The Development of an Infrastructure to Facilitate the Use of Whole Genome Sequencing for Population Health
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Selection
2.2. HerediGene Sequencing Program
2.3. Clinical Validation of Research Data
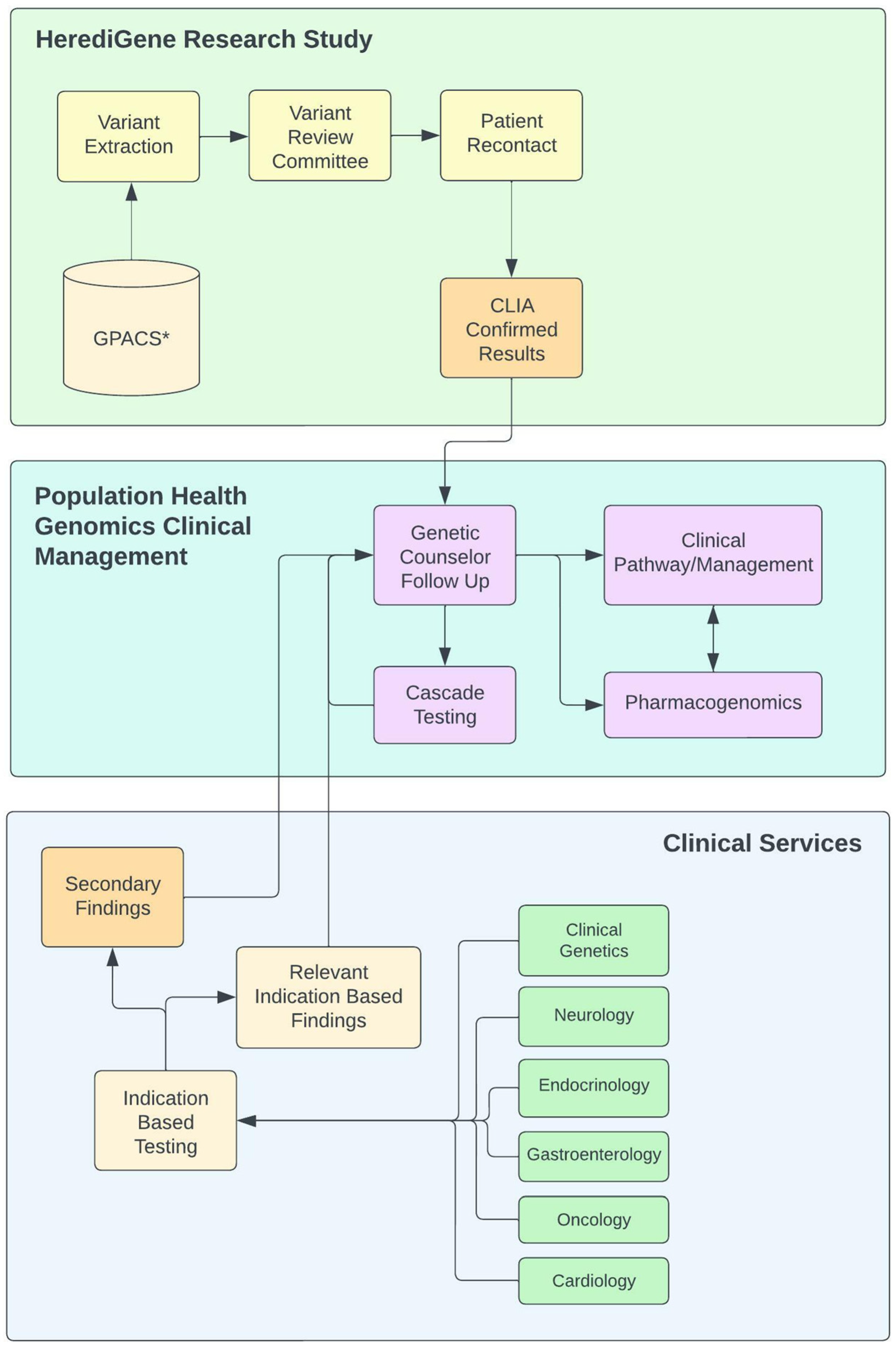
2.4. Population Health Framework
2.5. Clinical Domain Management Team
2.6. Primary Patient and Provider Contacts
2.7. Variant Review Committee (VRC)
2.8. Variant Selection Guidelines
2.9. Standardized Patient- and Provider-Facing Information
2.10. Pharmacogenomics Implications
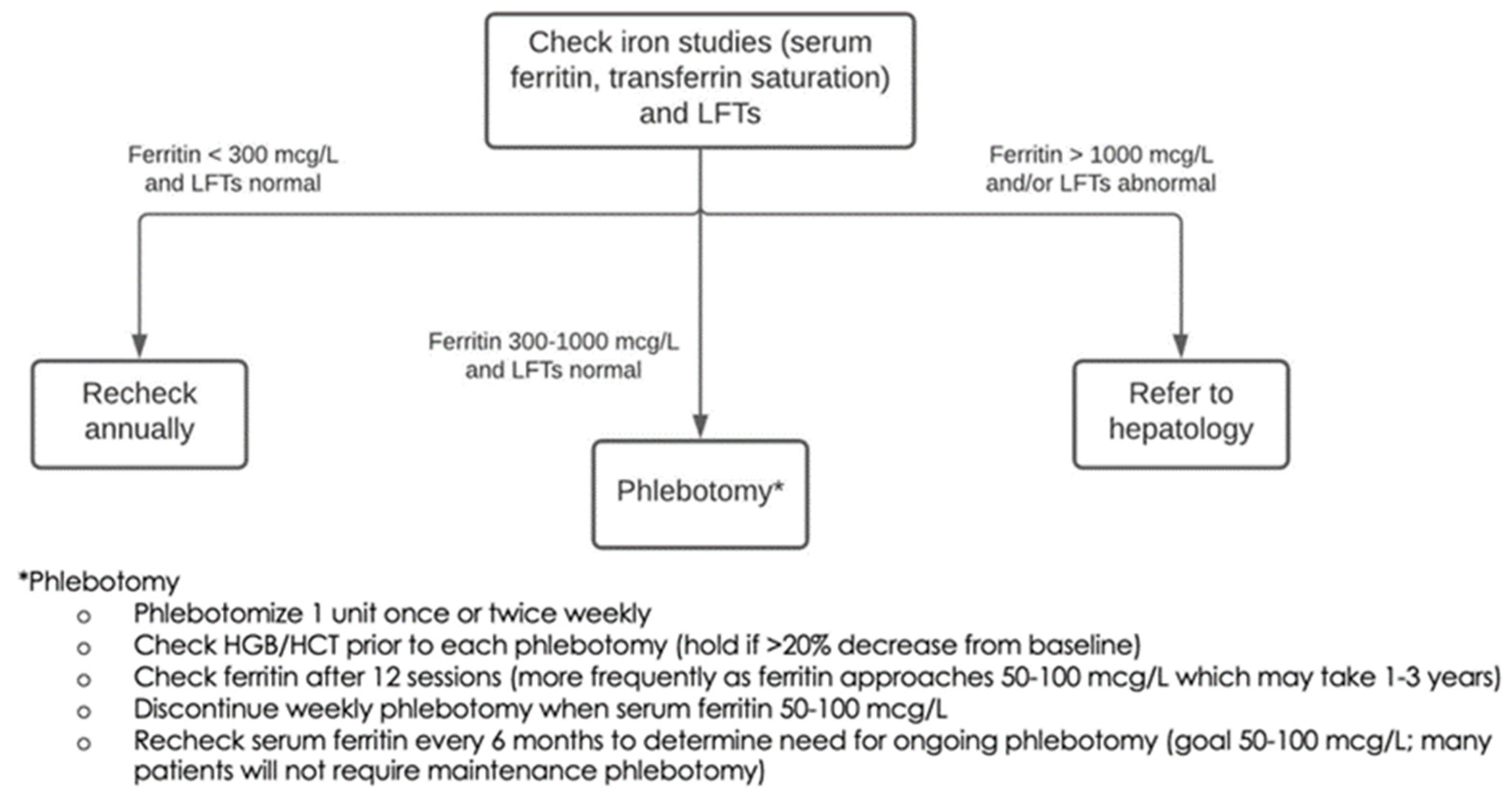
2.11. Defined Clinical Care Pathways
2.12. Clinical Decision Support (CDS)
2.13. Outcomes Definition and Measurement
2.14. Genetic Counseling Note Template
2.15. Informational Videos
3. Results
3.1. Initial Implementation
3.2. Scaling the Framework
3.3. Clinical Integration
3.4. Patient Reaction
3.5. Pediatric Considerations
3.6. Informatics Framework
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Tier 1 | ||
| Familial hypercholesterolemia | APOB | |
| LDLR | ||
| PCSK9 | ||
| Hereditary breast and ovarian cancer | BRCA1 | |
| BRCA2 | ||
| Hereditary hemochromatosis | HFE | |
| Lynch Syndrome | MLH1 | |
| MSH2 | ||
| MSH6 | ||
| PMS2 | ||
| Tier 2A | ||
| Aortopathies | ACTA2 | |
| FBN1 | ||
| MYH11 | ||
| SMAD3 | ||
| TGFBR1 | ||
| TGFBR2 | ||
| Arrhythmogenic right ventricular cardiomyopathy | DSC2 | |
| DSG2 | ||
| DSP | ||
| PKP2 | ||
| TMEM43 | ||
| Biotinidase deficiency | BTD | |
| Cardiomyopathies | ACTC1 | |
| FLNC | ||
| LMNA | ||
| MYBPC3 | ||
| MYH7 | ||
| MYL2 | ||
| MYL3 | ||
| PRKAG2 | ||
| TNNI3 | ||
| TPM1 | ||
| TTN | ||
