Audiological Evidence of Frequent Hereditary Mild, Moderate and Moderate-to-Severe Hearing Loss
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Audiological Examination
2.3. Genetic Investigation
2.4. Statistics
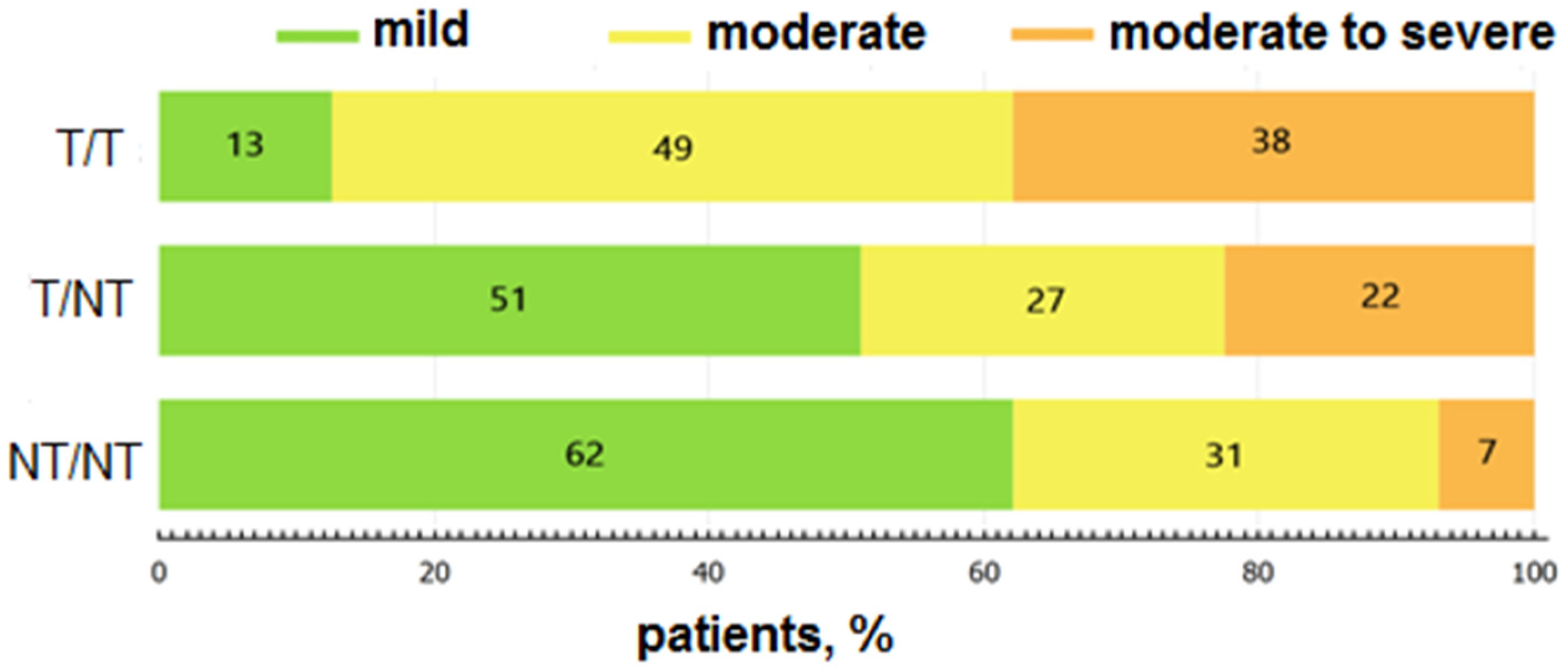
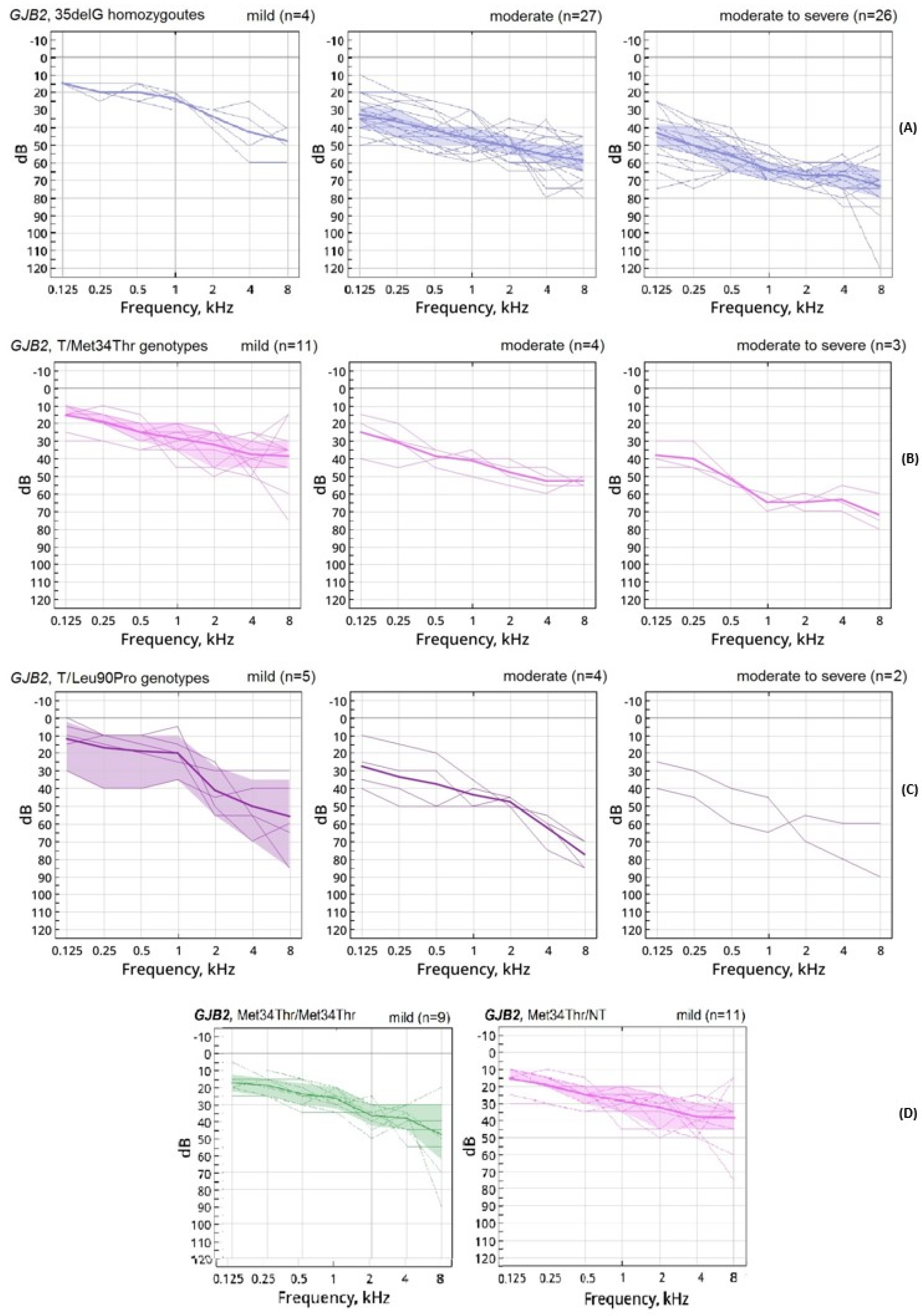
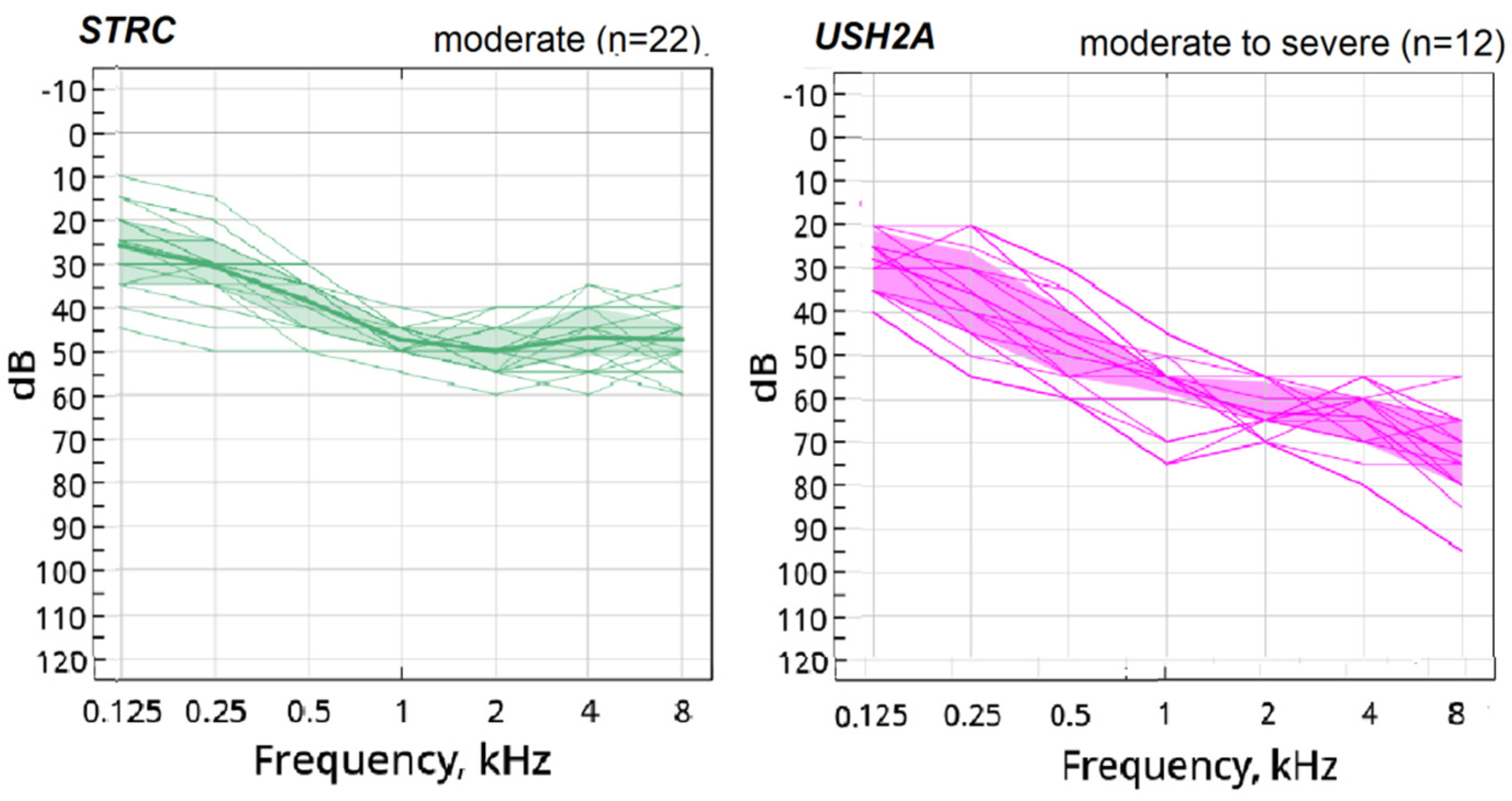
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morton, C.C.; Nance, W.E. Newborn Hearing Screening—A Silent Revolution. N. Engl. J. Med. 2006, 354, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.S.; Smith, K.Y.; Widen, J.E.; Chertoff, M.E. Effects of universal newborn hearing screening on an early intervention program for children with hearing loss, birth to 3 yr of age. J. Am. Acad. Audiol. 2010, 21, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Butcher, E.; Dezateux, C.; Cortina-Borja, M.; Knowles, R.L. Prevalence of permanent childhood hearing loss detected at the universal newborn hearing screen: Systematic review and meta-analysis. PLoS ONE 2019, 14, e0219600. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Huang, L.H.; Wang, G.J.; Gao, X.; Qu, C.Y.; Chen, X.W.; Ma, F.R.; Zhang, J.; Xing, W.L.; Xi, S.Y.; et al. Concurrent Hearing and Genetic Screening of 180,469 Neonates with Follow-up in Beijing, China. Am. J. Hum. Genet. 2019, 105, 803–812. [Google Scholar] [CrossRef]
- Guo, L.; Xiang, J.; Sun, L.; Yan, X.; Yang, J.; Wu, H.; Guo, K.; Peng, J.; Xie, X.; Yin, Y.; et al. Concurrent hearing and genetic screening in a general newborn population. Hum. Genet. 2020, 139, 521–530. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef]
- Ito, T.; Kawashima, Y.; Fujikawa, T.; Honda, K.; Makabe, A.; Kitamura, K.; Tsutsumi, T. Rapid screening of copy number variations in STRC by droplet digital PCR in patients with mild-to-moderate hearing loss. Hum. Genome Var. 2019, 6, 41. [Google Scholar] [CrossRef]
