Exploring the Metabolic Heterogeneity of Cancers: A Benchmark Study of Context-Specific Models
Abstract
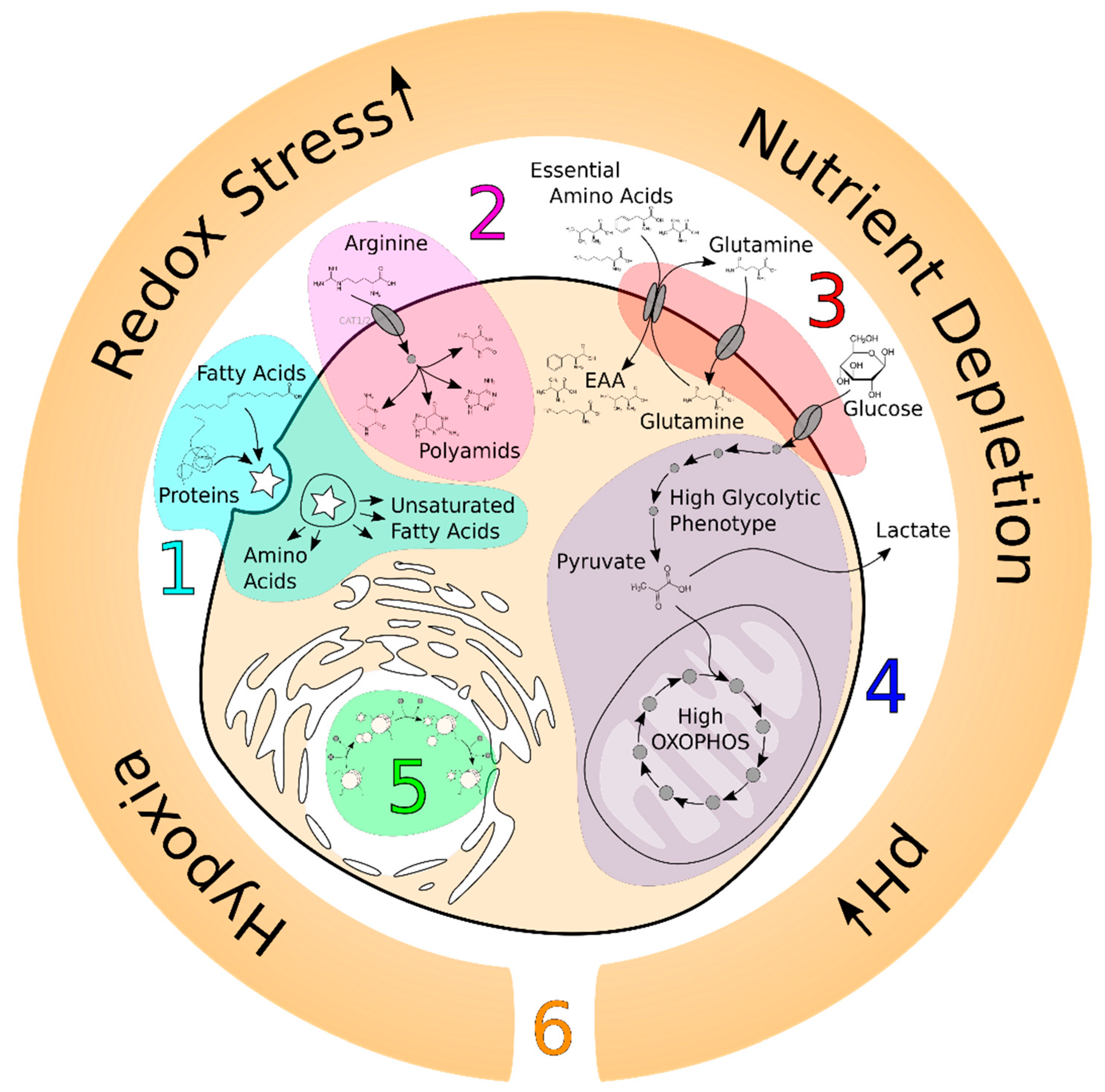
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.1.1. Human Cancer Cell Line Data
Human Protein Atlas (HPA)
EMTAB-37
NCI-60 Proteome
2.1.2. Human Cancer Patient Data
TCGA
GSE2109
ProteomeXchange (PX)
