Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians
Abstract
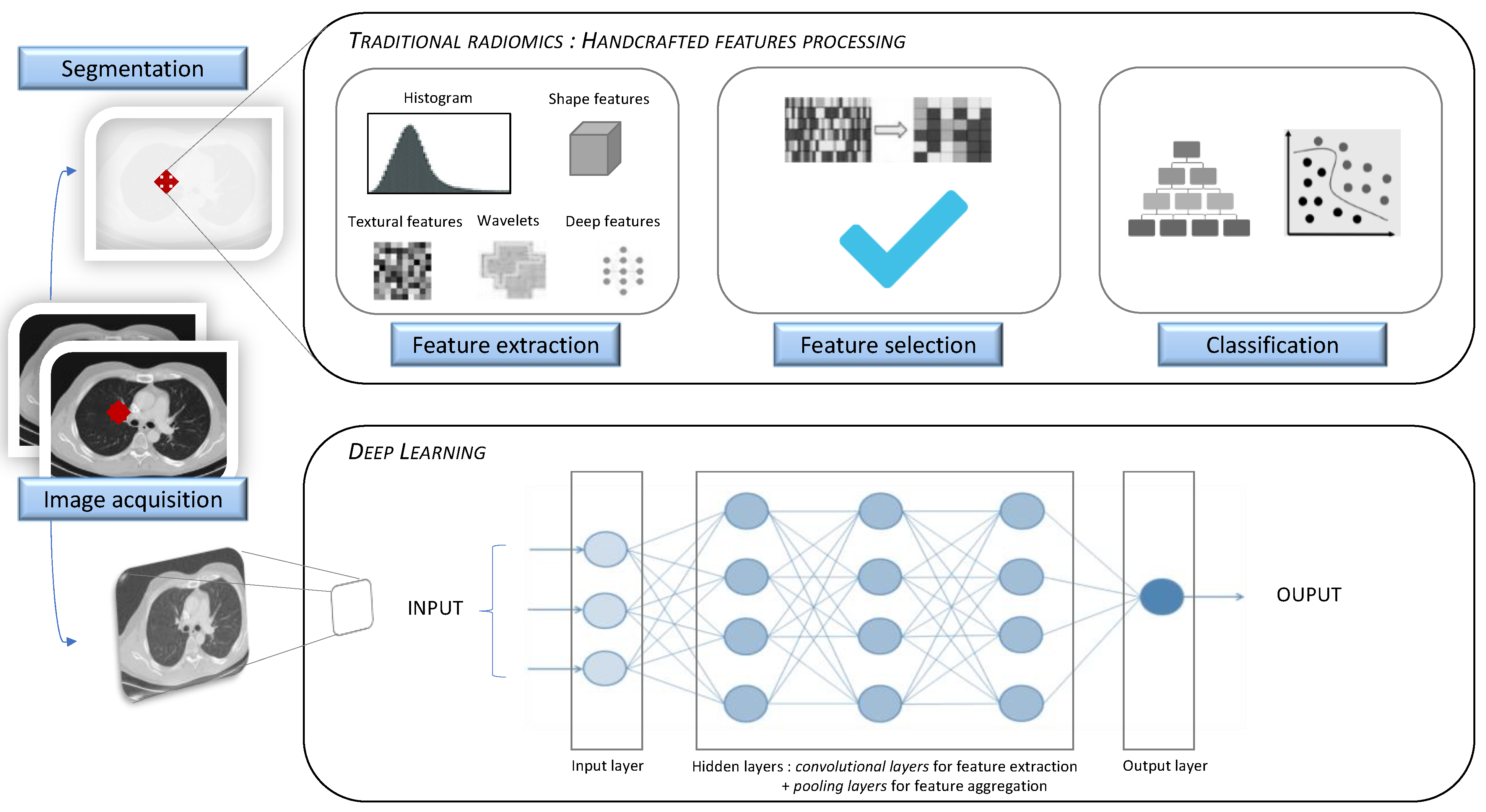
:1. Introduction
2. The Role of Radiomics in Lung Diseases
2.1. Lung Nodules
2.2. Cancer
2.3. Obstructive Lung Diseases
2.3.1. COPD
2.3.2. Asthma
2.4. Interstitial Lung Diseases
2.5. Vascular Lung Diseases
2.6. Pleural Diseases
3. Challenges and Limitations
- -
- Image acquisition: There is a disparity in acquisition parameters, as there are no real standardized imaging protocols. For example, there are differences in dose administration, reconstruction kernels, section thickness between the different imaging centers, and modalities. Moreover, variations in inspiratory effort can modify lung attenuation and volume, possibly leading to misinterpretation, affecting both threshold- and histogram-based quantification.
- -
- Image segmentation: There is an inevitable intra- and inter-observer variability in manual segmentation methods, which could be improved by the use of semi- or fully automatic techniques. Similarly, intra-lesional heterogeneity can be a real challenge for accurate segmentation, as well as motion artifacts or noisy background due to low-dose scanning.
- -
- Feature extraction: There is a risk of confusion between the signal of the ROI and the background noise, which could be improved by the development of filtering techniques, or by resampling strategies. Moreover, feature selection is facing disparities imputable to human error, which could be improved by the implementation of deep learning methods such as CNN.
- -
- Statistical analysis: There is a disproportion between the tremendous number of possible features and the small population, generating a high rate of false positives (elegantly called “curse of dimensionality” [51]), which could be improved by the development of statistical corrections or cross-validation.
- -
- Implementation and reproducibility: There is a lack of reproducibility between research groups, which could be improved by increasing the access to full data, extraction software and statistical methods. Another example of a limitation is misinterpretation from a trained algorithm. Indeed, a specific algorithm can only define a disease for which it was trained, possibly leading to the false suggestion of diseases sharing some common features.