- Plevova, P.; Paprskarova, M.; Tvrda, P.; Turska, P.; Slavkovsky, R.; Mrazkova, E. STRC Deletion is a Frequent Cause of Slight to Moderate Congenital Hearing Impairment in the Czech Republic. Otol. Neurotol. 2017, 38, e393–e400. [Google Scholar] [CrossRef]
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997, 387, 80–83. [Google Scholar] [CrossRef]
- Marlin, S.; Garabédian, E.N.; Roger, G.; Moatti, L.; Matha, N.; Lewin, P.; Petit, C.; Denoyelle, F. Connexin 26 gene mutations in congenitally deaf children. Arch. Otolaryngol. Head Neck Surg. 2001, 127, 927–933. [Google Scholar] [CrossRef][Green Version]
- Snoeckx, R.L.; Huygen, P.L.; Feldmann, D.; Marlin, S.; Denoyelle, F.; Waligora, J.; Mueller-Malesinska, M.; Pollak, A.; Ploski, R.; Murgia, A.; et al. GJB2 mutations and degree of hearing loss: A multicenter study. Am. J. Hum. Genet. 2005, 77, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Marlin, S.; Feldmann, D.; Blons, H.; Loundon, N.; Rouillon, I.; Albert, S.; Chauvin, P.; Garabédian, E.N.; Couderc, R.; Odent, S.; et al. GJB2 and GJB6 mutations: Genotypic and phenotypic correlations in a large cohort of hearing-impaired patients. Arch. Otolaryngol. Head Neck Surg. 2005, 131, 481–487. [Google Scholar] [CrossRef][Green Version]
- del Castillo, F.J.; del Castillo, I. DFNB1 Non-syndromic Hearing Impairment: Diversity of Mutations and Associated Phenotypes. Front. Mol. Neurosci. 2017, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Hilgert, N.; Huentelman, M.J.; Thorburn, A.Q.; Fransen, E.; Dieltjens, N.; Malesinska, M.M.; Pollak, A.; Skorka, A.; Waligora, J.; Ploski, R.; et al. Phenotypic variability of patients homozygous for the GJB2 mutation 35delG cannot be explained by the influence of one major modifier gene. Eur. J. Hum. Genet. 2009, 17, 517–524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Green, G.E.; Smith, R.J.H.; Bent, J.P.; Cohn, E.S. Genetic testing to identify deaf newborns. JAMA 2000, 284, 1245. [Google Scholar] [CrossRef] [PubMed]
