Genetic Markers as Risk Factors for the Development of Impulsive-Compulsive Behaviors in Patients with Parkinson’s Disease Receiving Dopaminergic Therapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Ethical Principles
2.3. Methods
- ACE (rs4646994)
- BDNF (rs2049046, rs6265)
- COMT (rs4680)
- DBH (rs141116007, rs2097629, rs1611115)
- DRD1 (rs686)
- DRD2 (rs1799732, rs6275, rs2283265, rs12364283, rs1076560)
- MAOA (VNTR)
- SLC6A3 (rs27072)
- SLC6A4 (rs38130034)
2.3.1. DNA Isolation
2.3.2. PCR Testing
2.3.3. Restriction Analysis
2.4. Statistical Analysis
2.5. Principal Component Analysis
3. Results
3.1. Association between the Genetic Markers in PD Patients without ICD (PD2 Group)
- rs2097629 substitution in the DBH gene (9q34.2, 1434 + 1579A > G, 3′ region) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the G allele with PD (p = 0.016, OR = 2.97, CI95% [1.17–8.97]). The mode of inheritance was found to be dominant.
- rs1611115 substitution in the DBH gene (9q34.2, 1021T > C, 5′ region) is associated with the disease. Analysis of the frequencies of alleles of this substitution also showed an association of the allele with PD (p = 2.8 × 10−3, OR = 3.71, CI95% [1.46–8.77]). The mode of inheritance was found to be recessive.
- rs6265 substitution in the BDNF gene (11p14.1, 196G > A, Val66Met, Exon 2) it is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of Allele A with PD (p = 6.7 × 10−3, OR = 2.83, CI95% [1.27–6.29]). The mode of inheritance was found to be dominant.
- rs6275 substitution in the DRD2 gene (11q23.2, 939T > C, His313His, Exon 7) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 2.9 × 10−8, OR = 9.00, CI95% [3.97–20.14]). The mode of inheritance was found to be recessive.
- rs12364283 substitution in the DRD2 gene (11q23.2, 4047A > G, 5′ region) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 0.012, OR = 3.30, CI95% [1.25–10.19]). The mode of inheritance was found to be recessive.
- rs686 substitution in the DRD1 gene (5q35.1, 7464G > A, 3′ region) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of Allele A with PD (p = 0.017, OR = ∞, CI95% [1.3583–∞]). The mode of inheritance was found to be dominant.
3.2. Association between the Genetic Markers and ICD in PD Patients (PD1 Group)
- rs1611115 substitution in the DBH gene (9q34.2, 1021T > C, 5′ region) is associated with the disease. Analysis of the frequencies of alleles of this substitution also showed an association of the allele with PD (p = 2.8 × 10−3, OR = 3.71, CI95% [1.46–8.77]). The mode of inheritance was found to be dominant.
- rs6265 substitution in the BDNF gene (11p14.1, 196G > A, Val66Met, Exon 2) it is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of Allele A with PD (p = 6.7 × 10−3, OR = 2.83, CI95% [1.27–6.29]). The mode of inheritance was found to be dominant.
- rs6275 substitution in the DRD2 gene (11q23.2, 939T > C, His313His, Exon 7) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 2.9 × 10−8, OR = 9.00, CI95% [3.97–20.14]). The mode of inheritance was found to be dominant.
- rs12364283 substitution in the DRD2 gene (11q23.2, 4047A > G, 5′ region) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 0.012, OR = 3.30, CI95% [1.25–10.19]). The mode of inheritance was found to be recessive.
- rs1076560 substitution in the DRD2 gene (11q23.2, 67314C > A, Intron 6) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 0.012, OR = 3.30, CI95% [1.25–10.19]). The mode of inheritance was found to be dominant.
- rs4646994 substitution in ACE gene (11q23.2, I/D 289bp, Intron 16) is associated with the disease. Analysis of allele frequencies of this substitution also showed an association of the T allele with PD (p = 0.024, OR = 2.64, CI95% [1.12–7.22]). The mode of inheritance was found to be dominant.