| Ehlers-Danlos syndrome, vascular type | COL3A1 | |
| Fabry’s disease | GLA | |
| Familial medullary thyroid cancer | RET | |
| Hereditary breast and ovarian cancer | PALB2 | |
| Inherited cardiac Arrhythmias | CASQ2 | |
| KCNH2 | ||
| KCNQ1 | ||
| RYR2 | ||
| SCN5A | ||
| TRDN | ||
| Hereditary colorectal cancer | APC | |
| BMPR1A | ||
| MUTYH | ||
| SMAD4 | ||
| Hereditary hemorrhagic telangiectasia | ACVRL1 | |
| ENG | ||
| Hereditary paraganglioma-pheochromocytoma syndrome | MAX | |
| SDHAF2 | ||
| SDHB | ||
| SDHC | ||
| SDHD | ||
| TMEM127 | ||
| Left ventricular noncompaction | TNNT2 | |
| Li-Fraumeni syndrome | TP53 | |
| Malignant hyperthermia | CACNA1S | |
| RYR1 | ||
| Maturity-onset of diabetes of the young | HNF1A | |
| Multiple endocrine neoplasia | MEN1 | |
| Neurofibromatosis, type 2 | NF2 | |
| Ornithine carbamoyltransferase deficiency | OTC | |
| Peutz-Jeghers syndrome | STK11 | |
| Pompe disease | GAA | |
| PTEN hamartoma tumor syndrome | PTEN | |
| Retinoblastoma | RB1 | |
| RPE65-related retinopathy | RPE65 | |
| Tuberous sclerosis complex | TSC1 | |
| TSC2 | ||
| Von Hippel-Lindau syndrome | VHL | |
| WT1-related Wilms tumor | WT1 | |
| Wilson disease | ATP7B | |
| Tier 2B | ||
| Aortopathies | MYLK | |
| LOX | ||
| PRKG1 | ||
| SMAD2 | ||
| TGFB2 | ||
| TGFB3 | ||
| Arrhythmogenic right ventricular cardiomyopathy | JUP | |
| PKP2 | ||
| BAP1 tumor predisposition syndrome | BAP1 | |
| Birt-Hogg-Dubé syndrome | FLCN | |
| Cardiomyopathies | ACTN2 | |
| BAG3 | ||
| CSRP3 | ||
| DES | ||
| DMD | ||
| LAMP2 | ||
| PLN | ||
| RBM20 | ||
| DICER1 tumor predisposition | DICER1 | |
| Familial hypercholesterolemia | LDLRAP1 | |