- -
- Robustness and Explainability of Artificial Intelligence: There are numerous issues to be addressed, concerning the application of AI in real life. For example, the use of extensive data in the development of machine learning models does not imply the automatic understanding of underlying mechanisms linking data. Moreover, AI systems can face concerns regarding reliability, as they may accumulate edge cases that are not taken into account by the algorithm. Lastly, the question of data protection must be raised, as potential matters concerning confidentiality can surface [92].
4. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| PET | Positron Emission Tomography |
| DL | Deep Learning |
| IPF | Idiopathic Pulmonary Fibrosis |
| CPFE | Combined Pulmonary Fibrosis and Emphysema |
| QIN | Quantitative Imaging Network |
| VTD | Volume Doubling Time |
| SSPNs | Small Solitary Pulmonary Nodules |
| AUC | Area Under the Curve |
| HRCT | High-Resolution Computed Tomography |
| CNN | Convolutional Neural Networks |
| CAD | Computer-Aided Detection/Diagnosis |
| NLST | National Lung Screening Trial |
| EGFR | Epidermal Growth Factor Receptor |
| NSCLC | Non-Small Cell Lung Carcinoma |
| GTV | Gross-Tumor Volume |
| TKI | Tyrosine Kinase Inhibitors |
| COPD | Chronic Obstructive Pulmonary Disease |
| HU | Hounsfield Unit |
| PRM | Parametric Response Mapping |
| PFTs | Pulmonary Function Tests |
| LVR | Lung Volume Reduction |
| ICS | Inhaled Corticosteroids |
| ILD | Interstitial Lung Diseases |
| NSIP | Non-Specific Interstitial Pneumonia |
| CALIPER | Computer-Aided Lung Informatics for Pathology Evaluation and Rating |
| FVC | Forced Vital Capacity |
| QLF | Quantitative Lung Fibrosis |
| PH | Pulmonary Hypertension |
| PAH | Pulmonary Arterial Hypertension |
| RHC | Right Heart Catheterization |
| ROI | Region of Interest |
References
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D.O.; Ryan, A.; Fuhrman, C.; Schuchert, M.; Shapiro, S.; Siegfried, J.M.; Weissfeld, J. Doubling Times and CT Screen–Detected Lung Cancers in the Pittsburgh Lung Screening Study. Am. J. Respir. Crit. Care Med. 2012, 185, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, M.; Sone, S.; Takashima, S.; Li, F.; Yang, Z.G.; Maruyama, Y.; Watanabe, T. Growth rate of small lung cancers detected on mass CT screening. Br. J. Radiol. 2000, 73, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.P.; Berman, E.J.; Kaur, M.; Babb, J.S.; Bomsztyk, E.; Greenberg, A.K.; Naidich, D.P.; Rusinek, H. Pulmonary Nodules: Growth Rate Assessment in Patients by Using Serial CT and Three-dimensional Volumetry. Radiology 2012, 262, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, R.J. The Quantitative Imaging Network in Precision Medicine. Tomography 2016, 2, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Ather, S.; Kadir, T.; Gleeson, F. Artificial intelligence and radiomics in pulmonary nodule management: Current status and future applications. Clin. Radiol. 2020, 75, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-H.; Chang, C.-K.; Tu, C.-Y.; Liao, W.-C.; Wu, B.-R.; Chou, K.-T.; Chiou, Y.-R.; Yang, S.-N.; Zhang, G.; Huang, T.-C. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS ONE 2018, 13, e0192002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horeweg, N.; van der Aalst, C.M.; Vliegenthart, R.; Zhao, Y.; Xie, X.; Scholten, E.T.; Mali, W.; Thunnissen, E.; Weenink, C.; Groen, H.J.M.; et al. Volumetric computed tomography screening for lung cancer: Three rounds of the NELSON trial. Eur. Respir. J. 2013, 42, 1659–1667. [Google Scholar] [CrossRef] [Green Version]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Q.; Ren, Y.; Hu, H.; Zhao, J. Automatic lung nodule classification with radiomics approach. In Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations; Zhang, J., Cook, T.S., Eds.; International Society for Optics and Photonics: San Diego, CA, USA, 2016; p. 978906. Available online: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2220768 (accessed on 21 June 2021).
- Hawkins, S.; Wang, H.; Liu, Y.; Garcia, A.; Stringfield, O.; Krewer, H.; Li, Q.; Cherezov, D.; Gatenby, R.A.; Balagurunathan, Y.; et al. Predicting Malignant Nodules from Screening CT Scans. J. Thorac. Oncol. 2016, 11, 2120–2128. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Park, S.; Yan, R.; Lee, J.; Chu, L.C.; Lin, C.T.; Hussien, A.; Rathmell, J.; Thomas, B.; Chen, C.; et al. Added Value of Computer-aided CT Image Features for Early Lung Cancer Diagnosis with Small Pulmonary Nodules: A Matched Case-Control Study. Radiology 2018, 286, 286–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uthoff, J.; Stephens, M.J.; Newell, J.D.; Hoffman, E.A.; Larson, J.; Koehn, N.; De Stefano, F.A.; Lusk, C.M.; Wenzlaff, A.S.; Watza, D.; et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med. Phys. 2019, 46, 3207–3216. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Chen, H.; Liang, M.; Li, K.; Gao, J.; Qin, P.; Ding, X.; Li, X.; Liu, X. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant. Imaging Med. Surg. 2019, 9, 263–272. [Google Scholar] [CrossRef] [PubMed]
- ACR. Lung CT Screening Reporting & Data System (Lung-RADS). Available online: https://www.acr.org (accessed on 20 April 2021).