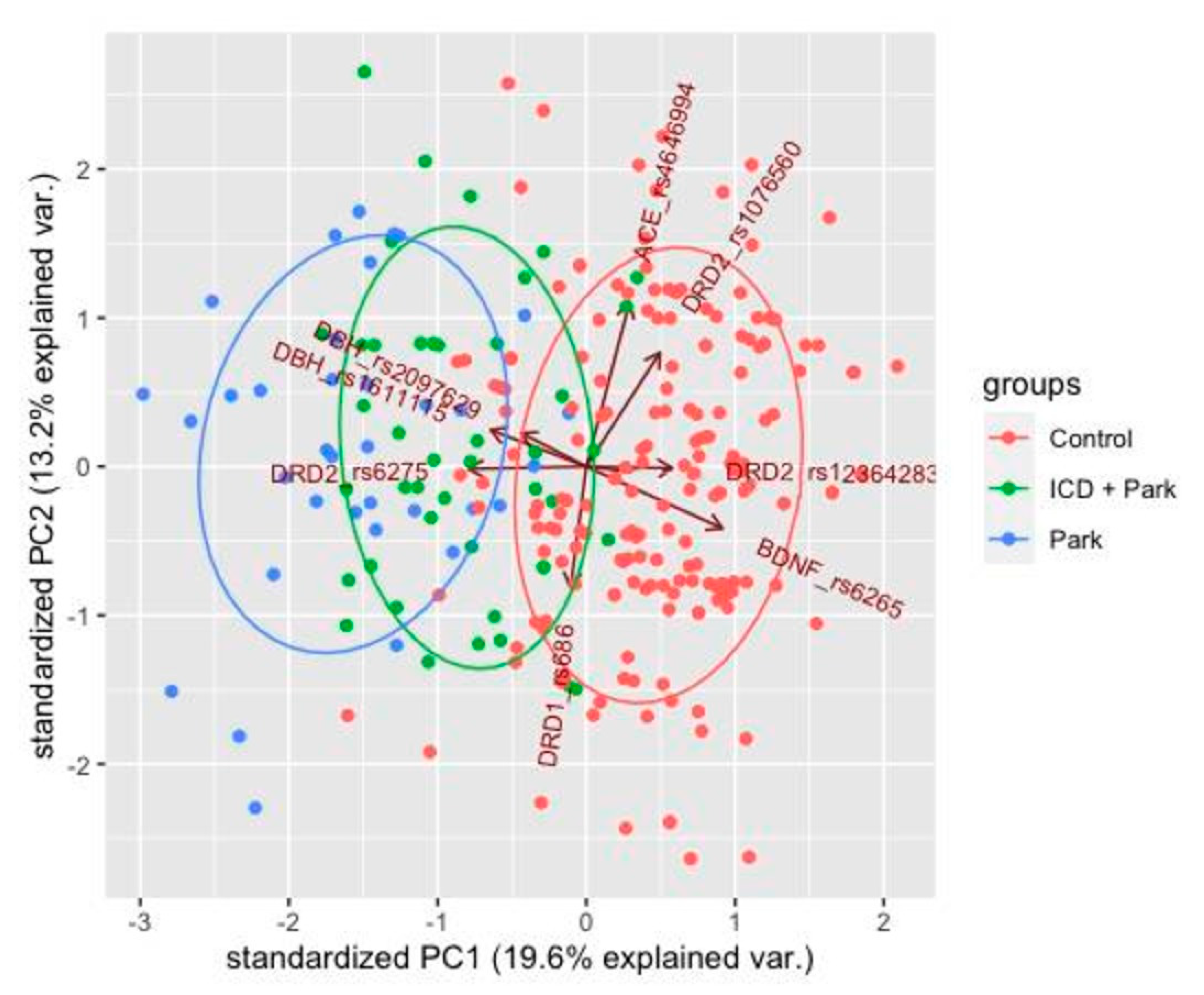
3.3. Principal Component Analysis
4. Discussion
4.1. Association between the Genetic Markers and PD
4.2. Analysis of Complex Genotype Associations in PD Patients
4.3. Association between the Genetic Markers and ICD in PD Patients
4.4. Analysis of Complex Genotype Associations in PD Patients with ICD
4.5. Association between the Genetic Markers and ICD in PD Patients
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Titova, N.; Padmakumar, C.; Lewis, S.J.G.; Chaudhuri, K.R. Parkinson’s: A syndrome rather than a disease? J. Neural Transm. 2017, 124, 907–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magistrelli, L.; Ferrari, M.; Furgiuele, A.; Milner, A.V.; Contaldi, E.; Comi, C.; Marino, F. Polymorphisms of Dopamine Receptor Genes and Parkinson’s Disease: Clinical Relevance and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 3781. [Google Scholar] [CrossRef]
- Vilas, D.; Pont-Sunyer, C.; Tolosa, E. Impulse control disorders in Parkinson’s disease. Parkinsonism Relat. Disord. 2012, 18 (Suppl. 1), S80–S84. [Google Scholar] [CrossRef]
- Antonini, A.; Barone, P.; Bonuccelli, U.; Annoni, K.; Asgharnejad, M.; Stanzione, P. ICARUS study: Prevalence and clinical features of impulse control disorders in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2017, 88, 317–324. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Kwon, D.Y.; Seo, W.K.; Kim, J.H.; Baik, J.S.; Koh, S.B. Clinical characteristics of impulse control and repetitive behavior disorders in Parkinson’s disease. J. Neurol. 2013, 260, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Sarathchandran, P.; Soman, S.; Sarma, G.; Krishnan, S.; Kishore, A. Impulse control disorders and related behaviors in Indian patients with Parkinson’s disease. Mov. Disord. 2013, 28, 1901–1902. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, D.; Potenza, M.N. Impulse control disorders in Parkinson’s disease. Curr. Neurol. Neurosci. Rep. 2006, 6, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Voon, V.; Hassan, K.; Zurowski, M.; de Souza, M.; Thomsen, T.; Fox, S.; Lang, A.E.; Miyasaki, J. Prevalence of repetitive and reward-seeking behaviors in Parkinson’s disease. Neurology 2006, 67, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, D.; Koester, J.; Potenza, M.N.; Siderowf, A.D.; Stacy, M.; Voon, V.; Whetteckey, J.; Wunderlich, G.R.; Lang, A.E. Impulse control disorders in Parkinson’s disease: A cross-sectional study of 3090 patients. Arch. Neurol. 2010, 67, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; He, A.Q.; Li, L.; Chen, W.; Liu, Z.G. Clinical characteristics of impulse control and related disorders in Chinese Parkinson’s disease patients. BMC Neurol. 