen, E.; McKee, L.; Haites, N.; Matthews, E. To tell or not to tell: Barriers and facilitators in family communication about genetic risk. Clin. Genet. 2003, 64, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Hardcastle, S.J.; Legge, E.; Laundy, C.S.; Egan, S.J.; French, R.; Watts, G.; Hagger, M.S. Patients’ Perceptions and Experiences of Familial Hypercholesterolemia, Cascade Genetic Screening and Treatment. Int. J. Behav. Med. 2014, 22, 92–100. [Google Scholar] [CrossRef] [PubMed]


| Total (n = 354) | |
|---|---|
| Age at result receipt | |
| Mean (SD) | 57.6 (15.72) |
| Median (IQR) | 60.6 (47.1, 69.6) |
| Range | 21.7, 89.5 |
| Sex, n (%) | |
| Female | 221 (62.6%) |
| Male | 132 (37.4%) |
| Missing | 1 |
| Race, n (%) | |
| White | 310 (87.6%) |
| Asian | 2 (0.6%) |
| Black Or African American | 2 (0.6%) |
| Native Hawaiian or Other Pacific Islander | 1 (0.3%) |
| Unknown | 39 (11.0%) |
| Ethnicity, n (%) | |
| Hispanic or Latino | 5 (1.4%) |
| Not Hispanic or Latino | 310 (87.6%) |
| Unknown | 39 (11.0%) |
| What is your current marital status, n (%) | |
| Married/Living with partner | 235 (66.8%) |
| Divorced/Separated | 43 (12.2%) |
| Widowed | 34 (9.7%) |
| Never Married | 40 (11.4%) |
| Missing | 2 |
| What is the highest grade or year of school you completed, n (%) | |
| Less than high school | 15 (4.3%) |
| Completed high school/GED | 90 (25.6%) |
| Some College | 119 (33.8%) |
| College grad or Advanced degree | 128 (36.4%) |
| Missing | 2 |
| What is your annual HOUSEHOLD income from all sources, n (%) | |
| Less than $49,999 | 103 (29.1%) |
| $50,000–$99,999 | 104 (29.4%) |
| $100,000 or more | 69 (19.5%) |
| I prefer not to answer | 78 (22.0%) |
| Have you ever been told that you were at risk for a genetic condition before getting a call from the My Code team? (6 weeks only) n (%) | |
| Yes | 61 (21.9%) |
| No | 217 (78.1%) |
| Missing | 76 |
| Has anyone in your family been told of a risk for THIS genetic condition? (6 weeks only) n (%) | |
| Yes | 95 (34.2%) |
| No | 176 (63.3%) |
| Don’t know | 7 (2.5%) |
| Missing | 76 |
| How would you say your HEALTH is? (6 months), n (%) | |
| Mean (SD) | 2.8 (1.00) |
| Median (IQR) | 3.0 (2.0, 3.0) |