- Lee, S.H.; Lee, S.M.; Goo, J.M.; Kim, K.-G.; Kim, Y.J.; Park, C.M. Usefulness of Texture Analysis in Differentiating Transient from Persistent Part-solid Nodules(PSNs): A Retrospective Study. PLoS ONE 2014, 9, e85167. [Google Scholar]
- Autrusseau, P.-A.; Labani, A.; De Marini, P.; Leyendecker, P.; Hintzpeter, C.; Ortlieb, A.-C.; Calhoun, M.; Goldberg, I.; Roy, C.; Ohana, M. Radiomics in the evaluation of lung nodules: Intrapatient concordance between full-dose and ultra-low-dose chest computed tomography. Diagn. Interv. Imaging 2021, 102, 233–239. [Google Scholar] [CrossRef]
- Maldonado, F.; Varghese, C.; Rajagopalan, S.; Duan, F.; Balar, A.B.; Lakhani, D.A.; Antic, S.L.; Massion, P.P.; Johnson, T.F.; Karwoski, R.A.; et al. Validation of the BRODERS classifier (Benign versus aggRessive nODule Evaluation using Radiomic Stratification), a novel HRCT-based radiomic classifier for indeterminate pulmonary nodules. Eur. Respir. J. 2021, 57, 2002485. [Google Scholar] [CrossRef]
- Murphy, A.; Skalski, M.; Gaillard, F. The utilisation of convolutional neural networks in detecting pulmonary nodules: A review. Br. J. Radiol. 2018, 91, 20180028. [Google Scholar] [CrossRef]
- da Silva, G.L.F.; Valente, T.L.A.; Silva, A.C.; de Paiva, A.C.; Gattass, M. Convolutional neural network-based PSO for lung nodule false positive reduction on CT images. Comput. Methods Programs Biomed. 2018, 162, 109–118. [Google Scholar] [CrossRef]
- Li, S.; Xu, P.; Li, B.; Chen, L.; Zhou, Z.; Hao, H.; Duan, Y.; Folkert, M.; Ma, J.; Huang, S.; et al. Predicting lung nodule malignancies by combining deep convolutional neural network and handcrafted features. Phys. Med. Biol. 2019, 64, 175012. [Google Scholar] [CrossRef] [Green Version]
- Mehta, K.; Jain, A.; Mangalagiri, J.; Menon, S.; Nguyen, P.; Chapman, D.R. Lung Nodule Classification Using Biomarkers, Volumetric Radiomics, and 3D CNNs. J. Digit. Imaging 2021. [Google Scholar] [CrossRef]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J.W.L. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front. Oncol. 2016, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Digumarthy, S.R.; Padole, A.M.; Rastogi, S.; Price, M.; Mooradian, M.J.; Sequist, L.V.; Kalra, M.K. Predicting malignant potential of subsolid nodules: Can radiomics preempt longitudinal follow up CT? Cancer Imaging 2019, 19, 36. [Google Scholar] [CrossRef] [Green Version]
- Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features. Tomography 2016, 2, 430–437. [CrossRef] [PubMed]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A.; et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- The National Lung Screening Trial Research Team Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [CrossRef] [PubMed] [Green Version]
- National Lung Screening Trial Research Team Lung Cancer Incidence and Mortality with Extended Follow-up in the National Lung Screening Trial. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2019, 14, 1732–1742.
- Refaee, T.; Wu, G.; Ibrahim, A.; Halilaj, I.; Leijenaar, R.T.H.; Rogers, W.; Gietema, H.A.; Hendriks, L.E.L.; Lambin, P.; Woodruff, H.C. The Emerging Role of Radiomics in COPD and Lung Cancer. Respiration 2020, 99, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Maldonado, F.; Boland, J.M.; Raghunath, S.; Aubry, M.C.; Bartholmai, B.J.; Deandrade, M.; Hartman, T.E.; Karwoski, R.A.; Rajagopalan, S.; Sykes, A.-M.; et al. Noninvasive characterization of the histopathologic features of pulmonary nodules of the lung adenocarcinoma spectrum using computer-aided nodule assessment and risk yield (CANARY)—A pilot study. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2013, 8, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Balagurunathan, Y.; Atwater, T.; Antic, S.; Li, Q.; Walker, R.C.; Smith, G.T.; Massion, P.P.; Schabath, M.B.; Gillies, R.J. Radiological Image Traits Predictive of Cancer Status in Pulmonary Nodules. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 1442–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Tao, G.; Zhu, L.; Wang, G.; Li, Z.; Ye, J.; Chen, Q. Prediction of pathologic stage in non-small cell lung cancer using machine learning algorithm based on CT image feature analysis. BMC Cancer 2019, 19, 464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Chen, B.; Liu, X.; Song, J.; Fang, M.; Hu, C.; Dong, D.; Li, W.; Tian, J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl. Oncol. 2018, 11, 94–101. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448.e6. [Google Scholar] [CrossRef] [Green Version]
- Rios Velazquez, E.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.J.; Ganeshan, B.; Miles, K.A.; Campbell, D.H.; Cheung, P.Y.; Frank, S.; Korn, R.L. Noninvasive Image Texture Analysis Differentiates K-ras Mutation from Pan-Wildtype NSCLC and Is Prognostic. PLoS ONE 2014, 9, e100244. [Google Scholar]