| Hereditary breast and ovarian cancer | ATM | |
| BRIP1 | ||
| CHEK2 | ||
| RAD51C | ||
| RAD51D | ||
| Hereditary colorectal cancer | POLE | |
| Hereditary diffuse gastric cancer | CDH1 | |
| Hereditary skin cancer | CDKN2A | |
| Hereditary transthyretin amyloidosis | TTR | |
| Heritable pulmonary arterial hypertension | BMPR2 | |
| TBX4 | ||
| Hypertrophic cardiomyopathy | TNNC1 | |
| Inherited cardiac arrhythmias | CALM1 | |
| CALM2 | ||
| CALM3 | ||
| KCNE1 | ||
| Lynch syndrome | EPCAM | |
| Metachromatic leukodystrophy | ARSA | |
| Neurofibromatosis, type 1 | NF1 | |
| Prostate cancer | HOXB13 | |
| Pulmonary venoocclusive disease | EIF2AK4 | |
| X-Linked adrenoleukodystrophy | ABCD1 | |
| Tier 3 | ||
| Aortopathies | COL5A2 | |
| EFEMP2 | ||
| FBN2 | ||
| FOXE3 | ||
| GATA1D | ||
| MAT2A | ||
| MFAP5 | ||
| NOTCH1 | ||
| PLOD1 | ||
| SKI | ||
| SLC2A10 | ||
| Bloom Syndrome | RECQL3 | |
| Capillary malformation-ateriovenous malformation | RASA1 | |
| Cardiac Arrhythmias | ANK2 | |
| CACNA1C | ||
| CACNB2 | ||
| GPD1L | ||
| HCN4 | ||
| KCNA5 | ||
| KCNE2 | ||
| KCNE3 | ||
| KCNJ2 | ||
| NKX2-5 | ||
| SCN1B | ||
| SCNB3 | ||
| SNTA1 | ||
| Cardiomyopathies | ABCC9 | |
| CAV3 | ||
| CRYAB | ||
| DOLK | ||
| EYA4 | ||
| FKRP | ||
| HRAS | ||
| RAF1 | ||
| SGCD | ||
| TAZ | ||
| TCAP | ||
| VCL | ||
| Carney complex | PRKAR1A | |
| Glycogen storage disease | AGL | |
| Hereditary Multiple Osteochondromas | EXT1 | |
| EXT2 | ||
| Hereditary colorectal cancer | AXIN2 | |
| GREM1 | ||
| MSH3 | ||
| NTHL1 | ||
| POLD1 | ||
| Hereditary paraganglioma-pheochromocytoma syndrome | SDHA | |
| Maturity-onset of diabetes of the young | ABCC8 | |