Data Preprocessing
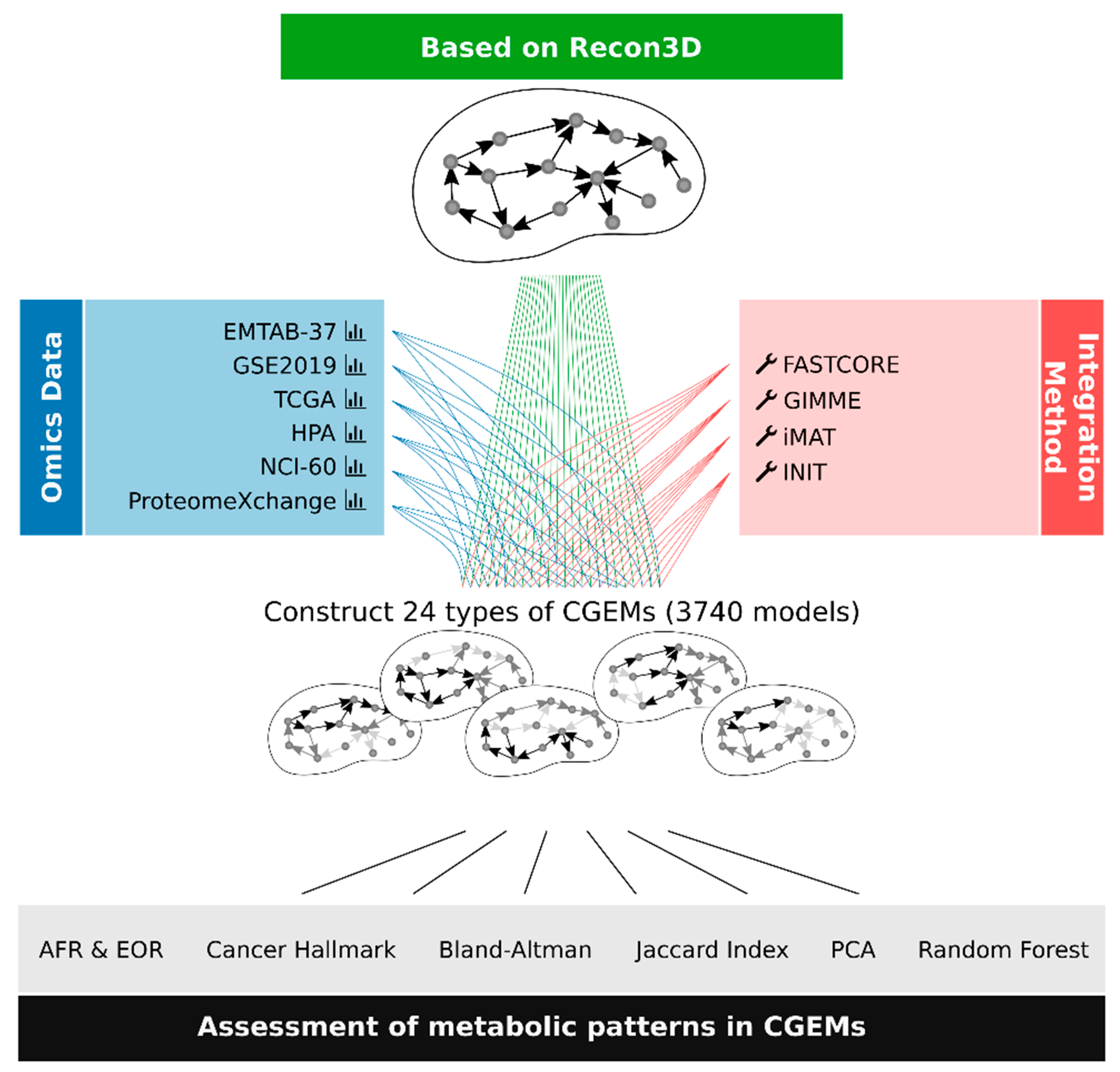
2.2. Genome-Scale Metabolic Models (GEMs)
Data Processing and Extraction of Context-Specific Model
2.3. Validation of CGEMs by Two Known Hallmarks of Cancer
2.3.1. Metabolic Hallmark of CGEMs (Warburg Score)
2.3.2. The Gene Set Hallmark of Cancer
2.4. Characterization of Metabolic Pattern of CGEMs
2.4.1. Jaccard Index
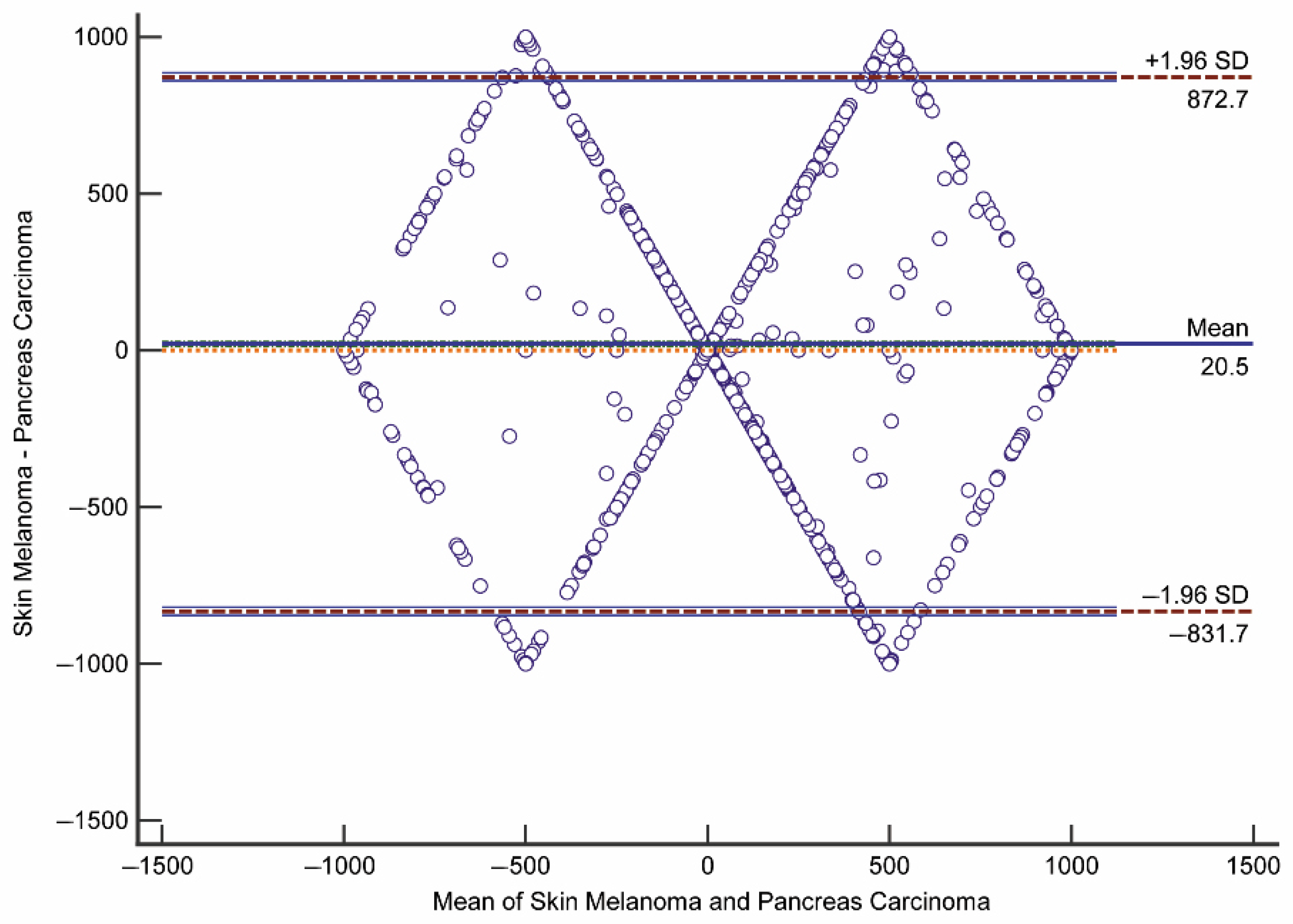
2.4.2. Bland–Altman Plot
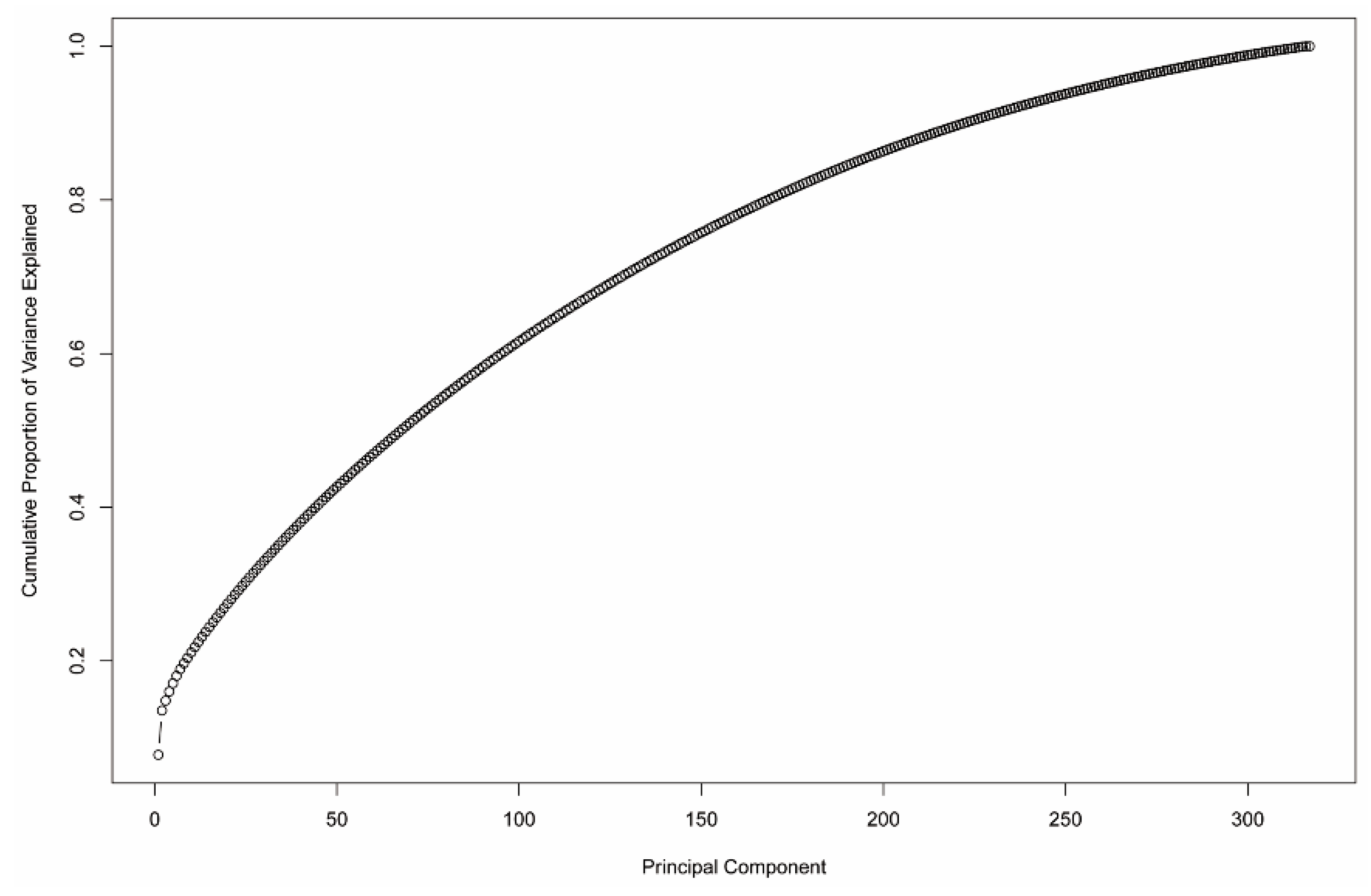
2.4.3. Principal Component Analysis (PCA)
2.4.4. Machine Learning Method (Random Forest)
2.5. Implementation
3. Results
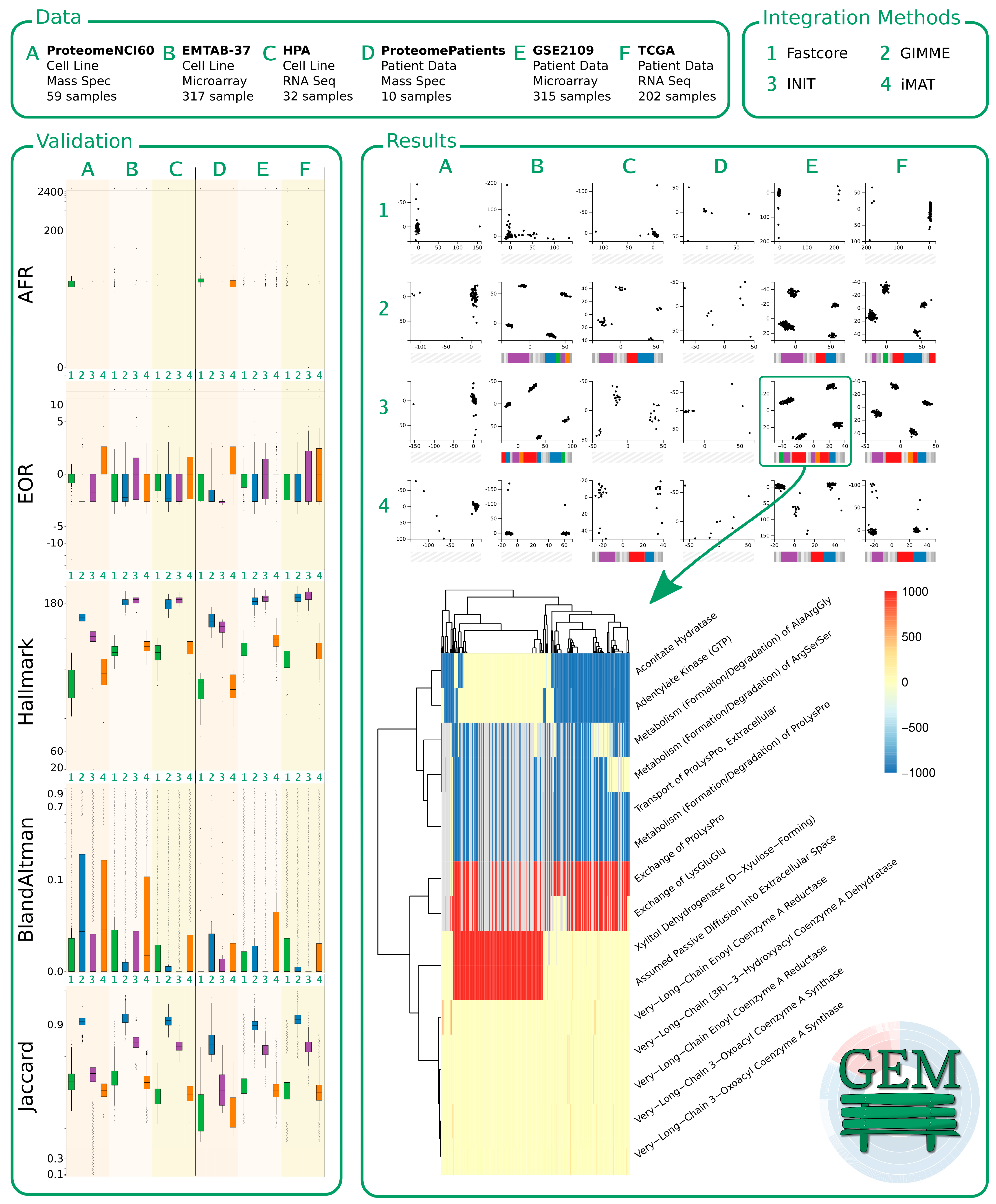
3.1. Construction of Cancer-Specific GEMs (CGEMs)
3.2. Evaluation of the Quality of a Context-Specific Model of Cancer
3.2.1. Assessment of Metabolic Hallmarks of Cancer in all CGEMs
3.2.2. Assessment of a Gene Set Hallmark of Cancer in all CGEMs
3.3. Evaluation of the Impact of Integration Algorithms and Omics Data on the Behavior of CGEMs
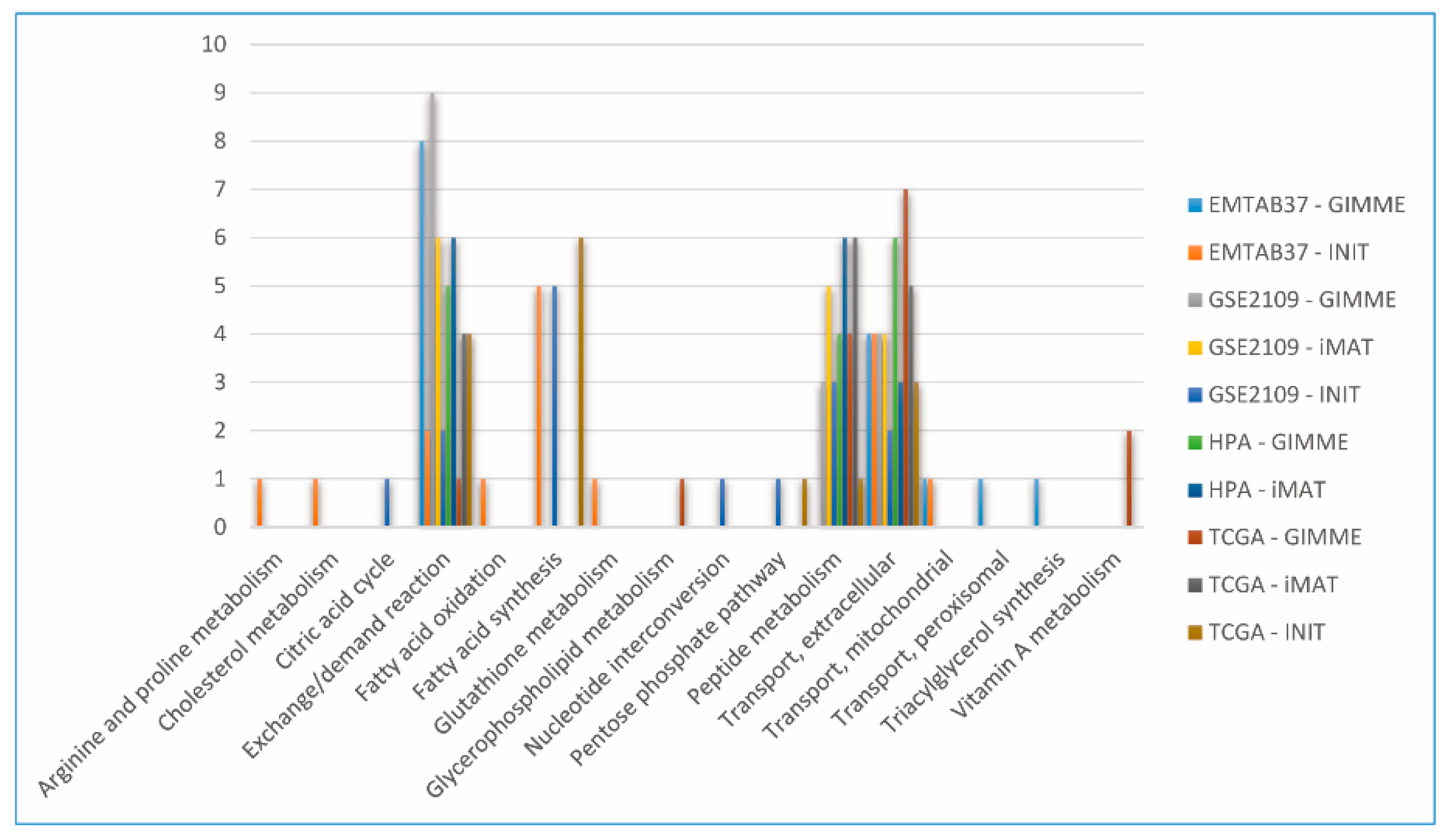
3.4. Characterization of Metabolic Pattern (FBA-Based Feature) in CGEMs
3.4.1. Number of Active Reactions (Carrying Flux Reactions)
