COVID-19 Pandemic Outbreak in the Subcontinent: A Data Driven Analysis
Abstract
:1. Introduction
2. Related Work
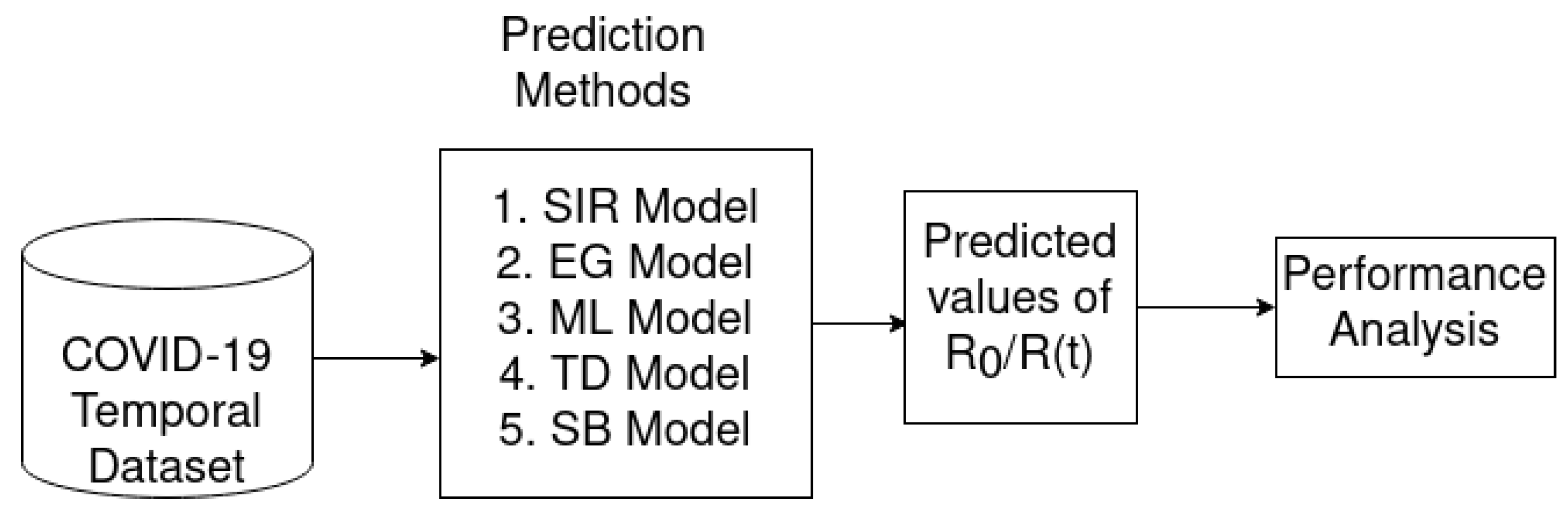
3. Overall Architecture
4. The Reproduction Number
5. Epidemic Forecasting Models
5.1. SIR Model
5.2. Exponential Growth (EG)
5.3. Sequential Bayesian Method (SB)
5.4. Maximum Likelihood Estimation (ML)
5.5. Time Dependent Estimation (TD)
6. Data Source
7. Experimental Results
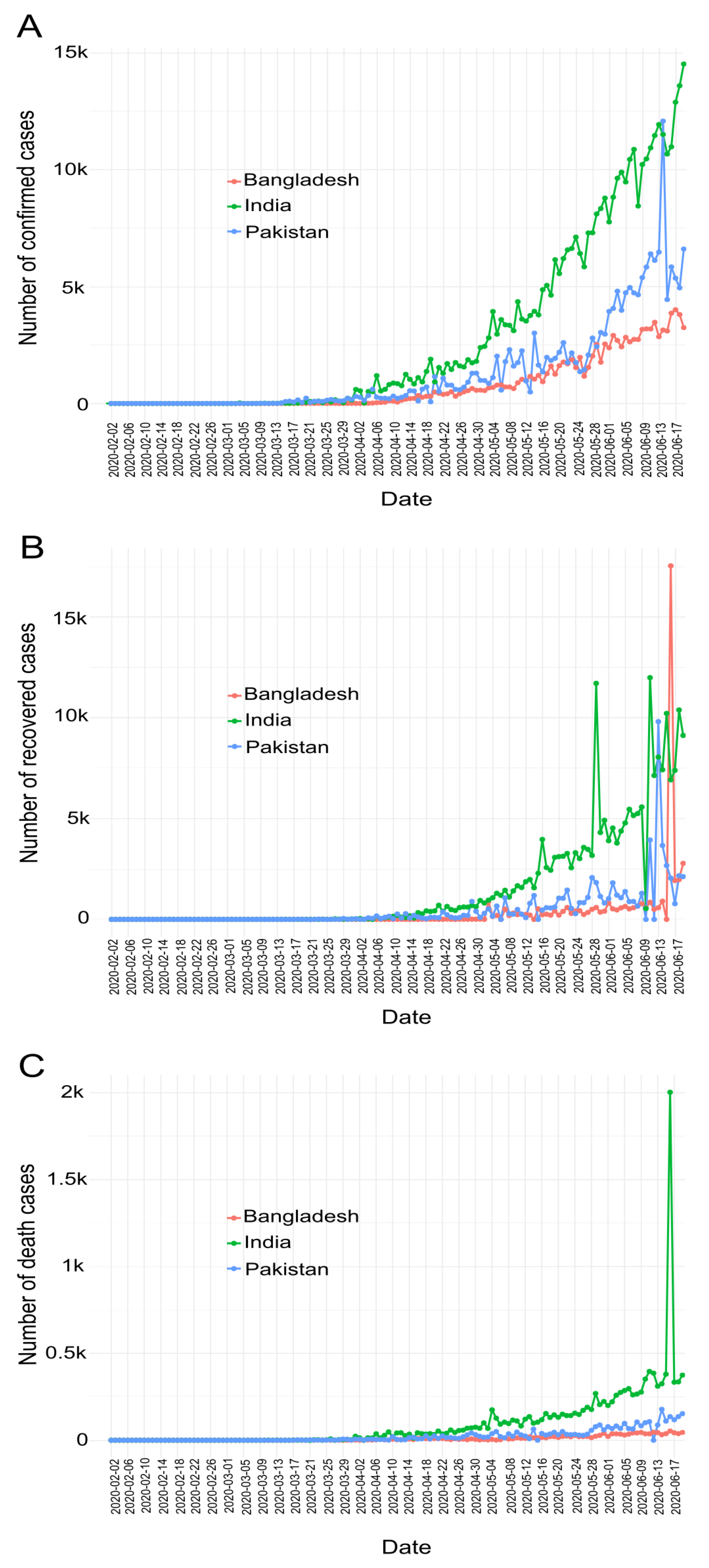
7.1. COVID-19 Cases
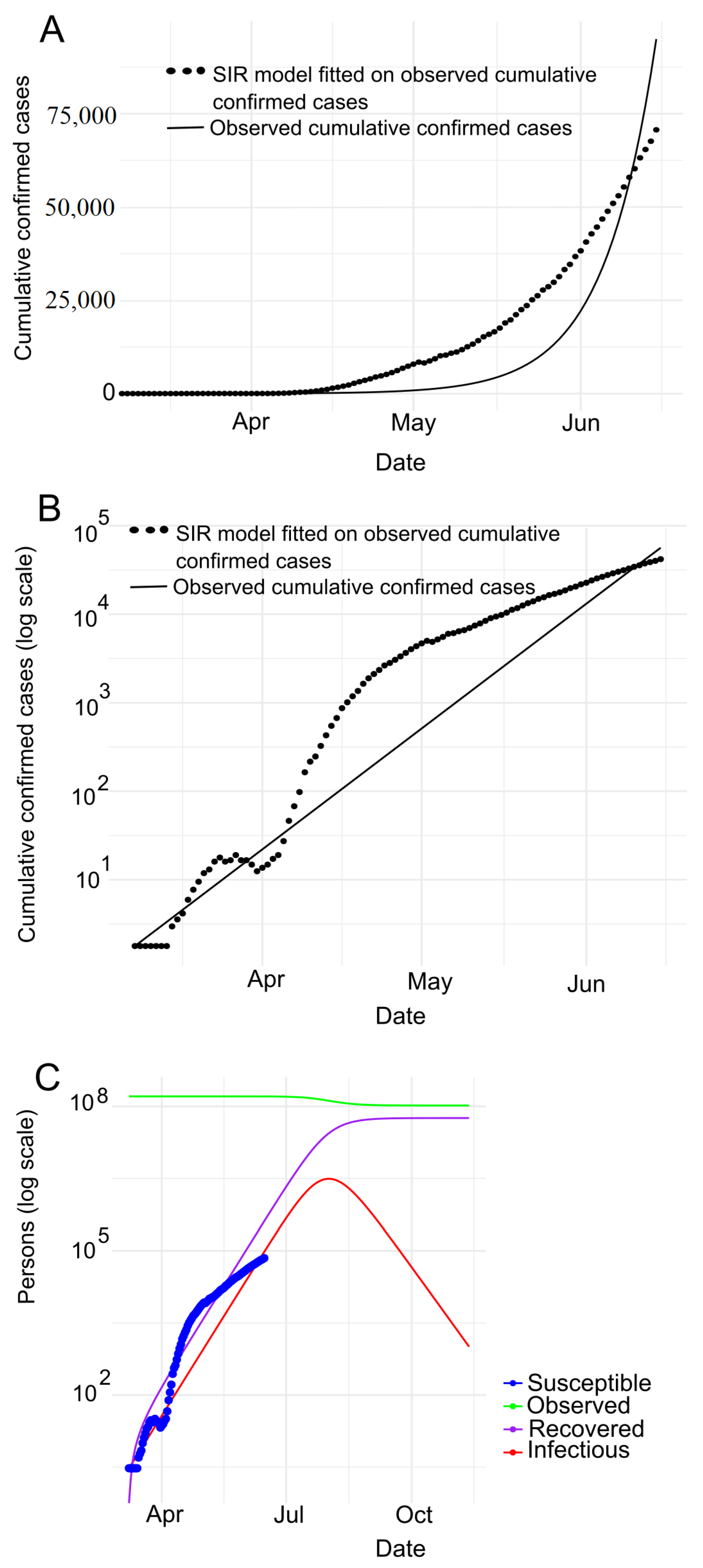
7.2. Prediction with SIR Model
7.3. COVID-19 Reproduction Number ( / ) Estimation
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Pneumonia of Unknown Cause—China. 2020. Available online: https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en (accessed on 30 June 2020).
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- WHO. Novel Coronavirus—China. 2020. Available online: https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en (accessed on 30 June 2020).
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- WHO. Naming the Coronavirus Disease (COVID-19) and the Virus that Causes It. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 30 June 2020).