| HNF1B | ||
| HNF4A | ||
| KCNJ11 | ||
| Nijmegen breakage syndrome | NBN | |
| Nevoid basal cell carcinoma syndrome | PTCH1 | |
| SUFU | ||
| Parathyroid Cancer | CDC73 | |
| Pulmonary Arterial Hypertension | CAV1 | |
| SMAD9 | ||
| Rhabdoid tumor predisposition syndrome | SMARCA4 | |
| SMARCB1 | ||
| Rothmund-Thomson syndrome, type 2 | RECQL4 | |
| RUNX1 familial platelet disorder with associated myeloid malignancies | RUNX1 | |
| Skin Cancer | CYLD | |
| ERCC1 | ||
| ERCC2 | ||
| ERCC3 | ||
| ERCC4 | ||
| ERCC5 | ||
| MITF | ||
| Systemic Primary Carnitine Deficiency | SLC22A5 | |
| Thrombosis | CBS | |
| Werner Syndrome | WRN |
| CPIC Pharmacogenes | |||||||
|---|---|---|---|---|---|---|---|
| ABCG2 | BCHE | CYP2C19 | DPYD | HLA-B | MT-RNR1 | OTC | SLC6A4 |
| ABL2 | CACNA1S | CYP2C9 | G6PD | HLA-DRB1 | MTHFR | POLG | SLCO1B1 |
| ADRB1 | CFTR | CYP2D6 | GBA | HPRT1 | NAGS | RYR1 | TPMT |
| ASL | CPS1 | CYP3A5 | GRK5 | IFNL3 | NAT2 | SCN1A | UGT1A1 |
| ASS1 | CYP2B6 | CYP4F2 | HLA-A | IFNL4 | NUDT15 | SLC28A3 | VKORC1 |
References
- Hancock, S.; Taber, K.J.; Goldberg, J.D. Fetal screening and whole genome sequencing: Where are the limits? Expert Rev. Mol. Diagn. 2021, 21, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Buxton, J. Whole Genome Sequencing at Birth: Genomic Data, a Resource from Cradle to Grave? BioNews, 21 February 2022. [Google Scholar]
- Kingsmore, S.F.; Smith, L.D.; Kunard, C.M.; Bainbridge, M.; Batalov, S.; Benson, W.; Blincow, E.; Caylor, S.; Chambers, C.; Del Angel, G.; et al. A genome sequencing system for universal newborn screening, diagnosis, and precision medicine for severe genetic diseases. Am. J. Hum. Genet. 