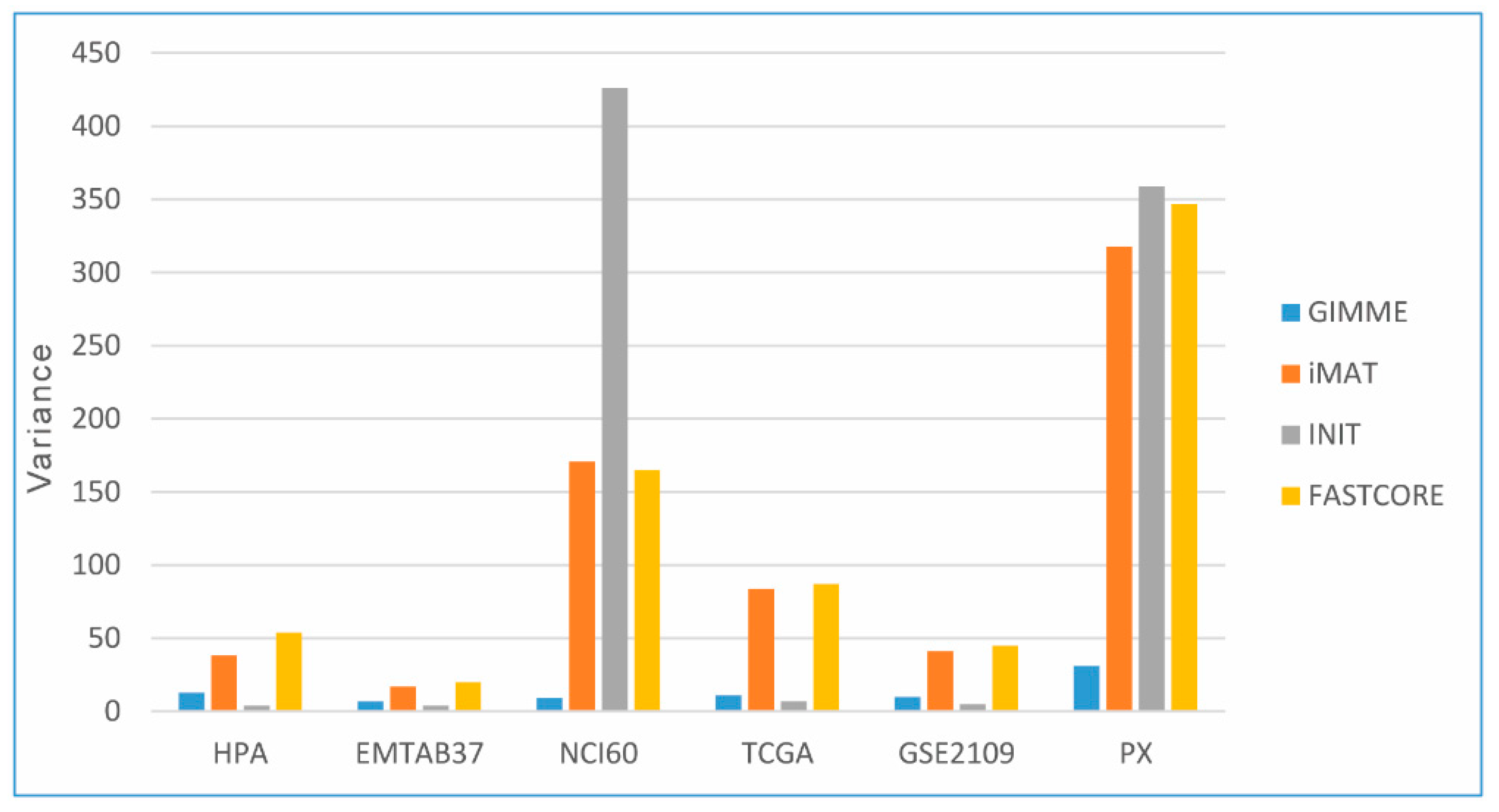
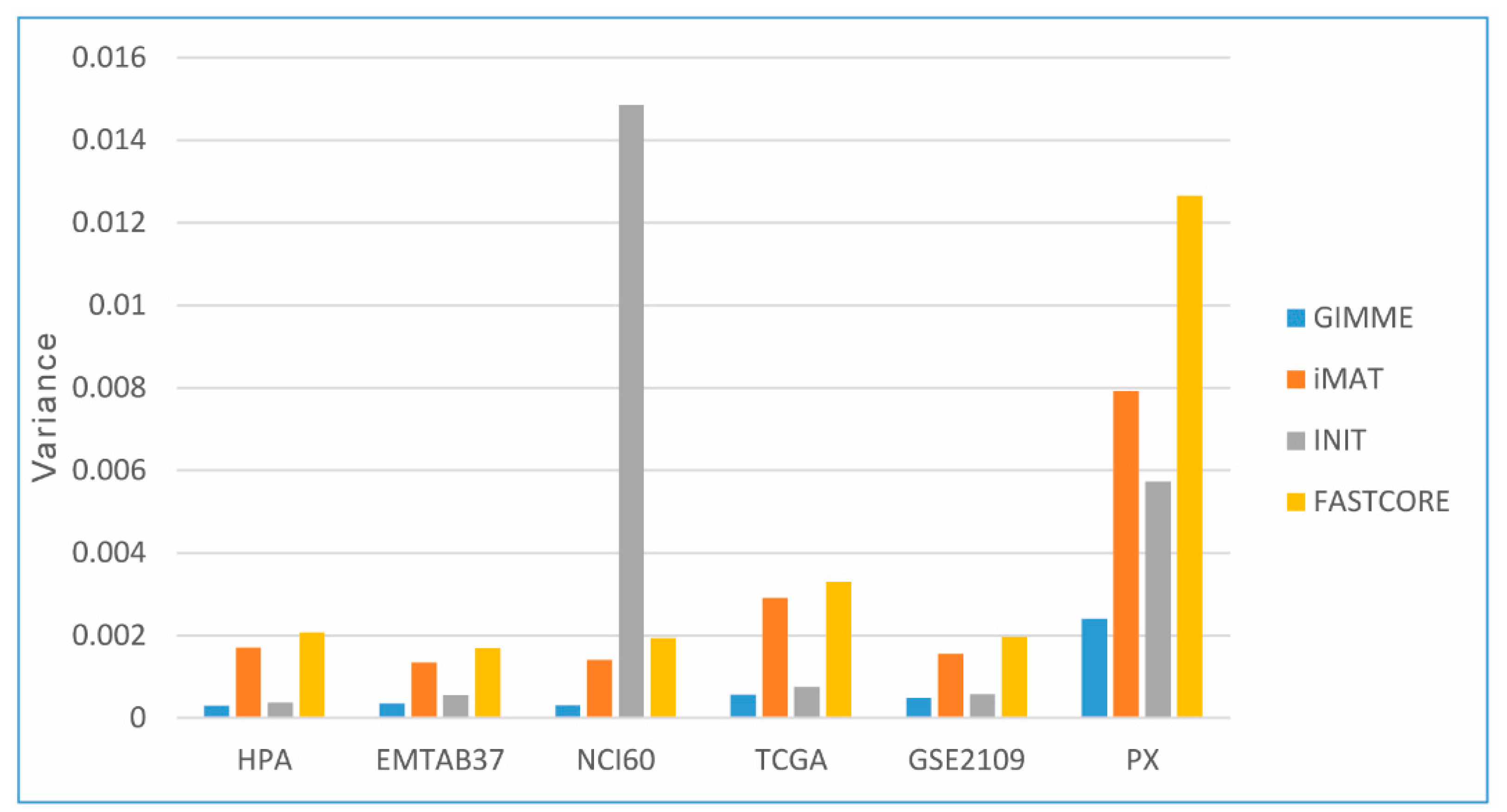
3.4.2. Flux States of CGEMs (Magnitude Flux of Active Reactions)
3.5. Flux States Are an Appropriate Index to Distinguish CGEMs
3.6. Are CGEMs Distinguishable Based on Their Flux States?
3.7. Main FBA-Based Features to Distinguish CGEMs
3.8. Development of the GEMbench Interactive Website
3.9. Limitations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Reznik, E.; Luna, A.; Aksoy, B.A.; Liu, E.M.; La, K.; Ostrovnaya, I.; Creighton, C.J.; Hakimi, A.A.; Sander, C. A Landscape of Metabolic Variation across Tumor Types. Cell Syst. 2018. [Google Scholar] [CrossRef]
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019. [Google Scholar] [CrossRef]
- Okegawa, T.; Morimoto, M.; Nishizawa, S.; Kitazawa, S.; Honda, K.; Araki, H.; Tamura, T.; Ando, A.; Satomi, Y.; Nutahara, K.; et al. Intratumor Heterogeneity in Primary Kidney Cancer Revealed by Metabolic Profiling of Multiple Spatially Separated Samples within Tumors. EBioMedicine 2017. [Google Scholar] [CrossRef]