| How would you say your QUALITY OF LIFE is? (6 months), n (%) | |
| Mean (SD) | 2.3 (0.97) |
| Median (IQR) | 2.0 (2.0, 3.0) |
| Gene group, n (%) | |
| Cancer | 157 (44.4%) |
| Cardiac | 116 (32.8%) |
| Miscellaneous phenotype (HFE, SMAD4, FBN1, TGFBR1, TGFBR2, SMAD3, OTC, PTEN, TSC1, TSC2, COL3A1, ATP7B, RYR1, GLA) | 81 (22.9%) |
| Completed GC visits, n (%) | 182 (51.4%) |
| Before T1 | 166 (91.2%) |
| Between T1-T2 | 16 (8.8%) |
| 6 Weeks (n = 354) | 6 Months (n = 354) | p-Value | Adjusted p-Value | |
|---|---|---|---|---|
| FACToR scales | ||||
| Positive subscale (0–16) | ||||
| Mean (SD) | 8.5 (3.56) | 8.9 (3.70) | 0.2422 | |
| Range | 0.0, 16.0 | 0.0, 16.0 | ||
| Negative subscale (0–12) | ||||
| Mean (SD) | 2.9 (3.22) | 2.0 (3.02) | 0.0004 | (Estimate (SE): −0.82 (0.140), p-value < 0.0001) * |
| Range | 0.0, 12.0 | 0.0, 12.0 | ||
| Privacy subscale (0–8) | ||||
| Mean (SD) | 0.6 (1.34) | 0.7 (1.52) | 0.4315 | |
| Range | 0.0, 8.0 | 0.0, 8.0 | ||
| Uncertain subscale (0–8) | ||||
| Mean (SD) | 2.9 (2.25) | 2.5 (2.27) | 0.0126 | (Estimate (SE): −0.42 (0.136), p-value = 0.0020) ** |
| Range | 0.0, 8.0 | 0.0, 8.0 | ||
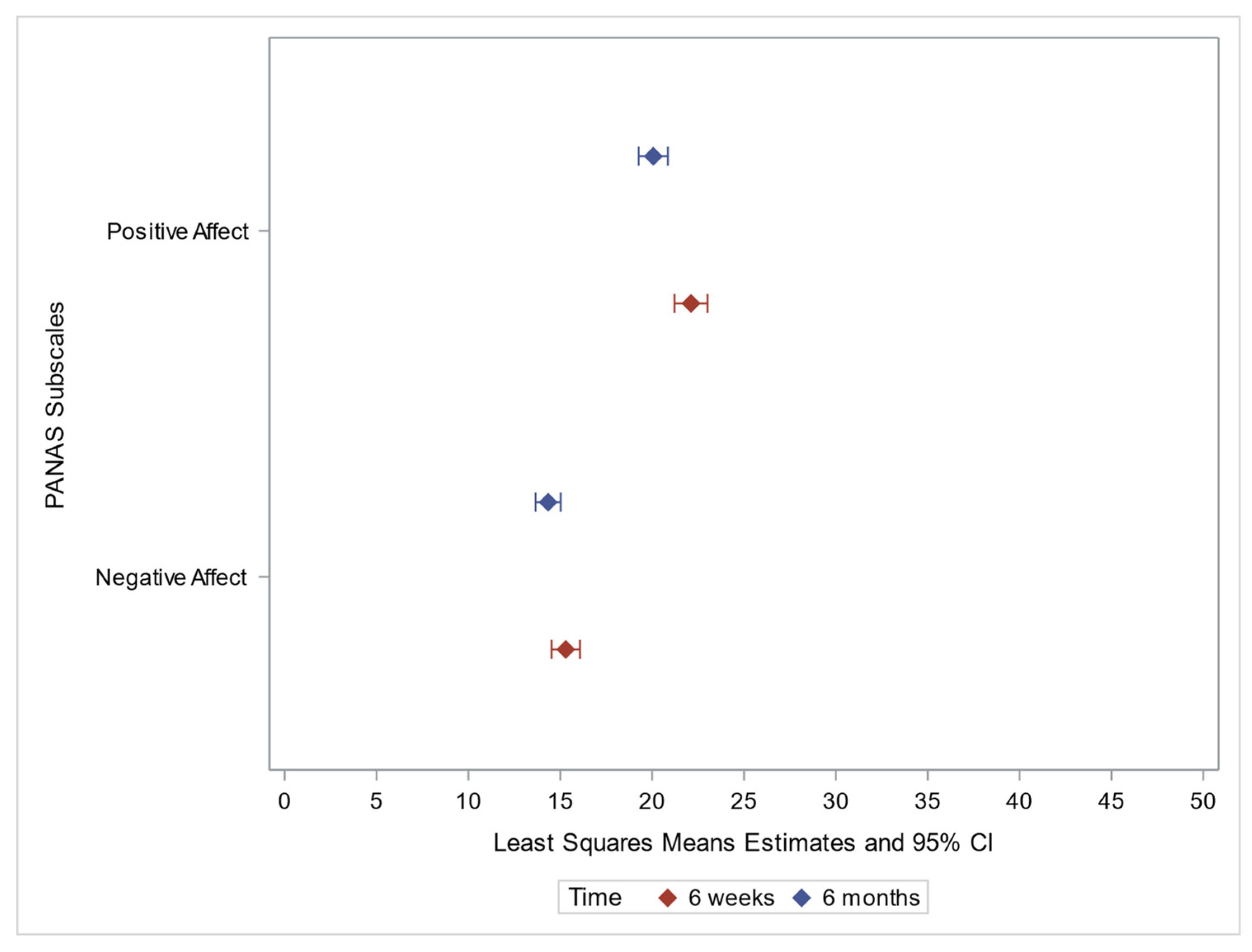
| PANAS Scale | ||||
| Positive Affect (0–50) | ||||
| Mean (SD) | 22.1 (7.91) | 20.1 (7.38) | 0.0008 | (Estimate (SE): −2.15 (0.449), p-value < 0.0001) *** |
| Range | 10.0, 49.0 | 10.0, 44.0 | ||
| Negative Affect (0–50) | ||||
| Mean (SD) | 15.3 (6.72) | 14.3 (6.42) | 0.0681 | |
| Range | 10.0, 47.0 | 6.0, 47.0 | ||
| Decision Regret Scale (0–100) | ||||
| Mean (SD) | 12.4 (12.84) | 11.0 (13.59) | 0.1640 | |
| Range | 0.0, 90.0 | 0.0, 75.0 |
| Six Weeks | Six Months | |||||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Privacy | Uncertain | Positive | Negative | Privacy | Uncertain | |
| p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | |
| GC visit | 0.1865 | 0.0092 | 0.0827 | 0.2183 | 0.5406 | 0.0702 | 0.0636 | 0.3974 |
| Yes | −0.50 (0.378) | 0.89 (0.340) | 0.25 (0.142) | 0.30 (0.239) | −0.24 (0.394) | 0.58 (0.320) | 0.30 (0.161) | −0.20 (0.241) |
| No (ref) | - | - | - | - | - | - | - | |
| Age at result receipt | <0.0001 | 0.0855 | 0.1555 | 0.9987 | 0.0004 | 0.0028 | 0.0015 | 0.4419 |