- WHO. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). 2020. Available online: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-theoutbreak-of-novel-coronavirus-(2019-ncov) (accessed on 30 June 2020).
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 30 June 2020).
- Liu, Y.C.; Kuo, R.L.; Shih, S.R. COVID-19: The first documented coronavirus pandemic in history. Biomed. J. 2020. [Google Scholar] [CrossRef]
- Worldometer. COVID-19 CORONAVIRUS PANDEMIC. 2020. Available online: https://www.worldometers.info/coronavirus (accessed on 30 June 2020).
- Sulaiman, A. On Dynamical Analysis of the Data-Driven SIR model (COVID-19 Outbreak in Indonesia). medRxiv 2020. [Google Scholar]
- Zhao, S.; Musa, S.S.; Lin, Q.; Ran, J.; Yang, G.; Wang, W.; Lou, Y.; Yang, L.; Gao, D.; He, D.; et al. Estimating the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of January 2020: A data-driven modelling analysis of the early outbreak. J. Clin. Med. 2020, 9, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roda, W.C.; Varughese, M.B.; Han, D.; Li, M.Y. Why is it difficult to accurately predict the COVID-19 epidemic? Infect. Dis. Model. 2020, 5, 271–281. [Google Scholar] [CrossRef]
- Lin, Q.; Zhao, S.; Gao, D.; Lou, Y.; Yang, S.; Musa, S.S.; Wang, M.H.; Cai, Y.; Wang, W.; Yang, L.; et al. A conceptual model for the outbreak of Coronavirus disease 2019 (COVID-19) in Wuhan, China with individual reaction and governmental action. Int. J. Infect. Dis. 2020, 93, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Wang, X.; Li, Q.; Bragazzi, N.L.; Tang, S.; Xiao, Y.; Wu, J. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J. Clin. Med. 2020, 9, 462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Zeng, Z.; Wang, K.; Wong, S.S.; Liang, W.; Zanin, M.; Liu, P.; Cao, X.; Gao, Z.; Mai, Z.; et al. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J. Thorac. Dis. 2020, 12, 165. [Google Scholar] [CrossRef]
- Fanelli, D.; Piazza, F. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos Solitons Fractals 2020, 134, 109761. [Google Scholar] [CrossRef]
- Salgotra, R.; Gandomi, M.; Gandomi, A.H. Time Series Analysis and Forecast of the COVID-19 Pandemic in India using Genetic Programming. Chaos Solitons Fractals 2020, 138, 109945. [Google Scholar] [CrossRef] [PubMed]
- Djilali, S.; Ghanbari, B. Coronavirus pandemic: A predictive analysis of the peak outbreak epidemic in South Africa, Turkey, and Brazil. Chaos Solitons Fractals 2020, 138, 109971. [Google Scholar] [CrossRef]
- Roosa, K.; Lee, Y.; Luo, R.; Kirpich, A.; Rothenberg, R.; Hyman, J.; Yan, P.; Chowell, G. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect. Dis. Model. 2020, 5, 256–263. [Google Scholar] [CrossRef]
- Li, L.; Yang, Z.; Dang, Z.; Meng, C.; Huang, J.; Meng, H.; Wang, D.; Chen, G.; Zhang, J.; Peng, H.; et al. Propagation analysis and prediction of the COVID-19. Infect. Dis. Model. 2020, 5, 282–292. [Google Scholar] [CrossRef]
- Acuña-Zegarra, M.A.; Santana-Cibrian, M.; Velasco-Hernandez, J.X. Modeling behavioral change and COVID-19 containment in Mexico: A trade-off between lockdown and compliance. Math. Biosci. 2020, 325, 108370. [Google Scholar] [CrossRef]
- Jung, S.m.; Akhmetzhanov, A.R.; Hayashi, K.; Linton, N.M.; Yang, Y.; Yuan, B.; Kobayashi, T.; Kinoshita, R.; Nishiura, H. Real-time estimation of the risk of death from novel coronavirus (COVID-19) infection: Inference using exported cases. J. Clin. Med. 2020, 9, 523. [Google Scholar] [CrossRef] [Green Version]
- Powell, D.R.; Fair, J.; LeClaire, R.J.; Moore, L.M.; Thompson, D. Sensitivity analysis of an infectious disease model. In Proceedings of the International System Dynamics Conference, Boston, MA, USA, 17–21 July 2005. [Google Scholar]
- Feng, Z.; Glasser, J.W.; Hill, A.N. On the benefits of flattening the curve: A perspective. Math. Biosci. 2020, 326, 108389. [Google Scholar] [CrossRef] [PubMed]
- Dhanwant, J.N.; Ramanathan, V. Forecasting COVID 19 growth in India using Susceptible-Infected-Recovered (SIR) model. arXiv 2020, arXiv:2004.00696. [Google Scholar]
- Bertozzi, A.L.; Franco, E.; Mohler, G.; Short, M.B.; Sledge, D. The challenges of modeling and forecasting the spread of COVID-19. arXiv 2020, arXiv:2004.04741. [Google Scholar] [CrossRef]
- De Castro, C.A. SIR Model for COVID-19 calibrated with existing data and projected for Colombia. arXiv 2020, arXiv:2003.11230. [Google Scholar]
- Qi, C.; Karlsson, D.; Sallmen, K.; Wyss, R. Model studies on the COVID-19 pandemic in Sweden. arXiv 2020, arXiv:2004.01575. [Google Scholar]
- Shim, E.; Tariq, A.; Choi, W.; Lee, Y.; Chowell, G. Transmission potential and severity of COVID-19 in South Korea. Int. J. Infect. Dis. 2020, 93, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, D.; Giovanetti, M.; Vassallo, L.; Angeletti, S.; Ciccozzi, M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief 2020, 29, 105340. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins University Center for Systems Science and Engineering. Available online: https://github.com/CSSEGISandData/COVID-19 (accessed on 21 July 2020).