- Tang, C.; Hobbs, B.; Amer, A.; Li, X.; Behrens, C.; Canales, J.R.; Cuentas, E.P.; Villalobos, P.; Fried, D.; Chang, J.Y.; et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 1922. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Woodruff, H.C.; Shen, J.; Refaee, T.; Sanduleanu, S.; Ibrahim, A.; Leijenaar, R.T.H.; Wang, R.; Xiong, J.; Bian, J.; et al. Diagnosis of Invasive Lung Adenocarcinoma Based on Chest CT Radiomic Features of Part-Solid Pulmonary Nodules: A Multicenter Study. Radiology 2020, 297, E282. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.H.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J.W.L. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Song, J.; Liu, Z.; Zhong, W.; Huang, Y.; Ma, Z.; Dong, D.; Liang, C.; Tian, J. Non-small cell lung cancer: Quantitative phenotypic analysis of CT images as a potential marker of prognosis. Sci. Rep. 2016, 6, 38282. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Huang, Y.; Yan, L.; Zheng, J.; Liang, C.; Liu, Z. China Radiomics-based predictive risk score: A scoring system for preoperatively predicting risk of lymph node metastasis in patients with resectable non-small cell lung cancer. Chin. J. Cancer Res. 2019, 31, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J.W.L. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLOS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira Junior, J.R.; Koenigkam-Santos, M.; Cipriano, F.E.G.; Fabro, A.T.; Azevedo-Marques, P.M. de Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput. Methods Programs Biomed. 2018, 159, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Mattonen, S.A.; Palma, D.A.; Johnson, C.; Louie, A.V.; Landis, M.; Rodrigues, G.; Chan, I.; Etemad-Rezai, R.; Yeung, T.P.C.; Senan, S.; et al. Detection of Local Cancer Recurrence After Stereotactic Ablative Radiation Therapy for Lung Cancer: Physician Performance Versus Radiomic Assessment. Int. J. Radiat. Oncol. 2016, 94, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Chetan, M.R.; Gleeson, F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: Current status, challenges and future perspectives. Eur. Radiol. 2021, 31, 1049–1058. [Google Scholar] [CrossRef]
- Bera, K.; Velcheti, V.; Madabhushi, A. Novel Quantitative Imaging for Predicting Response to Therapy: Techniques and Clinical Applications. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 1008–1018. [Google Scholar] [CrossRef]
- Coroller, T.P.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother. Oncol. 2016, 119, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Park, C.M.; Keam, B.; Park, S.J.; Kim, M.; Kim, T.M.; Kim, D.-W.; Heo, D.S.; Goo, J.M. The prognostic value of CT radiomic features for patients with pulmonary adenocarcinoma treated with EGFR tyrosine kinase inhibitors. PLoS ONE 2017, 12, e0187500. [Google Scholar] [CrossRef] [Green Version]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Lafata, K.J.; Zhou, Z.; Liu, J.-G.; Hong, J.; Kelsey, C.R.; Yin, F.-F. An Exploratory Radiomics Approach to Quantifying Pulmonary Function in CT Images. Sci. Rep. 2019, 9, 11509. [Google Scholar] [CrossRef] [Green Version]
- Gevenois, P.A.; de Maertelaer, V.; De Vuyst, P.; Zanen, J.; Yernault, J.C. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. Am. J. Respir. Crit. Care Med. 1995, 152, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Zanen, J.; de Maertelaer, V.; Gevenois, P.A. Pulmonary emphysema: Objective quantification at multi-detector row CT--comparison with macroscopic and microscopic morphometry. Radiology 2006, 238, 1036–1043. [Google Scholar] [CrossRef]
- de Jong, P.A.; Müller, N.L.; Paré, P.D.; Coxson, H.O. Computed tomographic imaging of the airways: Relationship to structure and function. Eur. Respir. J. 2005, 26, 140–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madani, A.; Van Muylem, A.; Gevenois, P.A. Pulmonary Emphysema: Effect of Lung Volume on Objective Quantification at Thin-Section CT. Radiology 2010, 257, 260–268. [Google Scholar] [CrossRef]
- Bankier, A.A.; De Maertelaer, V.; Keyzer, C.; Gevenois, P.A. Pulmonary Emphysema: Subjective Visual Grading versus Objective Quantification with Macroscopic Morphometry and Thin-Section CT Densitometry. Radiology 1999, 211, 851–858. [Google Scholar] [CrossRef]
- Mohamed Hoesein, F.A.A.; de Hoop, B.; Zanen, P.; Gietema, H.; Kruitwagen, C.L.J.J.; van Ginneken, B.; Isgum, I.; Mol, C.; van Klaveren, R.J.; Dijkstra, A.E.; et al. CT-quantified emphysema in male heavy smokers: Association with lung function decline. Thorax 2011, 66, 782–787. [Google Scholar] [CrossRef] [Green Version]