2017, 17, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharjee, S. Impulse control disorders in Parkinson’s disease: Review of pathophysiology, epidemiology, clinical features, management, and future challenges. Neurol. India 2018, 66, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Latella, D.; Maggio, M.G.; Maresca, G.; Saporoso, A.F.; Le Cause, M.; Manuli, A.; Milardi, D.; Bramanti, P.; De Luca, R.; Calabrò, R.S. Impulse control disorders in Parkinson’s disease: A systematic review on risk factors and pathophysiology. Neurol. Sci. 2019, 398, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Brewer, J.A.; Potenza, M.N. The neurobiology and genetics of impulse control disorders: Relationships to drug addictions. Biochem. Pharm. 2008, 75, 63–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comings, D.E.; Gade-Andavolu, R.; Gonzalez, N.; Wu, S.; Muhleman, D.; Chen, C.; Koh, P.; Farwell, K.; Blake, H.; Dietz, G.; et al. The additive effect of neurotransmitter genes in pathological gambling. Clin. Genet. 2001, 60, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, E.K.; Park, S.S.; Lim, J.Y.; Kim, H.J.; Kim, J.S.; Jeon, B.S. Association of DRD3 and GRIN2B with impulse control and related behaviors in Parkinson’s disease. Mov. Disord. 2009, 24, 1803–1810. [Google Scholar] [CrossRef] [PubMed]
- Cilia, R.; Benfante, R.; Asselta, R.; Marabini, L.; Cereda, E.; Siri, C.; Pezzoli, G.; Goldwurm, S.; Fornasari, D. Tryptophan hydroxylase type 2 variants modulate severity and outcome of addictive behaviors in Parkinson’s disease. Parkinsonism Relat. Disord. 2016, 29, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Zainal Abidin, S.; Tan, E.L.; Chan, S.C.; Jaafar, A.; Lee, A.X.; Abd Hamid, M.H.; Abdul Murad, N.A.; Pakarul Razy, N.F.; Azmin, S.; Ahmad Annuar, A.; et al. DRD and GRIN2B polymorphisms and their association with the development of impulse control behaviour among Malaysian Parkinson’s disease patients. BMC Neurol. 2015, 15, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Martínez, X.H.; García-Ruiz, P.J.; Martínez-García, C.; Martínez-Castrillo, J.C.; Vela, L.; Mata, M.; Hoenicka, J. Behavioral addictions in early-onset Parkinson disease are associated with DRD3 variants. Parkinsonism Relat. Disord. 2018, 49, 100–103. [Google Scholar] [CrossRef]
- Rieck, M.; Schumacher-Schuh, A.; Altmann, V.; Callegari-Jacques, S.M.; Rieder, C.R.M.; Hutz, M.H. Association between DRD2 and DRD3 gene polymorphisms and gastrointestinal symptoms induced by levodopa therapy in Parkinson’s disease. Pharm. J. 2018, 18, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, S.; Rajan, R.; Banerjee, M.; Kumar, H.; Sarma, G.; Krishnan, S.; Sarma, S.; Kishore, A. Dopamine D3 receptor Ser9Gly variant is associated with impulse control disorders in Parkinson’s disease patients. Parkinsonism Relat. Disord. 