| 0.05 (0.012) | −0.02 (0.011) | −0.01 (0.004) | −0.00001 (0.007) | 0.04 (0.012) | −0.03 (0.010) | −0.02 (0.005) | −0.006 (0.008) | |
| Sex | 0.0007 | 0.0061 | 0.1348 | 0.0235 | 0.0222 | 0.0030 | 0.0551 | 0.3098 |
| Female | −1.32 (0.386) | 0.97 (0.351) | 0.22 (0.147) | 0.56 (0.245) | −0.93 (0.405) | 0.98 (0.329) | 0.32 (0.166) | 0.25 (0.250) |
| Male (ref) | - | - | - | - | - | - | - | - |
| Highest Education | 0.9173 | 0.5677 | 0.3485 | 0.0141 | 0.1780 | 0.1598 | 0.0070 | 0.0004 |
| Less than high school | −0.16 (0.997) | 0.76 (0.902) | −0.10 (0.375) | 0.0642 1.16 (0.622) | −1.42 (1.029) | 1.58 (0.837) | 0.0367 0.88 (0.418) | 0.9141 0.07 (0.618) |
| Completed high school/GED (ref) | - | - | - | - | - | - | - | - |
| Some College | −0.08 (0.499) | −0.38 (0.452) | −0.32 (0.188) | 0.9508 −0.02 (0.312) | −0.07 (0.515) | −0.18 (0.419) | 0.1120 −0.33 (0.210) | 0.0011 −1.02 (0.309) |
| College grad/Advanced degree | 0.21 (0.492) | −0.23 (0.445) | −0.26 (0.185) | 0.0582 −0.58 (0.307) | 0.57 (0.508) | −0.25 (0.413) | 0.0590 −0.39 (0.206) | 0.0002 −1.16 (0.305) |
| Has anyone in your family been told of a risk for THIS genetic condition? | 0.0016 | 0.8142 | 0.5661 | 0.7981 | N/A | N/A | N/A | N/A |
| Yes | 0.0003 −1.60 (0.443) | 0.26 (0.414) | 0.18 (0.173) | 0.01 (0.294) | N/A | N/A | N/A | N/A |
| No (ref) | - | - | - | - | N/A | N/A | N/A | N/A |
| Don’t know | 0.8292−0.29 (1.342) | 0.02 (1.254) | −0.01 (0.524) | −0.59 (0.889) | N/A | N/A | N/A | N/A |
| Gene group | 0.0120 | 0.0044 | 0.1044 | 0.4717 | 0.5772 | 0.0001 | 0.5358 | 0.1417 |
| Cancer (ref) | - | - | - | - | - | - | - | - |
| Cardio | 0.0358 | 0.0945 | 0.0031 | |||||
| 0.91 (0.431) | −0.65 (0.390) | −0.24 (0.164) | 0.23 (0.276) | 0.16 (0.453) | −1.08 (0.362) | −0.04 (0.186) | −0.36 (0.277) | |
| Miscellaneous | 0.2451 | 0.0011 | <0.0001 | |||||
| −0.56 (0.482) | −1.43 (0.435) | −0.36 (0.183) | −0.16 (0.308) | −0.40 (0.507) | −1.60 (0.404) | −0.23 (0.208) | −0.58 (0.309) | |
| Six Weeks | Six Months | |||||
|---|---|---|---|---|---|---|
| PANAS Positive Affect | PANAS Negative Affect | Decision Regret | PANAS Positive Affect | PANAS Negative Affect | Decision Regret | |