- Feng, J.; Gao, H.; Zhang, Q.; Zhou, Y.; Li, C.; Zhao, S.; Hong, L.; Yang, J.; Hao, S.; Hong, W.; et al. Metabolic profiling reveals distinct metabolic alterations in different subtypes of pituitary adenomas and confers therapeutic targets. J. Transl. Med. 2019. [Google Scholar] [CrossRef]
- Vermeersch, K.; Styczynski, M. Applications of metabolomics in cancer research. J. Carcinog. 2013. [Google Scholar] [CrossRef]
- Lewis, N.E.; Abdel-Haleem, A.M. The evolution of genome-scale models of cancer metabolism. Front. Physiol. 2013, 4, 237. [Google Scholar] [CrossRef]
- Cho, J.S.; Gu, C.; Han, T.H.; Ryu, J.Y.; Lee, S.Y. Reconstruction of context-specific genome-scale metabolic models using multiomics data to study metabolic rewiring. Curr. Opin. Syst. Biol. 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Palsson, B. Metabolic systems biology. FEBS Lett. 2009, 583, 3900–3904. [Google Scholar] [CrossRef]
- Fouladiha, H.; Marashi, S.A. Biomedical applications of cell- and tissue-specific metabolic network models. J. Biomed. Inform. 2017, 68, 35–49. [Google Scholar] [CrossRef]
- Machado, D.; Herrgård, M. Systematic Evaluation of Methods for Integration of Transcriptomic Data into Constraint-Based Models of Metabolism. PLoS Comput. Biol. 2014. [Google Scholar] [CrossRef]