2016, 30, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Todt, U.; Netzer, C.; Toliat, M.; Heinze, A.; Goebel, I.; Nürnberg, P.; Göbel, H.; Freudenberg, J.; Kubisch, C. New genetic evidence for involvement of the dopamine system in migraine with aura. Hum. Genet. 2009, 125, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Zabetian, C.P.; Anderson, G.M.; Buxbaum, S.G.; Elston, R.C.; Ichinose, H.; Nagatsu, T.; Kim, K.S.; Kim, C.H.; Malison, R.T.; Gelernter, J.; et al. A quantitative-trait analysis of human plasma–dopamine β-hydroxylase activity: Evidence for a major functional polymorphism at the DBH locus. Am. J. Hum. Genet. 2001, 68, 515–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.Y.; Patel, P.D.; Sant, G.; Meng, C.X.; Teng, K.K.; Hempstead, B.L.; Lee, F.S. Variant brain-derived neurotrophic factor (BDNF)(Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J. Neurosci. 2004, 24, 4401–4411. [Google Scholar] [CrossRef]
- Cheng, L.; Ge, Q.; Xiao, P.; Sun, B.; Ke, X.; Bai, Y.; Lu, Z. Association study between BDNF gene polymorphisms and autism by three-dimensional gel-based microarray. Int. J. Mol. Sci. 2009, 10, 2487–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boots, E.A.; Schultz, S.A.; Clark, L.R.; Racine, A.M.; Darst, B.F.; Koscik, R.L.; Carlsson, C.M.; Gallagher, C.L.; Hogan, K.J.; Bendlin, B.B.; et al. BDNF Val66Met predicts cognitive decline in the Wisconsin Registry for Alzheimer’s Prevention. Neurology 2017, 88, 2098–2106. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Wainwright, M.S.; Comeron, J.M.; Saitou, N.; Sanders, A.R.; Gelernter, J.; Gejman, P.V. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003, 12, 205–216. [Google Scholar] [CrossRef]
- Davis, C.; Levitan, R.D.; Yilmaz, Z.; Kaplan, A.S.; Carter, J.C.; Kennedy, J.L. Binge eating disorder and the dopamine D2 receptor: Genotypes and sub-phenotypes. Prog. Neuropsychopharmacol. Biol. Psychiatry 2012, 38, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, K.M.; Pereira-Morales, A.J.; Forero, D.A. A functional polymorphism in the DRD1 gene, that modulates its regulation by miR-504, is associated with depressive symptoms. Psychiatry Investig. 2018, 15, 402–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, E.C.; Heath, A.C.; Lynskey, M.T.; Agrawal, A.; Henders, A.K.; Bowdler, L.M.; Todorov, A.A.; Madden, P.A.; Moore, E.; Degenhardt, L.; et al. PTSD risk associated with a functional DRD2 polymorphism in heroin-dependent cases and controls is limited to amphetamine-dependent individuals. Addict. Biol. 2014, 19, 700–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucht, M.; Samochowiec, A.; Samochowiec, J.; Jasiewicz, A.; Grabe, H.J.; Geissler, I.; Rimmbach, C.; Rosskopf, D.; Grzywacz, A.; Wysiecka, J.P.; et al. Influence of DRD2 and ANKK1 genotypes on apomorphine-induced growth hormone (GH) response in alcohol-dependent patients. Prog. Neuropsychopharmacol. Biol. Psychiatry 2010, 34, 45–49. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, L.; Cao, C.; Li, G.; Fang, R.; Liu, P.; Luo, S.; Zhang, X.; Liberzon, I. A DRD2/ANNK1-COMT Interaction, Consisting of Functional Variants, Confers Risk of Post-traumatic Stress Disorder in Traumatized Chinese. Front. Psychiatry 2018, 9, 170. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Liu, B.; Wu, C.; Gao, X.; Lu, Y.; Lian, Y.; Liu, J. Dopamine Receptor D2 Gene (DRD2) Polymorphisms, Job Stress, and Their Interaction on Sleep Dysfunction. Int. J. Environ. Res. Public Health 2020, 17, 8174. [Google Scholar] [CrossRef]
- Sayed-Tabatabaei, F.A.; Oostra, B.A.; Isaacs, A.; van Duijn, C.M.; Witteman, J.C. ACE polymorphisms. Circ. Res. 2006, 98, 1123–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmorzynski, S.; Szudy-Szczyrek, A.; Popek-Marciniec, S.; Korszen-Pilecka, I.; Wojcierowska-Litwin, M.; Luterek, M.; Chocholska, S.; Styk, W.; Swiderska-Kołacz, G.; Januszewska, J.; et al. ACE insertion/deletion polymorphism (rs4646994) is associated with the increased risk of multiple myeloma. Front. Oncol. 2019, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Kieling, C.; Genro, J.P.; Hutz, M.H.; Rohde, L.A. The–1021 C/T DBH polymorphism is associated with neuropsychological performance among children and adolescents with ADHD. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147B, 485–490. [Google Scholar] [CrossRef]
- Gibb, W.R. Accuracy in the clinical diagnosis of parkinsonian syndromes. Postgrad. Med. J. 1988, 64, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Weintraub, D.; Hoops, S.; Shea, J.A.; Lyons, K.E.; Pahwa, R.; Driver-Dunckley, E.D.; Adler, C.H.; Potenza, M.N.; Miyasaki, J.; Siderowf, A.D.; et al. Validation of the questionnaire for impulsive-compulsive disorders in Parkinson’s disease. Mov. Disord. 2009, 24, 1461–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weintraub, D.; Mamikonyan, E.; Papay, K.; Shea, J.A.; Xie, S.X.; Siderowf, A. Questionnaire for impulsive-compulsive disorders in Parkinson’s Disease–Rating Scale. Mov. Disord. 2012, 27, 242–247. [Google Scholar] [CrossRef] [Green Version]
- McElroy, S.L.; Keck, P.E., Jr.; Pope, H.G., Jr.; Smith, J.M.; Strakowski, S.M. Compulsive buying: A report of 20 cases. J. Clin. Psychiatry 1994, 55, 242–248. [Google Scholar] [PubMed]