- Fattah, J.; Ezzine, L.; Aman, Z.; El Moussami, H.; Lachhab, A. Forecasting of demand using ARIMA model. Int. J. Eng. Bus. Manag. 2018, 10, 1847979018808673. [Google Scholar] [CrossRef] [Green Version]
- Calafiore, G.C.; Novara, C.; Possieri, C. A modified sir model for the covid-19 contagion in italy. arXiv 2020, arXiv:2003.14391. [Google Scholar]
- Kucharski, A.J.; Russell, T.W.; Diamond, C.; Liu, Y.; Edmunds, J.; Funk, S.; Eggo, R.M.; Sun, F.; Jit, M.; Munday, J.D.; et al. Early dynamics of transmission and control of COVID-19: A mathematical modelling study. Lancet Infect. Dis. 2020, 20, 553–558. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Yang, W.; Zhang, D.; Zhuge, C.; Hong, L. Epidemic analysis of COVID-19 in China by dynamical modeling. arXiv 2020, arXiv:2002.06563. [Google Scholar]
- Wangping, J.; Ke, H.; Yang, S.; Wenzhe, C.; Shengshu, W.; Shanshan, Y.; Jianwei, W.; Fuyin, K.; Penggang, T.; Jing, L.; et al. Extended SIR prediction of the epidemics trend of COVID-19 in Italy and compared with Hunan, China. Front. Med. 2020, 7, 169. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Chatterjee, K.; Kumar, A.; Shankar, S. Healthcare impact of COVID-19 epidemic in India: A stochastic mathematical model. Med J. Armed Forces India 2020, 76, 147–155. [Google Scholar] [CrossRef]