| p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | p-Value Est (SE) | |
| GC visit | 0.0107 | 0.0154 | 0.1941 | 0.3023 | 0.0499 | 0.0552 |
| Yes | 2.52 (0.983) | 2.04 (0.836) | −1.77 (1.364) | 0.81 (0.785) | 1.34 (0.680) | −2.77 (1.440) |
| No (ref) | - | - | - | - | - | - |
| Age at result receipt | 0.1554 −0.04 (0.030) | 0.0440 −0.05 (0.025) | 0.0223 0.10 (0.043) | 0.6798 −0.01 (0.025) | 0.0033 −0.06 (0.022) | 0.2286 0.06 (0.046) |
| Sex | 0.1880 | 0.0005 | 0.0032 | 0.1188 | 0.0003 | 0.1051 |
| Female | 1.28 (0.966) | 2.88 (0.811) | −4.14 (1.397) | 1.27 (0.811) | 2.53 (0.695) | −2.42 (1.492) |
| Male (ref) | - | - | - | - | - | - |
| Highest Education | 0.9281 | 0.3638 | 0.0039 | 0.4707 | 0.2560 | 0.0255 |
| Less than high school | −1.00 (2.810) | 0.79 (2.389) | 0.1774 4.78 (3.535) | −0.57 (2.046) | 1.91 (1.780) | 0.0130 9.39 (3.762) |
| Completed high school/GED (ref) | - | - | - | - | - | - |
| Some College | −0.18 (1.264) | −1.32 (1.074) | 0.0106 −4.55 (1.771) | −0.27 (1.025) | −0.82 (0.892) | 0.5876 −1.02 (1.884) |
| College grad/Advanced degree | −0.71 (1.231) | −1.60 (1.046) | 0.0164 −4.21 (1.744) | −1.44 (1.009) | −1.11 (0.878) | 0.3523 −1.73 (1.856) |
| Has anyone in your family been told of a risk for THIS genetic condition? | 0.0063 | 0.2402 | 0.3121 | N/A | N/A | N/A |
| Yes | 0.0047 2.83 (0.992) | 1.30 (0.854) | −2.09 (1.679) | N/A | N/A | N/A |
| No (ref) | - | - | - | N/A | N/A | N/A |
| Don’t know | 0.0724 5.42 (3.004) | −1.46 (2.584) | 3.73 (5.082) | N/A | N/A | N/A |
| Gene group | 0.1747 | 0.0207 | 0.1953 | 0.2677 | 0.0141 | 0.0935 |
| Cancer (ref) | - | - | - | - | - | - |
| Cardio | −1.89 (1.080) | 0.0289 −2.00 (0.910) | −1.97 (1.569) | −1.18 (0.903) | 0.1077 −1.26 (0.779) | −0.64 (1.658) |
| Miscellaneous | −1.55 (1.252) | 0.0169 −2.54 (1.055) | −2.95 (1.753) | −1.40 (1.009) | 0.0042 −2.51 (0.870) | −3.95 (1.852) |
| PANAS Emotion | 6 weeks Survey, Mean (SD) (n = 278) | 6 months Survey, Mean (SD) (n = 278) | p-Value |
|---|---|---|---|
| Positive | |||
| Interested | 3.6 (1.13) | 3.3 (1.21) | <0.0001 |
| Excited | 1.5 (1.00) | 1.4 (0.87) | 0.0456 |
| Strong | 2.2 (1.29) | 1.9 (1.31) | 0.0254 |
| Enthusiastic | 1.5 (1.00) | 1.4 (0.95) | 0.2176 |