- Voon, V. Repetition, repetition, and repetition: Compulsive and punding behaviors in Parkinson’s disease. Mov. Disord. 2004, 19, 367–370. [Google Scholar] [CrossRef]
- Evans, A.H.; Katzenschlager, R.; Paviour, D.; O’Sullivan, J.D.; Appel, S.; Lawrence, A.D.; Lees, A.J. Punding in Parkinson’s disease: Its relation to the dopamine dysregulation syndrome. Mov. Disord. 2004, 19, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, G.; O’Sullivan, J.D.; Turner, K.; Manson, A.J.; Lees, A.J. Hedonistic homeostatic dysregulation in patients with Parkinson’s disease on dopamine replacement therapies. J. Neurol. Neurosurg. Psychiatry 2000, 68, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Papp, A.C.; Pinsonneault, J.K.; Cooke, G.; Sadée, W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques 2003, 34, 1068–1072. [Google Scholar] [CrossRef]
- Abramson, J.H. WINPEPI updated: Computer programs for epidemiologists, and their teaching potential. Epidemiol. Perspect. Innov. 2011, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favorov, A.V.; Andreewski, T.V.; Sudomoina, M.A.; Favorova, O.O.; Parmigiani, G.; Ochs, M.F. A Markov chain Monte Carlo technique for identification of combinations of allelic variants underlying complex diseases in humans. Genetics 2005, 171, 2113–2121. [Google Scholar] [CrossRef] [Green Version]
- Titova, N.; Chaudhuri, K.R. Personalized Medicine and Nonmotor Symptoms in Parkinson’s Disease. Int. Rev. Neurobiol. 2017, 134, 1257–1281. [Google Scholar] [CrossRef]
- Rota, S.; Boura, I.; Batzu, L.; Titova, N.; Jenner, P.; Falup-Pecurariu, C.; Chaudhuri, K.R. ‘Dopamine agonist Phobia’ in Parkinson’s disease: When does it matter? Implications for non-motor symptoms and personalized medicine. Expert Rev. Neurother. 2020, 20, 953–965. [Google Scholar] [CrossRef]
- Titova, N.; Qamar, M.A.; Chaudhuri, K.R. Biomarkers of Parkinson’s Disease: An Introduction. Int. Rev. Neurobiol. 2017, 132, 183–196. [Google Scholar] [CrossRef]
- Titova, N.; Jenner, P.; Chaudhuri, K.R. The Future of Parkinson’s Treatment—Personalised and Precision Medicine. Eur. Neurol. Rev. 2017, 12, 15. [Google Scholar] [CrossRef] [Green Version]
- Marras, C.; Chaudhuri, K.R.; Titova, N.; Mestre, T.A. Therapy of Parkinson’s Disease Subtypes. Neurotherapeutics 2020, 17, 1366–1377. [Google Scholar] [CrossRef]
| Parameters described | PD + ICD, PD1, n = 49 | PD2, n = 36 |
| Mean age, years | 65.8 ± 8 | 70.6 ± 5.9 |
| Number of subjects male | 23 | 18 |
| Number of subjects female | 26 | 18 |
| Education duration, years | 15.9 ± 3 | 15.8 ± 3.6 |
| Duration of the disease, years | 6.6 ± 4.94 | 7.53 ± 4.9 |
| Hoehn and Yahr stage | 2.2 ± 0.5 | 2.5 ± 0.5 |
| UPDRS, total score | 33.4 ± 11.9 | 36.3 ± 12.2 |
| LEDD, mg/day | 731.5 ± 454 | 762.4 ± 342.1 |