- Liang, K. Mathematical model of infection kinetics and its analysis for COVID-19, SARS and MERS. Infect. Genet. Evol. 2020, 82, 104306. [Google Scholar] [CrossRef]
- Ndairou, F.; Area, I.; Nieto, J.J.; Torres, D.F. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals 2020, 135, 109846. [Google Scholar] [CrossRef]
- Hellewell, J.; Abbott, S.; Gimma, A.; Bosse, N.I.; Jarvis, C.I.; Russell, T.W.; Munday, J.D.; Kucharski, A.J.; Edmunds, W.J.; Sun, F.; et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 2020, 8, e488–e496. [Google Scholar] [CrossRef] [Green Version]
- van den Driessche, P. Reproduction numbers of infectious disease models. Infect. Dis. Model. 2017, 2, 288–303. [Google Scholar] [CrossRef]
- Fraser, C.; Donnelly, C.A.; Cauchemez, S.; Hanage, W.P.; Van Kerkhove, M.D.; Hollingsworth, T.D.; Griffin, J.; Baggaley, R.F.; Jenkins, H.E.; Lyons, E.J.; et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science 2009, 324, 1557–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodpothong, P.; Auewarakul, P. Viral evolution and transmission effectiveness. World J. Virol. 2012, 1, 131. [Google Scholar] [CrossRef]
- Ma, J. Estimating epidemic exponential growth rate and basic reproduction number. Infect. Dis. Model. 2020, 5, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.; Heesterbeek, J. Model-consistent estimation of the basic reproduction number from the incidence of an emerging infection. J. Math. Biol. 2007, 55, 803. [Google Scholar] [CrossRef] [Green Version]