| Proud | 1.4 (0.94) | 1.3 (0.79) | 0.0920 |
| Alert | 2.6 (1.25) | 2.3 (1.35) | 0.0003 |
| Inspired | 1.8 (1.18) | 1.6 (1.11) | 0.0232 |
| Determined | 2.4 (1.29) | 2.2 (1.38) | 0.0212 |
| Attentive | 2.7 (1.31) | 2.5 (1.35) | 0.0169 |
| Active | 2.4 (1.36) | 2.0 (1.17) | <0.0001 |
| Negative | |||
| Distressed | 1.8 (1.10) | 1.6 (1.04) | 0.0179 |
| Upset | 1.8 (1.12) | 1.6 (1.02) | 0.0035 |
| Guilty | 1.2 (0.72) | 1.2 (0.76) | 0.4359 |
| Scared | 1.8 (1.05) | 1.7 (1.07) | 0.4433 |
| Hostile | 1.0 (0.36) | 1.1 (0.37) | 0.4237 |
| Irritable | 1.3 (0.71) | 1.3 (0.80) | 0.7841 |
| Ashamed | 1.1 (0.51) | 1.1 (0.49) | 0.8116 |
| Nervous | 2.0 (1.14) | 1.8 (1.08) | 0.0029 |
| Jittery | 1.5 (0.89) | 1.3 (0.78) | 0.0050 |
| Afraid | 1.8 (1.08) | 1.6 (1.03) | 0.0106 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCormick, C.Z.; Yu, K.D.; Johns, A.; Campbell-Salome, G.; Hallquist, M.L.G.; Sturm, A.C.; Buchanan, A.H. Investigating Psychological Impact after Receiving Genetic Risk Results—A Survey of Participants in a Population Genomic Screening Program. J. Pers. Med. 2022, 12, 1943. https://doi.org/10.3390/jpm12121943
McCormick CZ, Yu KD, Johns A, Campbell-Salome G, Hallquist MLG, Sturm AC, Buchanan AH. Investigating Psychological Impact after Receiving Genetic Risk Results—A Survey of Participants in a Population Genomic Screening Program. Journal of Personalized Medicine. 2022; 12(12):1943. https://doi.org/10.3390/jpm12121943
Chicago/Turabian StyleMcCormick, Cara Zayac, Kristen Dilzell Yu, Alicia Johns, Gemme Campbell-Salome, Miranda L. G. Hallquist, Amy C. Sturm, and Adam H. Buchanan. 2022. "Investigating Psychological Impact after Receiving Genetic Risk Results—A Survey of Participants in a Population Genomic Screening Program" Journal of Personalized Medicine 12, no. 12: 1943. https://doi.org/10.3390/jpm12121943
APA StyleMcCormick, C. Z., Yu, K. D., Johns, A., Campbell-Salome, G., Hallquist, M. L. G., Sturm, A. C., & Buchanan, A. H. (2022). Investigating Psychological Impact after Receiving Genetic Risk Results—A Survey of Participants in a Population Genomic Screening Program. Journal of Personalized Medicine, 12(12), 1943. https://doi.org/10.3390/jpm12121943