- Gopalarao, D.; Kimberling, W.J.; Jesteadt, W.; Kelley, P.M.; Beauchaine, K.L.; Cohn, E.S. Is hearing loss due to mutations in the connexin 26 gene progressive? Int. J. Audiol. 2008, 47, 11–20. [Google Scholar] [CrossRef]
- Kokotas, H.; Theodosiou, M.; Korres, G.; Grigoriadou, M.; Ferekidou, E.; Giannoulia-Karantana, A.; Petersen, M.B.; Korres, S. Sudden hearing loss in a family with GJB2 related progressive deafness. Int. J. Pediatr. Otorhinolaryngol. 2008, 72, 1735–1740. [Google Scholar] [CrossRef]
- Norris, V.W.; Arnos, K.S.; Hanks, W.D.; Xia, X.; Nance, W.E.; Pandya, A. Does universal newborn hearing screening identify all children with GJB2 (connexin 26) deafness? Penetrance of GJB2 deafness. Ear Hear. 2006, 27, 732–741. [Google Scholar] [CrossRef]
- Pagarkar, W.; Bitner-Glindzicz, M.; Knight, J.; Sirimanna, T. Late postnatal onset of hearing loss due to GJB2 mutations. Int. J. Pediatr. Otorhinolaryngol. 2006, 70, 1119–1124. [Google Scholar] [CrossRef]
- Minami, S.B.; Mutai, H.; Nakano, A.; Arimoto, Y.; Taiji, H.; Morimoto, N.; Sakata, H.; Adachi, N.; Masuda, S.; Sakamoto, H.; et al. GJB2-associated hearing loss undetected by hearing screening of newborns. Gene 2013, 532, 41–45. [Google Scholar] [CrossRef]
- Orzan, E.; Murgia, A. Connexin 26 deafness is not always congenital. Int. J. Pediatr. Otorhinolaryngol. 2007, 71, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Kenna, M.A.; Feldman, H.A.; Neault, M.W.; Frangulov, A.; Wu, B.-L.; Fligor, B.; Rehm, H.L. Audiologic phenotype and progression in GJB2 (Connexin 26) hearing loss. Arch. Otolaryngol. Head Neck Surg. 2010, 136, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Anichkina, A.; Kulenich, T.; Zinchenko, S.; Shagina, I.; Polyakov, A.; Ginter, E.; Evgrafov, E.; Viktorova, T.; Khusnitdonova, E. On the origin and frequency of the 35delG allele in GJB2-linked deafness in Europe. Eur. J. Hum. Genet. 