| Duration of the use of dopaminergic therapy, years | 6.6 ± 4.94 | 7.53 ± 4.9 |
| Breakdown of the types of dopaminergic therapy | Levodopa + DA (n = 13; 26.5%), Levodopa + DA + amantadine (n = 13; 26.5%), DA monotherapy (n = 7; 14.3%), Levodopa monotherapy (n = 4; 8.25%), DA + amantadine (n = 4; 8.25%), Levodopa + COMT inhibitor + DA + amantadine (n = 3; 6.1%), Levodopa + amantadine (n = 2; 4.1%), Levodopa + COMT inhibitor + DA (n = 1; 2%), Levodopa + MAO-B inhibitor (n = 1; 2%), Levodopa + DA + amantadine + MAO-B inhibitor (n = 1; 2%). | Levodopa + DA + amantadine (n = 14; 38.9%), Levodopa + DA (n = 12; 33.3%), DA + amantadine (n = 5; 13.9%), DA monotherapy (n = 4; 11.1%), Levodopa + COMT inhibitor + DA + amantadine (n = 1; 2.8%). |
| Gene | Substitution | PD2 | Control | Chi, p | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|---|---|---|
| DBH | rs141116007 | II + ID | 26 | 282 | 1.017 | 0.294 | 0.67 | 0.30–1.64 |
| DD | 10 | 73 | 0.313 | 1.49 | 0.61–3.36 | |||
| BDNF | rs2049046 | AA | 7 | 50 | 0.847 | 0.327 | 1.51 | 0.53–3.76 |
| AT + TT | 29 | 312 | 0.358 | 0.66 | 0.27–1.90 | |||
| DRD2 | rs1799732 | CC | 15 | 18 | 1.911 | 0.189 | 1.83 | 0.71–4.68 |
| CD + DD | 21 | 46 | 0.167 | 0.55 | 0.21–1.42 | |||
| MAOA | VNTR | SS + SL | 20 | 129 | 3.309 | 0.076 | 1.89 | 0.89–4.05 |
| LL | 16 | 195 | 0.069 | 0.53 | 0.25–1.12 | |||
| DRD2 | rs6275 | TT | 18 | 34 | 43.706 | 2.9 × 10−8 | 9.00 | 3.97–20.14 |
| CT + CC | 18 | 306 | 3.8 × 10−11 | 0.11 | 0.05–0.25 | |||
| DBH | rs2097629 | AA | 6 | 114 | 5.995 | 0.016 | 0.34 | 0.11–0.86 |
| AG + GG | 30 | 192 | 0.014 | 2.97 | 1.17–8.97 | |||
| BDNF | rs6265 | AA + AG | 16 | 94 | 8.308 | 6.7 × 10−3 | 2.83 | 1.27–6.29 |
| GG | 16 | 266 | 3.9 × 10−3 | 0.35 | 0.16–0.79 | |||
| DBH | rs1611115 | TT + CT | 26 | 328 | 11.256 | 2.8 × 10−3 | 0.27 | 0.11–0.68 |
| CC | 10 | 34 | 7.9 × 10−4 | 3.71 | 1.46–8.77 | |||
| COMT | rs4680 | AA + AG | 22 | 147 | 3.773 | 0.063 | 0.48 | 0.22–1.11 |
| GG | 14 | 45 | 0.052 | 2.08 | 0.90–4.65 | |||
| DRD2 | rs2283265 | TT + CT | 34 | 161 | 0.572 | 0.610 | 0.53 | 0.08–5.78 |
| CC | 2 | 5 | 0.449 | 1.89 | 0.17–12.13 | |||
| DRD2 | rs12364283 | TT | 30 | 100 | 6.877 | 0.012 | 3.30 | 1.25–10.19 |
| CT + CC | 6 | 66 | 8.7 × 10−3 | 0.30 | 0.10–0.80 | |||
| DRD2 | rs1076560 | TT + CT | 14 | 40 | 3.305 | 0.095 | 2.00 | 0.86–4.53 |
| CC | 22 | 126 | 0.069 | 0.50 | 0.22–1.16 | |||
| SLC6A4 | rs38130034 | TT | 13 | 43 | 2.762 | 0.140 | 1.89 | 0.81–4.28 |
| CT + CC | 23 | 144 | 0.097 | 0.53 | 0.23–1.24 | |||
| ACE | rs4646994 | II + ID | 26 | 228 | 0.367 | 0.708 | 1.27 | 0.57–3.05 |
| DD | 10 | 111 | 0.545 | 0.79 | 0.33–1.77 | |||
| SLC6A3 | rs27072 | CC | 24 | 86 | 2.634 | 0.139 | 1.86 | 0.83–4.36 |
| CT + TT | 12 | 80 | 0.105 | 0.54 | 0.23–1.21 | |||