- Opdam, S.; Richelle, A.; Kellman, B.; Li, S.; Zielinski, D.C.; Lewis, N.E. A Systematic Evaluation of Methods for Tailoring Genome-Scale Metabolic Models. Cell Syst. 2017. [Google Scholar] [CrossRef]
- Richelle, A.; Joshi, C.; Lewis, N.E. Assessing key decisions for transcriptomic data integration in biochemical networks. PLOS Comput. Biol. 2019. [Google Scholar] [CrossRef]
- Shlomi, T.; Benyamini, T.; Gottlieb, E.; Sharan, R.; Ruppin, E. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the warburg effect. PLoS Comput. Biol. 2011. [Google Scholar] [CrossRef]
- Folger, O.; Jerby, L.; Frezza, C.; Gottlieb, E.; Ruppin, E.; Shlomi, T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011. [Google Scholar] [CrossRef]
- Wang, Y.; Eddy, J.A.; Price, N.D. Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst. Biol. 2012. [Google Scholar] [CrossRef]
- Jerby, L.; Wolf, L.; Denkert, C.; Stein, G.Y.; Hilvo, M.; Oresic, M.; Geiger, T.; Ruppin, E. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 2012. [Google Scholar] [CrossRef]
- Yizhak, K.; Le Dévédec, S.E.; Rogkoti, V.M.; Baenke, F.; Boer, V.C.; Frezza, C.; Schulze, A.; Water, B.; Ruppin, E. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol. Syst. Biol. 2014. [Google Scholar] [CrossRef]
- Nam, H.; Campodonico, M.; Bordbar, A.; Hyduke, D.R.; Kim, S.; Zielinski, D.C.; Palsson, B.O. A Systems Approach to Predict Oncometabolites via Context-Specific Genome-Scale Metabolic Networks. PLoS Comput. Biol. 2014. [Google Scholar] [CrossRef]
- Asgari, Y.; Zabihinpour, Z.; Salehzadeh-Yazdi, A.; Schreiber, F.; Masoudi-Nejad, A. Alterations in cancer cell metabolism: The Warburg effect and metabolic adaptation. Genomics 2015. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015. [Google Scholar] [CrossRef]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress update - From bulk to single-cell expression data. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Gholami, A.M.; Hahne, H.; Wu, Z.; Auer, F.J.; Meng, C.; Wilhelm, M.; Kuster, B. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013, 4, 609–620. [Google Scholar] [CrossRef]
- Available online: https://www.cancer.gov/tcga (accessed on 23 September 2020).