- Bettencourt, L.M.; Ribeiro, R.M. Real time bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS ONE 2008, 3, e2185. [Google Scholar] [CrossRef] [Green Version]
- Forsberg White, L.; Pagano, M. A likelihood-based method for real-time estimation of the serial interval and reproductive number of an epidemic. Stat. Med. 2008, 27, 2999–3016. [Google Scholar] [CrossRef] [Green Version]
- Obadia, T.; Haneef, R.; Boëlle, P.Y. The R0 package: A toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med. Informatics Decis. Mak. 2012, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wallinga, J.; Teunis, P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004, 160, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fine, P.; Eames, K.; Heymann, D.L. Herd immunity: A rough guide. Clin. Infect. Dis. 2011, 52, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Ganyani, T.; Kremer, C.; Chen, D.; Torneri, A.; Faes, C.; Wallinga, J.; Hens, N. Estimating the generation interval for COVID-19 based on symptom onset data. MedRxiv 2020. [Google Scholar]
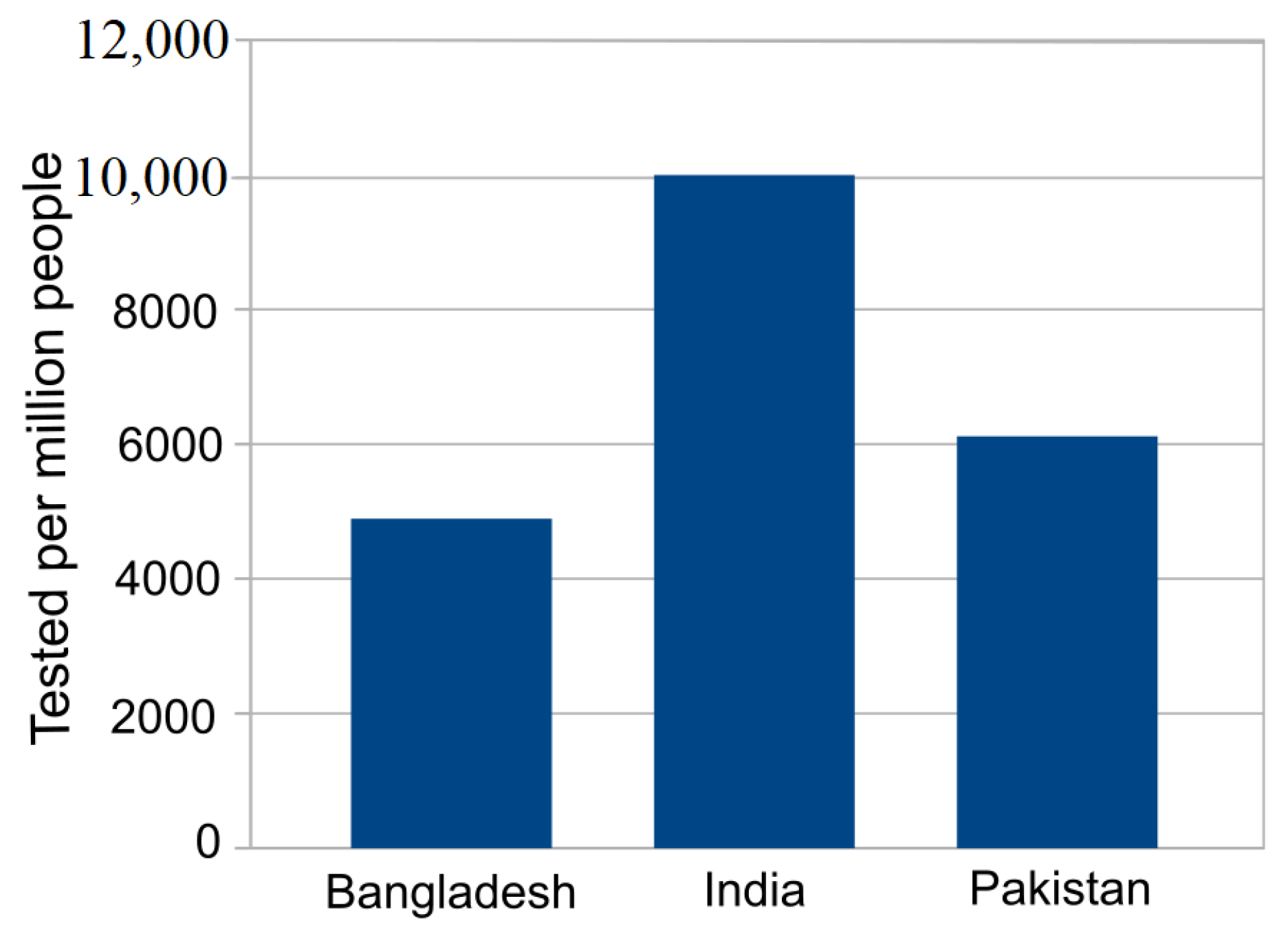
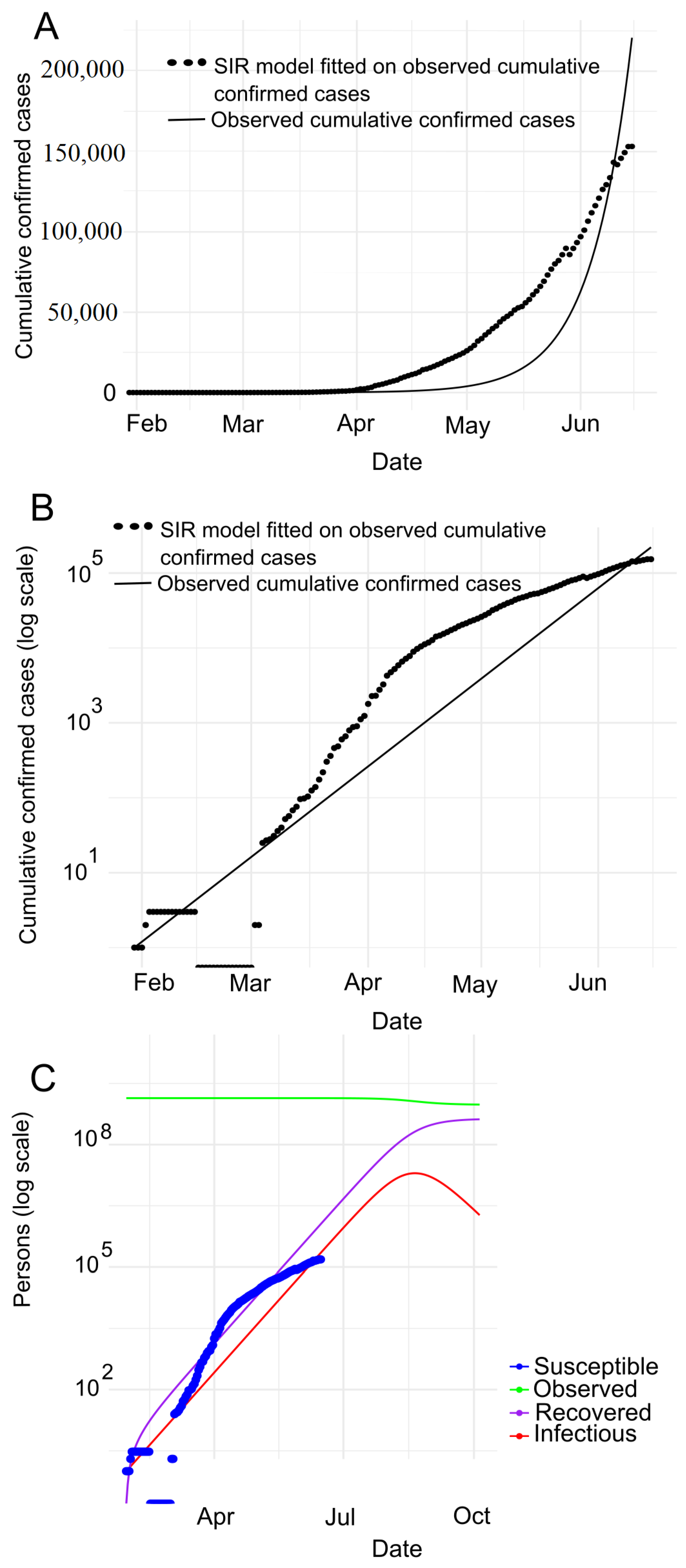
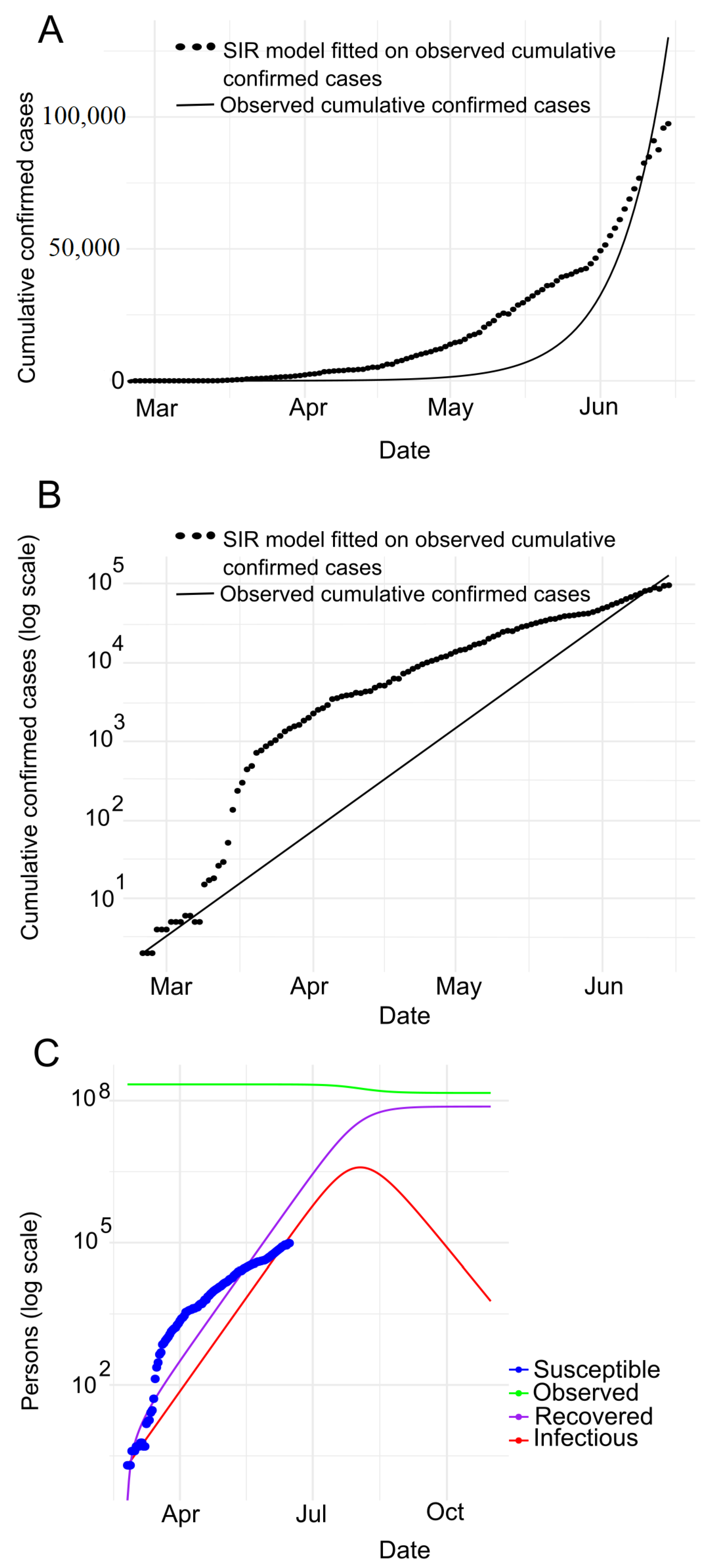
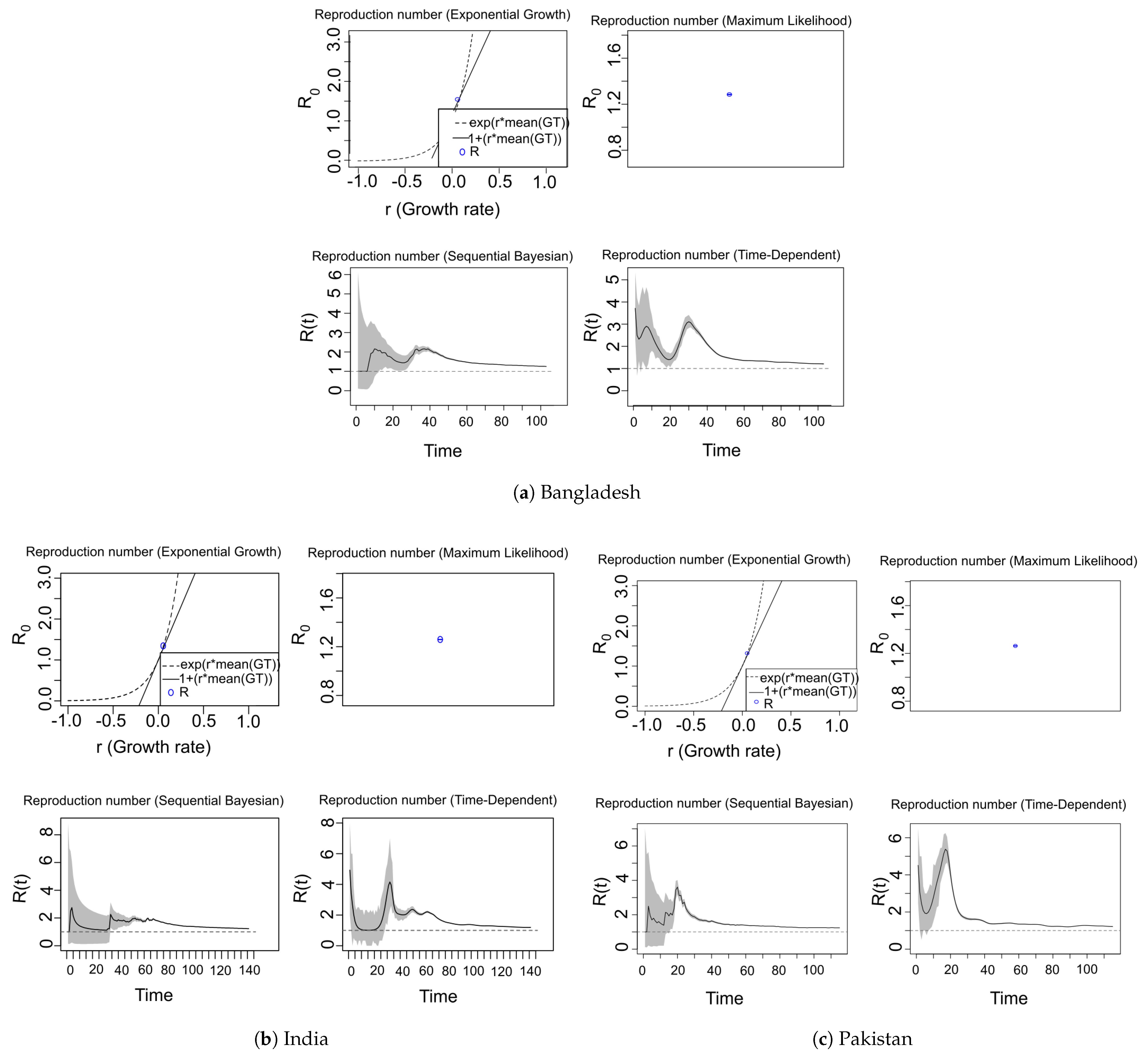
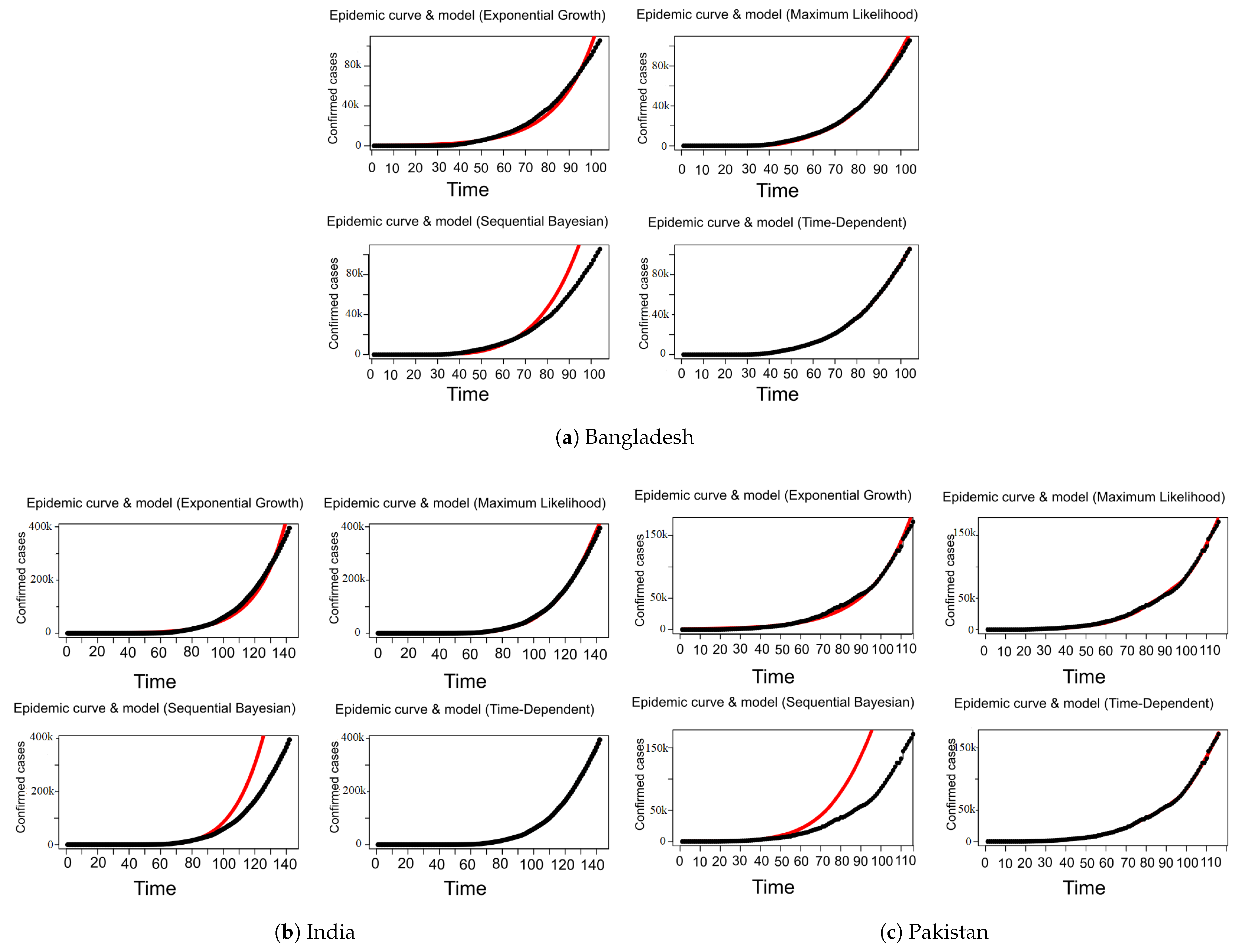