- Pompe, E.; van Rikxoort, E.M.; Schmidt, M.; Rühaak, J.; Estrella, L.G.; Vliegenthart, R.; Oudkerk, M.; de Koning, H.J.; van Ginneken, B.; de Jong, P.A.; et al. Parametric response mapping adds value to current computed tomography biomarkers in diagnosing chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2015, 191, 1084–1086. [Google Scholar] [CrossRef]
- Hackx, M.; Francotte, D.; Garcia, T.S.; Van Muylem, A.; Walsdorff, M.; Gevenois, P.A. Effect of total lung capacity, gender and height on CT airway measurements. Br. J. Radiol. 2017, 90, 20160898. [Google Scholar] [CrossRef]
- Ginsburg, S.B.; Lynch, D.A.; Bowler, R.P.; Schroeder, J.D. Automated Texture-based Quantification of Centrilobular Nodularity and Centrilobular Emphysema in Chest CT Images. Acad. Radiol. 2012, 19, 1241–1251. [Google Scholar] [CrossRef] [Green Version]
- Martini, K.; Frauenfelder, T. Advances in imaging for lung emphysema. Ann. Transl. Med. 2020, 8, 1467. [Google Scholar] [CrossRef]
- Lynch, D.A.; Austin, J.H.M.; Hogg, J.C.; Grenier, P.A.; Kauczor, H.-U.; Bankier, A.A.; Barr, R.G.; Colby, T.V.; Galvin, J.R.; Gevenois, P.A.; et al. CT-Definable Subtypes of Chronic Obstructive Pulmonary Disease: A Statement of the Fleischner Society. Radiology 2015, 277, 192–205. [Google Scholar] [CrossRef] [Green Version]
- Occhipinti, M.; Paoletti, M.; Bartholmai, B.J.; Rajagopalan, S.; Karwoski, R.A.; Nardi, C.; Inchingolo, R.; Larici, A.R.; Camiciottoli, G.; Lavorini, F.; et al. Spirometric assessment of emphysema presence and severity as measured by quantitative CT and CT-based radiomics in COPD. Respir. Res. 2019, 20, 101. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.H.; Seo, J.B.; Lee, S.M.; Kim, N.; Yun, J.; Hwang, J.E.; Lee, J.S.; Oh, Y.-M.; Do Lee, S.; Loh, L.-C.; et al. Radiomics approach for survival prediction in chronic obstructive pulmonary disease. Eur. Radiol. 2021. [Google Scholar] [CrossRef]
- Cho, Y.H.; Lee, S.M.; Seo, J.B.; Kim, N.; Bae, J.P.; Lee, J.S.; Oh, Y.-M.; Do-Lee, S. Quantitative assessment of pulmonary vascular alterations in chronic obstructive lung disease: Associations with pulmonary function test and survival in the KOLD cohort. Eur. J. Radiol. 2018, 108, 276–282. [Google Scholar] [CrossRef]
- Stockley, R.A.; Parr, D.G.; Piitulainen, E.; Stolk, J.; Stoel, B.C.; Dirksen, A. Therapeutic efficacy of alpha-1 antitrypsin augmentation therapy on the loss of lung tissue: An integrated analysis of 2 randomised clinical trials using computed tomography densitometry. Respir. Res. 2010, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Chandra, D.; Lipson, D.A.; Hoffman, E.A.; Hansen-Flaschen, J.; Sciurba, F.C.; DeCamp, M.M.; Reilly, J.J.; Washko, G.R. Perfusion Scintigraphy and Patient Selection for Lung Volume Reduction Surgery. Am. J. Respir. Crit. Care Med. 2010, 182, 937–946. [Google Scholar] [CrossRef] [Green Version]
- Milanese, G.; Silva, M.; Sverzellati, N. Lung volume reduction of pulmonary emphysema: The radiologist task. Curr. Opin. Pulm. Med. 2016, 22, 179–186. [Google Scholar] [CrossRef]
- Gupta, S.; Hartley, R.; Khan, U.T.; Singapuri, A.; Hargadon, B.; Monteiro, W.; Pavord, I.D.; Sousa, A.R.; Marshall, R.P.; Subramanian, D.; et al. Quantitative computed tomography–derived clusters: Redefining airway remodeling in asthmatic patients. J. Allergy Clin. Immunol. 2014, 133, 729–738.e18. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Hoffman, E.A.; Wenzel, S.E.; Castro, M.; Fain, S.; Jarjour, N.; Schiebler, M.L.; Chen, K.; Lin, C.-L. Quantitative computed tomographic imaging–based clustering differentiates asthmatic subgroups with distinctive clinical phenotypes. J. Allergy Clin. Immunol. 2017, 140, 690–700.e8. [Google Scholar] [CrossRef] [Green Version]
- Tunon-de-Lara, J.-M.; Laurent, F.; Giraud, V.; Perez, T.; Aguilaniu, B.; Meziane, H.; Basset-Merle, A.; Chanez, P. Air trapping in mild and moderate asthma: Effect of inhaled corticosteroids. J. Allergy Clin. Immunol. 2007, 119, 583–590. [Google Scholar] [CrossRef]
- Walsh, S.L.F.; Calandriello, L.; Silva, M.; Sverzellati, N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: A case-cohort study. Lancet Respir. Med. 2018, 6, 837–845. [Google Scholar] [CrossRef]
- Schniering, J.; Gabrys, H.; Brunner, M.; Distler, O.; Guckenberger, M.; Bogowicz, M.; Vuong, D.; Karava, K.; Müller, C.; Frauenfelder, T.; et al. Computed-tomography-based radiomics features for staging of interstitial lung disease—Transferability from experimental to human lung fibrosis—A proof-of-concept study. Eur. Respir. Soc. 2019, 54, PA4806. Available online: http://erj.ersjournals.com/lookup/doi/10.1183/13993003.congress-2019.PA4806 (accessed on 23 October 2020).