2001, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Bliznetz, E.A.; Galkina, V.A.; Matyushchenko, G.N.; Kisina, A.G.; Markova, T.G.; Polyakov, A.V. Changes in the connexin 26 gene (GJB2) in Russian patients with hearing loss: Results of long-term molecular diagnostics of hereditary nonsyndromic hearing loss. Russ. J. Genet. 2012, 48, 101–112. [Google Scholar] [CrossRef]
- Bliznetz, E.A.; Lalayants, M.R.; Markova, T.G.; Balanovsky, O.P.; Balanovska, E.V.; Skhalyakho, R.A.; Pocheshkhova, E.A.; Nikitina, N.V.; Voronin, S.V.; Kudryashova, E.K.; et al. Update of the GJB2/DFNB1 mutation spectrum in Russia: A founder Ingush mutation del(GJB2-D13S175) is the most frequent among other large deletions. J. Hum. Genet. 2017, 62, 789–795. [Google Scholar] [CrossRef][Green Version]
- Lalaiants, M.R.; Markova, T.G.; Bliznets, E.A.; Poliakov, A.V.; Tavartikiladze, G.A. The results of audiological examination of children presenting with sensorineural loss of hearing due to GJB2 gene mutations during the first year of life. Vestn. Otorinolaringol. 2011, 3, 31–35. [Google Scholar]
- Lalaiants, M.R.; Markova, T.G.; Bakhshinian, V.V.; Bliznets, E.A.; Poliakov, A.V.; Tavartikiladze, G.A. The audiological phenotype and the prevalence of GJB2-related sensorineural loss of hearing in the infants suffering acoustic disturbances. Vestn. Otorinolaringol. 2014, 2, 37–43. [Google Scholar]
- Markova, T.G.; Bliznets, E.A.; Poliakov, A.V.; Tavartikiladze, G.A. Twenty years of clinical study of GJB2-linked hearing loss in Russia. Vestn. Otorinolaringol. 2018, 4, 31–36. [Google Scholar] [CrossRef]
- Olusanya, B.O.; Davis, A.C.; Hoffman, H.J. Hearing loss grades and the International classification of functioning, disability and health. Bull. World Health Organ. 2019, 97, 725–728. [Google Scholar] [CrossRef]
- Humes, L.E. The World Health Organization’s hearing-impairment grading system: An evaluation for unaided communication in age-related hearing loss. Int. J. Audiol. 2019, 58, 12–20. [Google Scholar] [CrossRef]