2022, 109, 1605–1619. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, H.; An, K.; Liu, Z.; Hai, L.; Li, R.; Zhou, Y.; Zhao, W.; Jia, Y.; Wu, N.; et al. Whole-genome bisulfite sequencing analysis of circulating tumour DNA for the detection and molecular classification of cancer. Clin. Transl. Med. 2022, 12, e1014. [Google Scholar] [CrossRef] [PubMed]
- Wain, K.; Tolwinski, K.; Palen, E.; Heidlebaugh, A.; Holdren, K.; Walsh, L.; Oetjens, M.; Ledbetter, D.; Martin, C. Population Genomic Screening for Genetic Etiologies of Neurodevelopmental/Psychiatric Disorders Demonstrates Personal Utility and Positive Participant Responses. J. Pers. Med. 2021, 11, 365. [Google Scholar] [CrossRef] [PubMed]
- Service, R.F. The Race for the $1000 Genome. Science 2006, 311, 1544–1546. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Smith, K.R.; Martin, A.; Thomas, E.A.; McDonagh, E.M.; Cipriani, V.; Ellingford, J.M.; Arno, G.; Tucci, A.; Vandrovcova, J.; et al. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N. Engl. J. Med. 2021, 385, 1868–1880. [Google Scholar] [CrossRef]
- Carey, D.J.; Fetterolf, S.N.; Davis, F.D.; Faucett, W.A.; Kirchner, H.L.; Mirshahi, U.; Murray, M.F.; Smelser, D.T.; Gerhard, G.S.; Ledbetter, D.H. The Geisinger MyCode community health initiative: An electronic health record–linked biobank for precision medicine research. Genet. Med. 2016, 18, 906–913. [Google Scholar] [CrossRef] [PubMed]
- HerediGene Population Study. Intermountain Precision Genomics-Intermountain Healthcare. Available online: https://intermountainhealthcare.org/health-wellness-promotion/genomics/heredigene/ (accessed on 1 July 2022).