- Stefano, A.; Gioè, M.; Russo, G.; Palmucci, S.; Torrisi, S.E.; Bignardi, S.; Basile, A.; Comelli, A.; Benfante, V.; Sambataro, G.; et al. Performance of Radiomics Features in the Quantification of Idiopathic Pulmonary Fibrosis from HRCT. Diagnostics 2020, 10, 306. [Google Scholar] [CrossRef] [PubMed]
- Martini, K.; Baessler, B.; Bogowicz, M.; Blüthgen, C.; Mannil, M.; Tanadini-Lang, S.; Schniering, J.; Maurer, B.; Frauenfelder, T. Applicability of radiomics in interstitial lung disease associated with systemic sclerosis: Proof of concept. Eur. Radiol. 2021, 31, 1987–1998. [Google Scholar] [CrossRef]
- Ungprasert, P.; Wilton, K.M.; Ernste, F.C.; Kalra, S.; Crowson, C.S.; Rajagopalan, S.; Bartholmai, B.J. Novel Assessment of Interstitial Lung Disease Using the “Computer-Aided Lung Informatics for Pathology Evaluation and Rating” (CALIPER) Software System in Idiopathic Inflammatory Myopathies. Lung 2017, 195, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Brown, M.S.; Chong, D.; Gjertson, D.W.; Lu, P.; Kim, H.J.; Coy, H.; Goldin, J.G. Comparison of the Quantitative CT Imaging Biomarkers of Idiopathic Pulmonary Fibrosis at Baseline and Early Change with an Interval of 7 Months. Acad. Radiol. 2015, 22, 70–80. [Google Scholar] [CrossRef]
- De Giacomi, F.; Raghunath, S.; Karwoski, R.; Bartholmai, B.J.; Moua, T. Short-term Automated Quantification of Radiologic Changes in the Characterization of Idiopathic Pulmonary Fibrosis Versus Nonspecific Interstitial Pneumonia and Prediction of Long-term Survival. J. Thorac. Imaging 2018, 33, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.U.; Chong, S.; Choi, H.W.; Choi, J.C. Quantitative image analysis using chest computed tomography in the evaluation of lymph node involvement in pulmonary sarcoidosis and tuberculosis. PLoS ONE 2018, 13, e0207959. [Google Scholar] [CrossRef] [PubMed]
- Best, A.C.; Meng, J.; Lynch, A.M.; Bozic, C.M.; Miller, D.; Grunwald, G.K.; Lynch, D.A. Idiopathic pulmonary fibrosis: Physiologic tests, quantitative CT indexes, and CT visual scores as predictors of mortality. Radiology 2008, 246, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, F.; Moua, T.; Rajagopalan, S.; Karwoski, R.A.; Raghunath, S.; Decker, P.A.; Hartman, T.E.; Bartholmai, B.J.; Robb, R.A.; Ryu, J.H. Automated quantification of radiological patterns predicts survival in idiopathic pulmonary fibrosis. Eur. Respir. J. 2014, 43, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Jacob, J.; Bartholmai, B.J.; Rajagopalan, S.; Kokosi, M.; Nair, A.; Karwoski, R.; Walsh, S.L.F.; Wells, A.U.; Hansell, D.M. Mortality prediction in idiopathic pulmonary fibrosis: Evaluation of computer-based CT analysis with conventional severity measures. Eur. Respir. J. 2017, 49, 1601011. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Brown, M.S.; Elashoff, R.; Li, G.; Gjertson, D.W.; Lynch, D.A.; Strollo, D.C.; Kleerup, E.; Chong, D.; Shah, S.K.; et al. Quantitative texture-based assessment of one-year changes in fibrotic reticular patterns on HRCT in scleroderma lung disease treated with oral cyclophosphamide. Eur. Radiol. 2011, 21, 2455–2465. [Google Scholar] [CrossRef]
- Kiely, D.; Lawrie, A.; Doyle, O.; Salvatelli, V.; Daniels, F.; Drage, E.; Jenner, H.; Rigg, J.; Schmitt, C.; Samyshkin, Y.; et al. Real world data from hospital episode statistics can be used to determine patients at risk of idiopathic pulmonary arterial hypertension. Eur. Respir. Soc. 2018, 52, PA3082. Available online: http://erj.ersjournals.com/lookup/doi/10.1183/13993003.congress-2018.PA3082 (accessed on 23 March 2021).
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. Automatic segmentation of the right ventricle from cardiac MRI using a learning-based approach: Automatic Segmentation Using a Learning-Based Approach. Magn. Reson. Med. 2017, 78, 2439–2448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med. Image Anal. 2016, 30, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Kiely, D.G.; Levin, D.L.; Hassoun, P.M.; Ivy, D.; Jone, P.-N.; Bwika, J.; Kawut, S.M.; Lordan, J.; Lungu, A.; Mazurek, J.A.; et al. Statement on imaging and pulmonary hypertension from the Pulmonary Vascular Research Institute (PVRI). Pulm. Circ. 2019, 9, 204589401984199. [Google Scholar] [CrossRef] [Green Version]
- Lungu, A.; Swift, A.; Capener, D.; Kiely, D.; Hose, R.; Wild, J. Diagnosis of pulmonary hypertension from MR image based computational models of pulmonary vascular haemodynamics and decision tree analysis. Eur. Respir. Soc. 2015, 46, PA2109. Available online: http://erj.ersjournals.com/lookup/doi/10.1183/13993003.congress-2015.PA2109 (accessed on 6 December 2020).
- Yang, M.; Ren, Y.; She, Y.; Xie, D.; Sun, X.; Shi, J.; Zhao, G.; Chen, C. Imaging phenotype using radiomics to predict dry pleural dissemination in non-small cell lung cancer. Ann. Transl. Med. 2019, 7, 259. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Joint Research Centre. Robustness and Explainability of Artificial Intelligence: From Technical to Policy Solutions. Available online: https://data.europa.eu/doi/10.2760/57493 (accessed on 2 March 2021).