| Country | Date of the First Case | End Date | Total Confirmed Cases | Total Death Cases | Total Recovered Cases | Population | Tested per Million People |
|---|---|---|---|---|---|---|---|
| Bangladesh | 2020-03-08 | 2020-06-19 | 105,535 | 1388 | 42,945 | 161,376,708 | 4892 |
| India | 2020-01-30 | 2020-06-19 | 395,048 | 12,948 | 213,831 | 1,380,004,385 | 9995 |
| Pakistan | 2020-02-25 | 2020-06-19 | 171,666 | 3382 | 63,504 | 22,0695,321 | 6117 |
| The Predicted Values for the Following Parameters | Bangladesh | India | Pakistan |
|---|---|---|---|
| Infection Rate, | 0.5524 | 0.5449 | 0.550 |
| Recovery rate, | 0.4475 | 0.4550 | 0.449 |
| 1.234 | 1.197 | 1.22 | |
| Herd immunity threshold ( | 18.97 % of population | 16.49% of population | 18.18% of population |
| Peak of Pandemic | 2020-08-01 | 2020-08-20 | 2020-08-03 |
| Maximum Infected | 3,109,321 | 19,884,176 | 3,891,427 |
| Severe cases (assume 20% of Infected cases) | 621,864 | 3,976,835 | 778,285 |
| Patients need intensive care (assume 6% of Infected cases) | 186,560 | 1,193,051 | 233,485 |
| Deaths assumed for 3.5% fatality rate | 108,826 | 695,946 | 136,200 |
| Methods | SIR | EG | ML | TD | SB |
|---|---|---|---|---|---|
| Reproduction number | [CI.lower, CI.upper] | [CI.lower, CI.upper] | Rmean(t) [Rlow(t), Rhigh(t)] | Rmean(t)[Rlow(t), Rhigh(t)] | |
| Bangladesh | 1.234 | 1.380 [1.380, 1.381] | 1.288 [1.286, 1.290] | 1.746 [1.209, 3.715] | 1.555 [1.000, 2.16] |