- Parker, M.J.; Fortnum, H.M.; Young, I.D.; Davis, A.C.; Mueller, R.F. Population-based genetic study of Childhood hearing impairment in the Trent region of the United Kingdom. Audiology 2000, 39, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Markova, T.G.; Alekseeva, N.N.; Mironovich, O.L.; Galeeva, N.M.; Lalayants, M.R.; Bliznetz, E.A.; Chibisova, S.S.; Polyakov, A.V.; Tavartkiladze, G.A. Clinical features of hearing loss caused by STRC gene deletions/mutations in Russian population. Int. J. Pediatr. Otorhinolaryngol. 2020, 138, 110247. [Google Scholar] [CrossRef] [PubMed]
- Markova, T.G.; Lalayants, M.R.; Alekseeva, N.N.; Ryzhkova, O.P.; Shatokhina, O.L.; Galeeva, N.M.; Bliznetz, E.A.; Weener, M.E.; Belov, O.A.; Chibisova, S.S.; et al. Early audiological phenotype in patients with mutations in the USH2A gene. Int. J. Pediatr. Otorhinolaryngol. 2022, 157, 111140. [Google Scholar] [CrossRef]
- Kim, B.J.; Oh, D.Y.; Han, J.H.; Oh, J.; Kim, M.Y.; Park, H.R.; Seok, J.; Cho, S.D.; Lee, S.Y.; Kim, Y.; et al. Significant Mendelian genetic contribution to pediatric mild-to-moderate hearing loss and its comprehensive diagnostic approach. Genet Med. 2020, 22, 1119–1128. [Google Scholar] [CrossRef]
- Tekin, M.; Duman, T.; Boğoçlu, G.; Incesulu, A.; Cin, S.; Akar, N. Moderate hearing loss and pseudodominant inheritance due to L90P/35delG mutations in the GJB2 (connexin 26) gene. Genet. Couns. 2003, 14, 379–386. [Google Scholar]
- Hall, A.; Pembrey, M.; Lutman, M.; Steer, C.; Bitner-Glindzicz, M. Prevalence and audiological features in carriers of GJB2 mutations, c.35delG and c.101T>C (p.M34T), in a UK population study. BMJ Open 2012, 2, e001238. [Google Scholar] [CrossRef]