- Ibrahim, A.; Primakov, S.; Beuque, M.; Woodruff, H.C.; Halilaj, I.; Wu, G.; Refaee, T.; Granzier, R.; Widaatalla, Y.; Hustinx, R.; et al. Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods San Diego Calif. 2021, 188, 20–29. [Google Scholar] [CrossRef]
| Terminology Used in Radiomics and AI | |
|---|---|
| Artificial intelligence | Wide-ranging branch of computer science, generating complex software that perform tasks that would typically have required human intelligence, by sensing and responding to a feature of their environment. |
| CAD (Computer Aided Detection or Diagnosis) | Technology combining elements of artificial intelligence with radiological and pathology image processing. Its aim is to assist in the detection and/or diagnosis of diseases, improving the accuracy of radiologists with a reduction in time in the interpretation of images. |
| Radiomics | Method that extracts a large number of quantitative features from radiographic medical images using data-characterization algorithms, to help in disease diagnosis and prognosis. |
| Machine Learning | Field in artificial intelligence studying computer algorithms that improve automatically through experience, by building a model based on sample data, known as “training data”, in order to make predictions or decisions. Supervised learning: The computer receives example inputs and their foreseen outputs. Its goal is to learn a general and reproducible function that links inputs to outputs. Unsupervised learning: The computer receives no labels to the learning algorithm for previously undetected patterns in a data set, leaving it on its own to find structure in its input. |
| Convolutional neural networks | Class of deep neural networks, which have the particularity of being fully connected networks. It gives them the advantage of understanding the hierarchical pattern in data and assembling more complex patterns using smaller and simpler patterns. |
| Voxel | Single sample, or data point, on a regularly spaced, three-dimensional grid. In CT scans, the values of voxels are Hounsfield units. A voxel is a 3D pixel. |
| ROI (Region of Interest) | Image areas containing the information relevant to image processing. |
| Skew of histogram | Measure of the asymmetry of attenuation distribution. The lung normal attenuation histogram is skewed to the left. There is a decreased leftward skewness in IPF. |
| Kurtosis of histogram | Measurement of how sharp an attenuation distribution curve is. Kurtosis is abnormally low in idiopathic pulmonary fibrosis (IPF). |
| Threshold measurement | Total count of pixels/voxels above or below a specific attenuation value that determines a relative volume. Threshold measures in emphysema quantifies the extent of emphysema according to a specific index of −950 Hounsfield units (HU). |
| Texture analysis | Statistical methods that evaluate spatial relationship between voxels in an ROI, in order to characterize textural features of the parenchyma and give information about heterogeneity. |
| Study | Description | Cohort | Performance |
|---|---|---|---|
| Chen et al. (2018) [7] |
| 33 benign CT 42 malignant CT | Benign vs. malignant Accuracy 84% Sensitivity 92.85% Specificity 72.73% |
| De Koning et al. (2020) [9] |
| 15,792 patients | Benign vs. malignant: impact on mortality At 10 years, cancer mortality = 2.5 deaths/100,000 persons/years (screening group) vs. 3.3 deaths/100,000 (no-screening group) Cumulative ratio 0.76 (p = 0.01) |
| Ma et al. (2016) [10] |
| 36 benign CT 94 malignant CT | Benign vs. malignant Accuracy 82.7% Sensitivity 80% Specificity 85.5% |
| Hawkins et al. (2016) [11] |
| 328 benign CT 170 malignant CT | Benign vs. malignant Accuracy 80% |
| Huang et al. (2018) [12] |
| Training cohort 70 benign CT 70 malignant CT Validation cohort 26 benign CT 20 malignant CT | Benign vs. malignant Accuracy 91% Sensitivity 95% Specificity 88% |
| Uthoff et al. (2020) [13] |
| Training cohort 289 benign CT 74 malignant CT Validation cohort 50 benign CT 50 malignant CT | Benign vs. malignant Accuracy 98% Sensitivity 100% Specificity 96% |
| Mao et al. (2019) [14] |
| Training cohort 156 benign CT 40 malignant CT Validation cohort 75 benign CT 23 malignant CT | Benign vs. malignant Accuracy 89.8% Sensitivity 81% Specificity 92.2% |
| Maldonado et al. (2021) [18] |
| Validation cohort 91 malignant CT 79 benign CT | Benign vs. malignant AUC 0.90 Sensitivity 92.3% Specificity 62% |
| Mehta et al. (2021) [22] |
| 1018 CTs Malignancy rating from 1 to 5 | Benign vs. malignant AUC 0.87 on fully supervised 3D CNN + random forest model (images, biomarkers and volumetric features) AUC 0.93 on semi-supervised random forest (biomarkers only) |
| Digumarthy et al. (2019) [24] |
| 31 benign CT 77 malignant CT | Benign vs. malignant according to temporal changes AUC 0.741 |
| Lee et al. (2014) [16] |
| Transient PSNs 39 benign CT Persistent PSNs 17 benign CT 30 malignant CT | Prediction of persistent part-solid nodules AUC 0.93 if texture analysis was combined to clinical and CT features |
| Autrusseau et al. (2021) [17] |
| 99 lung nodules
| Concordance between FD and ULD chest CT in radiomic-guided nodule risk assessment ICC of 0.82, displaying a good agreement in malignancy similarity index between ULD and FD chest CT |
| Study | Description | Cohort | Performance |