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Dräger, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272. Available online: https://www.nature.com/articles/nbt.4072#supplementary-information (accessed on 23 September 2020). [CrossRef]
- Noronha, A.; Modamio, J.; Jarosz, Y.; Guerard, E.; Sompairac, N.; Preciat, G.; Daníelsdóttir, A.D.; Krecke, M.; Merten, D.; Haraldsdóttir, H.S.; et al. The Virtual Metabolic Human database: Integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Vlassis, N.; Pacheco, M.P.; Sauter, T. Fast reconstruction of compact context-specific metabolic network models. PLoS Comput. Biol. 2014, 10, e1003424. [Google Scholar] [CrossRef]
- Estévez, S.R.; Nikoloski, Z. Context-specific metabolic model extraction based on regularized least squares optimization. PLoS One 2015. [Google Scholar] [CrossRef]
- Angione, C. Human Systems Biology and Metabolic Modelling: A Review-From Disease Metabolism to Precision Medicine. Biomed Res. Int. 2019, 2019, 16. [Google Scholar] [CrossRef]
- Di Filippo, M.; Colombo, R.; Damiani, C.; Pescini, D.; Gaglio, D.; Vanoni, M.; Alberghina, L.; Mauri, G. Zooming-in on cancer metabolic rewiring with tissue specific constraint-based models. Comput. Biol. Chem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015. [Google Scholar] [CrossRef]
- Richelle, A.; Chiang, A.W.T.; Kuo, C.C.; Lewis, N.E. Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions. PLoS Comput. Biol. 2019. [Google Scholar] [CrossRef]
- Jaccard, P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bull. Soc. Vaudoise Sci. Nat. 1901, 37, 241–272. [Google Scholar]
- Bass, J.I.F.; Diallo, A.; Nelson, J.; Soto, J.M.; Myers, C.L.; Walhout, A.J.M. Using networks to measure similarity between genes: Association index selection. Nat. Methods 2013, 10, 1169. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 47, 931–936. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Packag. Vers. 2017, 1, 337–354. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Paluszynska, A.; Biecek, P. Randomforestexplainer: Explaining and Visualizing Random Forests in Terms of Variable Importance. randomForestExplainer 2017. [Google Scholar] [CrossRef]
- Schellenberger, J.; Que, R.; Fleming, R.M.T.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 2011. [Google Scholar] [CrossRef]
- Lieven, C.; Beber, M.E.; Olivier, B.G.; Bergmann, F.T.; Ataman, M.; Babaei, P.; Bartell, J.A.; Blank, L.M.; Chauhan, S.; Correia, K.; et al. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020, 38, 504. [Google Scholar] [CrossRef]
- Jamialahmadi, O.; Hashemi-Najafabadi, S.; Motamedian, E.; Romeo, S.; Bagheri, F. A benchmark-driven approach to reconstruct metabolic networks for studying cancer metabolism. PLoS Comput. Biol. 2019. [Google Scholar] [CrossRef]
- Salehzadeh-Yazdi, A.; Wolfien, M.; Wolkenhauer, O. Applications of Genome-Scale Metabolic Models and Data Integration in Systems Medicine. In Focus on Systems Theory Research; Casanova, M.F., Opris, I., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2019; pp. 131–150. ISBN 978-1-53614-561-8. [Google Scholar]
- Damaghi, M.; West, J.; Robertson-Tessi, M.; Xu, L.; Ferrall-Fairbanks, M.C.; Stewart, P.A.; Persi, E.; Fridley, B.L.; Altrock, P.M.; Gatenby, R.A.; et al. The harsh microenvironment in early breast cancer selects for a Warburg phenotype. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999. [Google Scholar] [CrossRef]
- Giavarina, D. Understanding Bland Altman analysis. Biochem. Medica 2015. [Google Scholar] [CrossRef]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014. [Google Scholar] [CrossRef]
- Sebestyén, E.; Singh, B.; Miñana, B.; Pagès, A.; Mateo, F.; Pujana, M.A.; Valcárcel, J.; Eyras, E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Eraslan, B.; Wieland, T.; Hallström, B.; Hopf, T.; Zolg, D.P.; Zecha, J.; Asplund, A.; Li, L.; Meng, C.; et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019. [Google Scholar] [CrossRef]
- Sun, S.Y.; Lotan, R. Retinoids and their receptors in cancer development and chemoprevention. Crit. Rev. Oncol. Hematol. 2002, 41, 41–55. [Google Scholar] [CrossRef]
- Upadhyay, M.; Samal, J.; Kandpal, M.; Singh, O.V.; Vivekanandan, P. The Warburg effect: Insights from the past decade. Pharmacol. Ther. 2013, 137, 318–330. [Google Scholar] [CrossRef]

| Reference | Study | Conclusions |
|---|---|---|
| Shlomi et al. [16] | Generation of cancer-specific GEM (CGEM) by omics data integration to investigate the metabolic reprogramming of 60 cell lines by employing stoichiometric and enzyme solvent capacity constraints. | Warburg effect is a direct consequence of the metabolic adaptation to maximize the growth rate of cancer cells. |
| Folger et al. [17] | Combination of microarray data from 59 cancer cell lines with Recon1 to generate a generic metabolic model of human cancer cells. | Generation of a generic metabolic model of human cancer cells. |