- Houseman, M.J.; Ellis, L.A.; Pagnamenta, A.; Di, W.L.; Rickard, S.; Osborn, A.H.; Dahl, H.H.; Taylor, G.R.; Bitner-Glindzicz, M.; Reardon, W.; et al. Genetic analysis of the connexin-26 M34T variant: Identification of genotype M34T/M34T segregating with mild-moderate non-syndromic sensorineural hearing loss. J. Med. Genet. 2001, 38, 20–25. [Google Scholar] [CrossRef][Green Version]
- Carew, P.; Mensah, F.K.; Rance, G.; Flynn, T.; Poulakis, Z.; Wake, M. Mild-moderate congenital hearing loss: Secular trends in outcomes across four systems of detection. Child Care Health Dev. 2018, 44, 71–82. [Google Scholar] [CrossRef]
- Burke, W.F.; Warnecke, A.; Schöner-Heinisch, A.; Lesinski-Schiedat, A.; Maier, H.; Lenarz, T. Prevalence and audiological profiles of GJB2 mutations in a large collective of hearing impaired patients. Hear. Res. 2016, 333, 77–86. [Google Scholar] [CrossRef]
- Janecke, A.R.; Hirst-Stadlmann, A.; Günther, B.; Utermann, B.; Müller, T.; Löffler, J.; Utermann, G.; Nekahm-Heis, D. Progressive hearing loss, and recurrent sudden sensorineural hearing loss associated with GJB2 mutations--phenotypic spectrum and frequencies of GJB2 mutations in Austria. Hum. Genet. 2002, 111, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.K.; Schrijver, I.; Chang, K.W. Connexin-26-associated deafness: Phenotypic variability and progression of hearing loss. Genet. Med. 2010, 12, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Varga, L.; Jovankovicova, A.; Huckova, M.; Demesova, L.; Gasperikova, D.; Sebova, I.; Profant, M. Hereditary bilateral sudden sensorineural hearing loss. Bratisl. Lek. Listy. 2019, 120, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Moeller, M.P.; McCleary, E.; Putman, C.; Tyler-Krings, A.; Hoover, B.; Stelmachowicz, P. Longitudinal development of phonology and morphology in children with late-identified mild-moderate sensorineural hearing loss. Ear Hear. 2010, 31, 625–635. [Google Scholar] [CrossRef]
- McCreery, R.W.; Walker, E.A.; Tomblin, J.B.; Oleson, J.J.; Ambrose, S.A.; Moeller, M.P. Mild hearing loss is a developmental risk: Response to Carew and colleagues. Child Care Health Dev. 2018, 44, 926–927. [Google Scholar] [CrossRef]