- Wilden, R. Hackathon–style Session: How Can We Maximize the Yield of Genetic Disease Diagnostic Evaluation and Testing? In Proceedings of the ACMGMtg22, Nashville, TN, USA, 23 March 2022. [Google Scholar]
- Miller, D.T.; Lee, K.; Chung, W.K.; Gordon, A.S.; Herman, G.E.; Klein, T.E.; Stewart, D.R.; Amendola, L.M.; Adelman, K.; Bale, S.J.; et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Genes-Drugs. Clinical Pharmacogenetics Implementation Consortium. 1 June 2022. Available online: https://cpicpgx.org/genes-drugs/ (accessed on 1 August 2022).
- Centers for Disease Control and Prevention. Tier 1 Genomics Applications and their Importance to Public Healthv. 6 March 2014. Available online: https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm (accessed on 8 July 2022).
- Barton, J.C.; Edwards, C.Q. HFE Hemochromatosis. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., L. Bean, J., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. Available online: http://www.ncbi.nlm.nih.gov/books/NBK1440/ (accessed on 5 August 2022).
- RxMatch. Intermountain Healthcare. Available online: https://intermountainhealthcare.org/services/genomics/providers/rxmatch/ (accessed on 1 August 2022).
- Walton, N.A.; Johnson, D.K.; Person, T.N.; Reynolds, J.C.; Williams, M.S. Pilot Implementation of Clinical Genomic Data into the Native Electronic Health Record: Challenges of Scalability. ACI Open 2020, 4, e162–e166. [Google Scholar] [CrossRef]
- Health Bot. A Managed Service Purpose-Built for Development of Virtual Healthcare Assistants, Microsoft. Available online: https://azure.microsoft.com/en-us/services/bot-services/health-bot/#overview (accessed on 1 August 2022).
- Levit, L.A.; Kim, E.S.; McAneny, B.L.; Nadauld, L.D.; Levit, K.; Schenkel, C.; Schilsky, R.L. Implementing Precision Medicine in Community-Based Oncology Programs: Three Models. J. Oncol. Pr. 2019, 15, 325–329. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walton, N.A.; Hafen, B.; Graceffo, S.; Sutherland, N.; Emmerson, M.; Palmquist, R.; Formea, C.M.; Purcell, M.; Heale, B.; Brown, M.A.; et al. The Development of an Infrastructure to Facilitate the Use of Whole Genome Sequencing for Population Health. J. Pers. Med. 2022, 12, 1867. https://doi.org/10.3390/jpm12111867
Walton NA, Hafen B, Graceffo S, Sutherland N, Emmerson M, Palmquist R, Formea CM, Purcell M, Heale B, Brown MA, et al. The Development of an Infrastructure to Facilitate the Use of Whole Genome Sequencing for Population Health. Journal of Personalized Medicine. 2022; 12(11):1867. https://doi.org/10.3390/jpm12111867
Chicago/Turabian StyleWalton, Nephi A., Brent Hafen, Sara Graceffo, Nykole Sutherland, Melanie Emmerson, Rachel Palmquist, Christine M. Formea, Maricel Purcell, Bret Heale, Matthew A. Brown, and et al. 2022. "The Development of an Infrastructure to Facilitate the Use of Whole Genome Sequencing for Population Health" Journal of Personalized Medicine 12, no. 11: 1867. https://doi.org/10.3390/jpm12111867
APA StyleWalton, N. A., Hafen, B., Graceffo, S., Sutherland, N., Emmerson, M., Palmquist, R., Formea, C. M., Purcell, M., Heale, B., Brown, M. A., Danford, C. J., Rachamadugu, S. I., Person, T. N., Shortt, K. A., Christensen, G. B., Evans, J. M., Raghunath, S., Johnson, C. P., Knight, S., ... McLeod, H. L. (2022). The Development of an Infrastructure to Facilitate the Use of Whole Genome Sequencing for Population Health. Journal of Personalized Medicine, 12(11), 1867. https://doi.org/10.3390/jpm12111867