| Wang et al. [18] | Reconstruction and pathway-level analyses of GEMs for 26 human cancer and counterpart normal tissues. | Eicosanoid metabolic pathway is a potential selective drug target. |
| Jerby et al. [19] | Development of a method to interpret metabolic phenotypes of breast cancer patients by the integration of transcriptomics and proteomics data into the GEMs. | Presentation of a novel metabolic hallmark of estrogen receptor in breast cancer. |
| Yizhak et al. [20] | Exploration of the role of the Warburg effect in supporting tumor migration by using GEMs. | Identification of metabolic targets inhibiting the cancer cell migration. |
| Nam et al. [21] | Deployment of genetic mutation information, transcriptomics data of nine cancer types as well as Recon2 for the reconstruction of CGEMs. | Prediction of oncometabolites that potentially dysregulate the epigenetic mechanisms. |
| Asgari et al. [22] | Construction of 13 CGEMs and their corresponding normal models by gene expression integration into Recon1 using the E-Flux method. | Results supporting the hypothesis that the Warburg effect is a consequence of metabolic adaptation. |
| Uhlen et al. [23] | Generation of more than 7000 patient-specific GEMs based on the integration of transcriptomics data into HMR2 by the tINIT method. | They disclosed that cancer patients show widespread metabolic heterogeneity, highlighting the need for personalized treatment. |
| Sources | Platforms | Samples |
|---|---|---|
| HPA | RNA-seq | 32 cell lines |
| EMTAB-37 | Microarray | 317 cell lines |
| NCI-60 | Mass spectrometry proteomics | 59 cell lines |
| TCGA | RNA-seq | 202 patients |
| GSE2019 | Microarray | 315 patients |
| PX | Mass spectrometry proteomics | 10 patients |
| GIMME | iMAT | INIT | FASTCORE | |||||
|---|---|---|---|---|---|---|---|---|
| AFR | EOR | AFR | EOR | AFR | EOR | AFR | EOR | |
| HPA | 1.00 | −0.31 | 1.03 | −0.05 | 1.00 | −0.44 | 1.32 | −0.14 |
| EMTAB-37 | 1.00 | −0.44 | 1.11 | −0.31 | 1.17 | −0.06 | 1.62 | −0.52 |
| NCI−60 | 1.00 | −0.76 | 1.10 | 0.63 | 1.07 | −0.41 | 1.65 | −0.50 |
| TCGA | 1.00 | −0.38 | 1.12 | 0.00 | 1.00 | −0.22 | 17.40 | −0.28 |
| GSE2109 | 1.01 | −0.48 | 1.33 | 0.06 | 1.20 | −0.10 | 1.27 | −0.21 |
| PX | 1.00 | −0.55 | 2.54 | 0.18 | 1.08 | −1.17 | 4.66 | −0.39 |
| GIMME | iMAT | INIT | FASTCORE | |
|---|---|---|---|---|
| HPA | 91.15 | 77.93 | 92.26 | 75.98 |
| EMTAB-37 | 91.39 | 78.67 | 92.12 | 76.84 |
| NCI-60 | 87.20 | 69.61 | 79.66 | 64.96 |
| TCGA | 92.69 | 76.84 | 93.11 | 73.85 |
| GSE2109 | 91.90 | 80.31 | 92.55 | 77.38 |
| PX | 86.14 | 62.18 | 80.76 | 61.52 |
| Dataset–Algorithm | GIMME | iMAT | INIT | FASTCORE | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPA | EMTAB-37 | NCI-60 | TCGA | GSE2019 | PX | HPA | EMTAB-37 | NCI-60 | TCGA | GSE2019 | PX | HPA | EMTAB-37 | NCI-60 | TCGA | GSE2019 | PX | HPA | EMTAB-37 | NCI-60 | TCGA | GSE2019 | PX | |
| Mean value | 0.91301 | 0.91918 | 0.90782 | 0.91436 | 0.89615 | 0.84622 | 0.65933 | 0.70365 | 0.67382 | 0.66188 | 0.67305 | 0.57403 | 0.83354 | 0.84521 | 0.71024 | 0.82953 | 0.82020 | 0.68047 | 0.65023 | 0.72104 | 0.70777 | 0.67031 | 0.69142 | 0.56654 |
| GIMME | iMAT | INIT | FASTCORE | |
|---|---|---|---|---|
| HPA | 88.71 | 82.46 | 88.91 | 83.87 |
| EMTAB-37 | 87.10 | 69.72 | 81.48 | 81.69 |
| NCI-60 | 61.31 | 62.71 | 82.29 | 84.98 |
| TCGA | 88.44 | 83.65 | 90.27 | 83.01 |
| GSE2109 | 84.05 | 77.91 | 89.16 | 85.54 |
| PX | 80.00 | 86.67 | 82.22 | 93.33 |
| GIMME | iMAT | INIT | FASTCORE | |
|---|---|---|---|---|
| HPA | 4 | 2 | - | 1 |
| EMTAB-37 | 4 | 2 | 4 | 1 |
| NCI-60 | 1 | 1 | 1 | 1 |
| TCGA | 4 | 2 | 4 | 1 |
| GSE2109 | 4 | 2 | 4 | 1 |
| PX | - | - | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jalili, M.; Scharm, M.; Wolkenhauer, O.; Damaghi, M.; Salehzadeh-Yazdi, A. Exploring the Metabolic Heterogeneity of Cancers: A Benchmark Study of Context-Specific Models. J. Pers. Med. 2021, 11, 496. https://doi.org/10.3390/jpm11060496
Jalili M, Scharm M, Wolkenhauer O, Damaghi M, Salehzadeh-Yazdi A. Exploring the Metabolic Heterogeneity of Cancers: A Benchmark Study of Context-Specific Models. Journal of Personalized Medicine. 2021; 11(6):496. https://doi.org/10.3390/jpm11060496
Chicago/Turabian StyleJalili, Mahdi, Martin Scharm, Olaf Wolkenhauer, Mehdi Damaghi, and Ali Salehzadeh-Yazdi. 2021. "Exploring the Metabolic Heterogeneity of Cancers: A Benchmark Study of Context-Specific Models" Journal of Personalized Medicine 11, no. 6: 496. https://doi.org/10.3390/jpm11060496
APA StyleJalili, M., Scharm, M., Wolkenhauer, O., Damaghi, M., & Salehzadeh-Yazdi, A. (2021). Exploring the Metabolic Heterogeneity of Cancers: A Benchmark Study of Context-Specific Models. Journal of Personalized Medicine, 11(6), 496. https://doi.org/10.3390/jpm11060496