- Čada, Z.; Šafka Brožková, D.; Balatková, Z.; Plevová, P.; Rašková, D.; Laštůvková, J.; Černý, R.; Bandúrová, V.; Koucký, V.; Hrubá, S.; et al. Moderate sensorineural hearing loss is typical for DFNB16 caused by various types of mutations affecting the STRC gene. Eur. Arch. Otorhinolaryngol. 2019, 276, 3353–3358. [Google Scholar] [CrossRef]
- Marková, S.P.; Brožková, D.Š.; Laššuthová, P.; Mészárosová, A.; Krůtová, M.; Neupauerová, J.; Rašková, D.; Trková, M.; Staněk, D.; Seeman, P. STRC Gene Mutations, Mainly Large Deletions, are a Very Important Cause of Early-Onset Hereditary Hearing Loss in the Czech Population. Genet. Test. Mol. Biomark. 2018, 22, 127–134. [Google Scholar] [CrossRef]
- Schrijver, I.; Gardner, P. Hereditary sensorineural hearing loss: Advances in molecular genetics and mutation analysis. Expert Rev. Mol. Diagn. 2006, 6, 375–386. [Google Scholar] [CrossRef]
- Kremer, H. Hereditary hearing loss; about the known and the unknown. Hear. Res. 2019, 376, 58–68. [Google Scholar] [CrossRef]
- Uehara, N.; Fujita, T.; Yamashita, D.; Yokoi, J.; Katsunuma, S.; Kakigi, A.; Nishio, S.Y.; Nibu, K.I.; Usami, S.I. Genetic background in late-onset sensorineural hearing loss patients. J. Hum. Genet. 2022, 67, 223–230. [Google Scholar] [CrossRef]
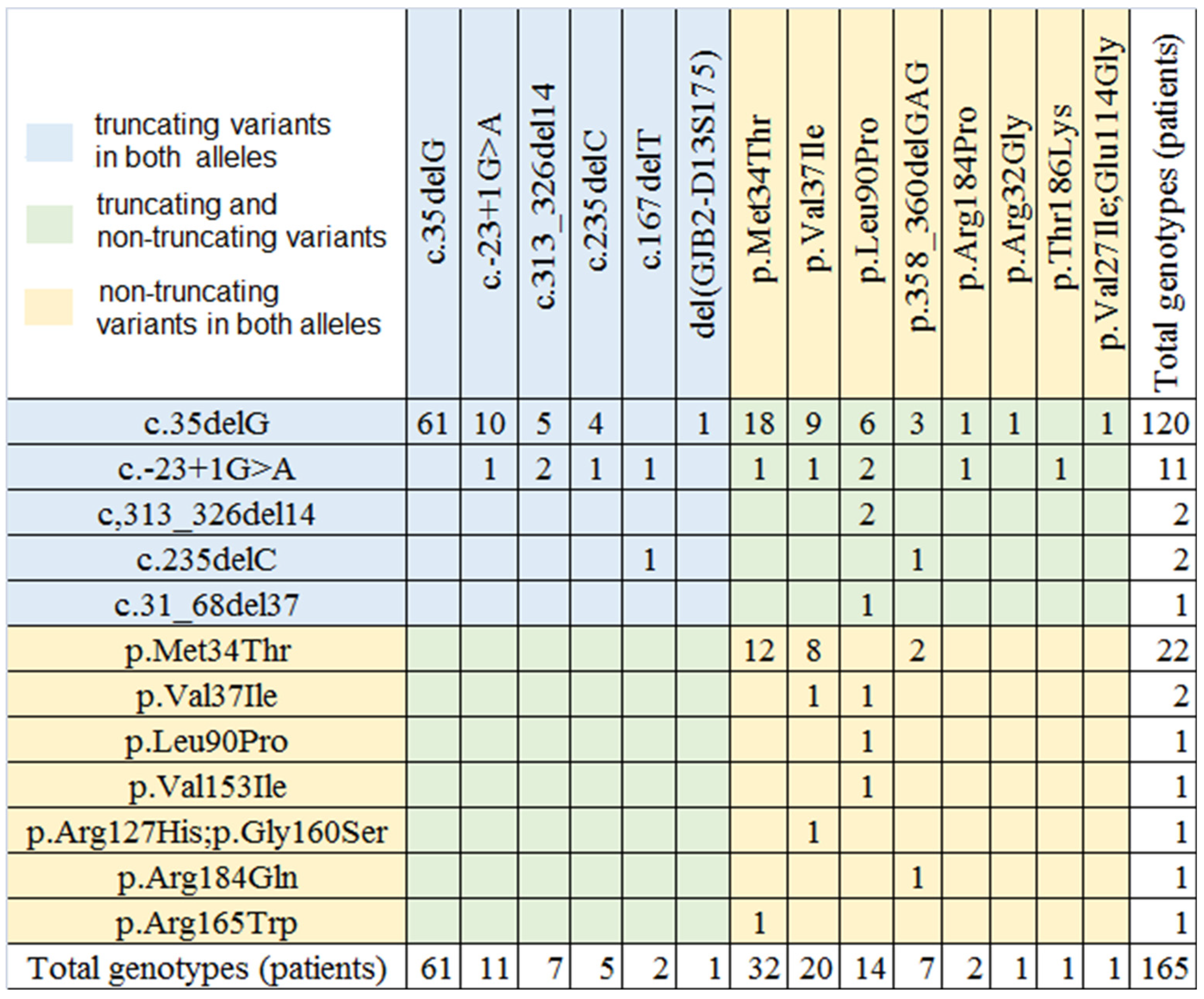
| Mutation | % (Number) of Alleles among 330 chrs. (Group with Mild, Moderate, Moderate-to-Severe Severity) | % (Number) of Alleles among 2141 chrs. (Whole Group with Different Severity) * |
|---|---|---|
| Truncating | ||
| c.35delG | 55.4 (181) | 77 (1700) |
| c.-23+1G > A | 6.4 (22) | 4.4 (97) |
| c.313_326del14 | 2.7 (9) | 4.7 (103) |
| c.235delC | 2.1 (7) | 2.3 (51) |
| c.167delT | 0.6 (2) | 1.5 (33) |
| c.31_68del | 0.3 (1) | <0.1 (1) |
| del(GJB2-D13S175) | 0.3 (1) | 0.5 (11) |
| 67.6 (223) | 93.2 (1996) | |
| Non-truncating | ||