| India | 1.197 | 1.344 [1.344, 1.344] | 1.259 [1.257, 1.260] | 1.668 [1.007, 4.928] | 1.507 [1.00, 2.75] |
| Pakistan | 1.220 | 1.319 [1.318, 1.319] | 1.264 [1.262, 1.266] | 1.774 [1.202, 5.381] | 1.560 [1.00, 3.61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, B.C.; Alom, Z.; Hu, H.; Rahman, M.M.; Baowaly, M.K.; Aung, Z.; Azim, M.A.; Moni, M.A. COVID-19 Pandemic Outbreak in the Subcontinent: A Data Driven Analysis. J. Pers. Med. 2021, 11, 889. https://doi.org/10.3390/jpm11090889
Singh BC, Alom Z, Hu H, Rahman MM, Baowaly MK, Aung Z, Azim MA, Moni MA. COVID-19 Pandemic Outbreak in the Subcontinent: A Data Driven Analysis. Journal of Personalized Medicine. 2021; 11(9):889. https://doi.org/10.3390/jpm11090889
Chicago/Turabian StyleSingh, Bikash Chandra, Zulfikar Alom, Haibo Hu, Mohammad Muntasir Rahman, Mrinal Kanti Baowaly, Zeyar Aung, Mohammad Abdul Azim, and Mohammad Ali Moni. 2021. "COVID-19 Pandemic Outbreak in the Subcontinent: A Data Driven Analysis" Journal of Personalized Medicine 11, no. 9: 889. https://doi.org/10.3390/jpm11090889
APA StyleSingh, B. C., Alom, Z., Hu, H., Rahman, M. M., Baowaly, M. K., Aung, Z., Azim, M. A., & Moni, M. A. (2021). COVID-19 Pandemic Outbreak in the Subcontinent: A Data Driven Analysis. Journal of Personalized Medicine, 11(9), 889. https://doi.org/10.3390/jpm11090889