|---|---|---|---|
| Wu et al. (2016) [23] |
| Training cohort 198 malignant CT Validation cohort 152 malignant CT | Tumor histology correlation AUC 0.72 |
| Yu et al. (2019) [34] |
| Training cohort 87 NSCLC CT Validation cohort 58 NSCLC CT | Diagnosis and staging in NSCLC AUC > 0.70, with predictive accuracy higher in lung adenocarcinoma than in lung squamous cell carcinoma |
| Liu et al. (2016) [36] |
| 298 malignant CT | Prediction of mutation status AUC EGFR+ status prediction 0.647, improved to 0.709 when adding a clinical model |
| Rios Velasquez et al. (2017) [37] |
| Training cohort 353 malignant CT Validation cohort 352 malignant CT | Prediction of mutation status AUC EGFR + versus EGFR− status 0.70 AUC KRAS + versus KRAS− status 0.63 AUC EGFR+ versus KRAS+ status 0.80 |
| Tang et al. (2018) [39] |
| Training cohort 114 malignant CT Validation cohort 176 malignant CT | Prediction of immune modulator status Favorable outcome in low CT intensity and high heterogeneity with low PDL 1 and high CD3 |
| Wu et al. (2020) [40] |
| Training cohort 229 NSCLC Validation cohort 68 NSCLC | Prediction of invasiveness AUC 0.98 for the model combining ground-glass and solid features Improvement of 0.14 in AUC when adding ground-glass radiomic features to solid features |
| Coroller et al. (2015) [41] |
| Training cohort 98 malignant CT Validation cohort 84 malignant CT | Prediction of distant metastasis A multivariate radiomic signature (3 features) yielded a high prognostic performance for distant metastasis (CI 0.61) |
| He et al. (2019) [43] |
| Training cohort 423 NSCLC CT Validation cohort 294 NSCLC CT | Prediction of lymph node metastasis Good discrimination for the model defining a radiomics-based predictive score (C index 0.785) |
| Ferreira et al. (2018) [45] |
| Training cohort 52 malignant CT Validation cohort 16 malignant CT | Histology and distant metastasis AUC lymph nodal metastasis 0.89 AUC distant metastasis 0.97 AUC histopathology 0.92 |
| Mattonen et al. (2016) [46] |
| 182 malignant CT | Prediction of recurrence after SBRT AUC 0.85 (radiomic signature of 5 features predicting local recurrence) |
| Coroller et al. (2016) [49] |
| 127 malignant CT Training cohort 80% Validation cohort 20% | Prediction of response after NCT AUC for pathologic gross residual disease prediction (7 features) > 0.6 AUC for pathologic complete response (1 feature) 0.63 AUC for poor response 0.63 (spherical disproportionality) or 0.61 (heterogeneous texture) |
| Kim et al. (2017) [50] |
| 48 malignant CT (NSCLC, EGFR mutant) | Prediction of response to TKI
|
| Lafata et al. (2019) [52] |
| 64 malignant CT (NSCLC) | Prediction of PFTs
|
| Study | Description | Cohort | Performance |
|---|---|---|---|
| Schniering et al. (2019) [74] |
| 66 ILD CT (20 mild ILD and 46 advanced ILD) | Staging of ILD (proof of concept) AUC 0.929 |
| Stefano et al. (2020) [75] |
| 32 IPF CT | Severity of IPF NL (normally attenuated lung) at -200 HU demonstrated the strongest correlation with disease severity (p = 0.009) |
| Martini et al. (2020) [76] |
| 66 SSc CT Training cohort 70% Validation cohort 30% | Severity and staging of SSc-ILD
|
| Ungprasert et al. (2017) [77] |
| 110 ILD CT
| Correlation with PFTs in IIM associated ILD
|
| Kim et al. (2015) [78] |
| 57 IPF patients
| Correlation with baseline lung function and prediction of evolution in IPF
|
| De Giacomi et al. (2017) [79] |
| 40 biopsy-confirmed UIP 20 biopsy-confirmed NSIP | Differentiation NSIP vs. IPF
|
| Lee et al. (2018) [80] |
| 26 CT from tubrcolosis patients, 21 CT from sarcoidosis patients. | Differentiation between tuberculosis and sarcoidosis LN
|
| Best et al. (2008) [81] |
| 167 IPF patients
| Prediction of mortality and progression in IPF
|
| Maldonado et al. (2014) [82] |
| 55 IPF patients | Correlation between CT changes and mortality in IPF
|
| Jacob et al. (2017) [83] |
| 283 IPF CT | Prediction of mortality in IPF
|
| Kim et al. (2011) [84] |
| 83 SSc-ILD CT
| Evaluate the effectiveness of cyclophosphamide in SSc-ILD
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frix, A.-N.; Cousin, F.; Refaee, T.; Bottari, F.; Vaidyanathan, A.; Desir, C.; Vos, W.; Walsh, S.; Occhipinti, M.; Lovinfosse, P.; et al. Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians. J. Pers. Med. 2021, 11, 602. https://doi.org/10.3390/jpm11070602
Frix A-N, Cousin F, Refaee T, Bottari F, Vaidyanathan A, Desir C, Vos W, Walsh S, Occhipinti M, Lovinfosse P, et al. Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians. Journal of Personalized Medicine. 2021; 11(7):602. https://doi.org/10.3390/jpm11070602
Chicago/Turabian StyleFrix, Anne-Noëlle, François Cousin, Turkey Refaee, Fabio Bottari, Akshayaa Vaidyanathan, Colin Desir, Wim Vos, Sean Walsh, Mariaelena Occhipinti, Pierre Lovinfosse, and et al. 2021. "Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians" Journal of Personalized Medicine 11, no. 7: 602. https://doi.org/10.3390/jpm11070602
APA StyleFrix, A.-N., Cousin, F., Refaee, T., Bottari, F., Vaidyanathan, A., Desir, C., Vos, W., Walsh, S., Occhipinti, M., Lovinfosse, P., Leijenaar, R. T. H., Hustinx, R., Meunier, P., Louis, R., Lambin, P., & Guiot, J. (2021). Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians. Journal of Personalized Medicine, 11(7), 602. https://doi.org/10.3390/jpm11070602