| c.101T > C, p.Met34Thr) | 16.7 (55) | 2.9 (65) |
| c.109G > A, p.Val37Ile | 6.4 (21) | 1.2 (27) |
| c.269T > C, p.Leu90Pro | 4.6 (15) | 1.0 (23) |
| c.358_360delGAG | 2.1 (7) | 0.9 (19) |
| c.550C > T, p.Arg184Pro | 0.6 (2) | 0.5 (10) |
| c.94C > G, p.Arg32Gly | 0.3 (1) | <0.1 (1) |
| p.Thr186Lys | 0.3 (1) | - |
| p.Val153Ile | 0.3 (1) | - |
| p.Arg127His;p.Gly160Ser | 0.3 (1) | - |
| p.Val27Ile;p.Glu114Gly | 0.3 (1) | - |
| p.Arg184Gln | 0.3 (1) | - |
| p.Arg165Trp | 0.3 (1) | - |
| 32.4 (107) | 6.7 (145) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markova, T.; Alekseeva, N.; Lalayants, M.; Ryzhkova, O.; Shatokhina, O.; Galeeva, N.; Bliznetz, E.; Belov, O.; Chibisova, S.; Polyakov, A.; et al. Audiological Evidence of Frequent Hereditary Mild, Moderate and Moderate-to-Severe Hearing Loss. J. Pers. Med. 2022, 12, 1843. https://doi.org/10.3390/jpm12111843
Markova T, Alekseeva N, Lalayants M, Ryzhkova O, Shatokhina O, Galeeva N, Bliznetz E, Belov O, Chibisova S, Polyakov A, et al. Audiological Evidence of Frequent Hereditary Mild, Moderate and Moderate-to-Severe Hearing Loss. Journal of Personalized Medicine. 2022; 12(11):1843. https://doi.org/10.3390/jpm12111843
Chicago/Turabian StyleMarkova, Tatiana, Natalia Alekseeva, Maria Lalayants, Oxana Ryzhkova, Olga Shatokhina, Nailya Galeeva, Elena Bliznetz, Oleg Belov, Svetlana Chibisova, Alexander Polyakov, and et al. 2022. "Audiological Evidence of Frequent Hereditary Mild, Moderate and Moderate-to-Severe Hearing Loss" Journal of Personalized Medicine 12, no. 11: 1843. https://doi.org/10.3390/jpm12111843
APA StyleMarkova, T., Alekseeva, N., Lalayants, M., Ryzhkova, O., Shatokhina, O., Galeeva, N., Bliznetz, E., Belov, O., Chibisova, S., Polyakov, A., & Tavartkiladze, G. (2022). Audiological Evidence of Frequent Hereditary Mild, Moderate and Moderate-to-Severe Hearing Loss. Journal of Personalized Medicine, 12(11), 1843. https://doi.org/10.3390/jpm12111843