| DRD1 | rs686 | CC | 0 | 23 | 5.629 | 0.017 | 0.00 | 0.0000–0.7362 |
| CT + TT | 36 | 143 | 0.018 | ∞ | 1.3583–∞ |
| Informative Allelic Pattern | Genotype Carriers | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|
| PD2 | Control | ||||
| DBH_rs2097629:G; DRD1_rs686:G; DRD2_rs12364283:A,A | 75.0% | 29.7% | 6.38 × 10−7 | 7.10 | 3.11–16.21 |
| DBH_rs2097629:G; DRD2_rs6275:T,T | 44.4% | 8.6% | 1.42 × 10−6 | 8.51 | 3.62–20.04 |
| BDNF_rs6265:A; DRD2_rs6275:T,T | 36.4% | 4.3% | 1.92 × 10−6 | 12.57 | 4.45–35.49 |
| DRD2_rs6275:T,T | 50% | 12.3% | 2.24 × 10−6 | 7.15 | 3.20–15.97 |
| Informative Allelic Pattern | Genotype Carriers | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|
| PD2 | Control | ||||
| BDNF_rs6265:G; DRD2_rs6275:C | 45.5% | 86.9% | 9.51 × 10−7 | 0.13 | 0.055–0.29 |
| DRD2_rs6275:C | 50% | 87.7% | 2.24 × 10−6 | 0.14 | 0.063–0.31 |
| Gene | Substitution | PD1 | Control | Chi, p | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|---|---|---|
| DBH | rs141116007 | II + ID | 38 | 282 | 0.312 | 0.580 | 0.82 | 0.39–1.81 |
| DD | 12 | 73 | 0.576 | 1.22 | 0.55–2.53 | |||
| BDNF | rs2049046 | AA | 11 | 50 | 2.335 | 0.138 | 1.76 | 0.76–3.79 |
| AT + TT | 39 | 312 | 0.126 | 0.57 | 0.26–1.32 | |||
| DRD2 | rs1799732 | CC | 20 | 18 | 2.506 | 0.156 | 1.89 | 0.79–4.52 |
| CD + DD | 27 | 46 | 0.113 | 0.53 | 0.22–1.26 | |||
| MAOA | VNTR | SS + SL | 27 | 129 | 3.585 | 0.065 | 1.77 | 0.93–3.39 |
| LL | 23 | 195 | 0.058 | 0.56 | 0.29–1.07 | |||
| DRD2 | rs6275 | TT | 13 | 34 | 10.528 | 3.8 × 10−3 | 3.16 | 1.40–6.81 |
| CT + CC | 37 | 306 | 1.1 × 10−3 | 0.32 | 0.15–0.72 | |||
| DBH | rs2097629 | AA + AG | 39 | 263 | 2.110 | 0.199 | 0.58 | 0.27–1.36 |
| GG | 11 | 43 | 0.146 | 1.73 | 0.74–3.76 | |||
| BDNF | rs6265 | AA + AG | 27 | 94 | 23.224 | 5.7 × 10−6 | 4.49 | 2.24–9.18 |
| GG | 17 | 266 | 1.4 × 10−6 | 0.22 | 0.11–0.45 | |||
| DBH | rs1611115 | TT | 14 | 209 | 15.644 | 1.1 × 10−4 | 0.28 | 0.14–0.56 |
| CT + CC | 36 | 153 | 7.6 × 10−5 | 3.51 | 1.77–7.29 | |||
| COMT | rs4680 | AA | 10 | 52 | 0.446 | 0.575 | 0.77 | 0.32–1.73 |
| AG + GG | 35 | 140 | 0.504 | 1.30 | 0.58–3.16 | |||
| DRD2 | rs2283265 | TT | 26 | 118 | 2.893 | 0.105 | 0.56 | 0.27–1.17 |
| CT + CC | 19 | 48 | 0.089 | 1.80 | 2.74–25.02 | |||
| DRD2 | rs12364283 | TT | 37 | 100 | 7.512 | 7.7 × 10−3 | 3.05 | 1.29–8.04 |
| CT + CC | 8 | 66 | 6.1 × 10−3 | 0.33 | 0.12–0.78 | |||
| DRD2 | rs1076560 | TT + CT | 19 | 40 | 5.774 | 0.024 | 2.30 | 1.08–4.83 |
| CC | 26 | 126 | 0.016 | 0.43 | 0.21–0.93 | |||
| SLC6A4 | rs38130034 | TT | 15 | 43 | 2.068 | 0.179 | 1.67 | 0.76–3.56 |
| CT + CC | 30 | 144 | 0.150 | 0.60 | 0.28–1.31 | |||
| ACE | rs4646994 | II + ID | 38 | 228 | 5.513 | 0.024 | 2.64 | 1.12–7.22 |
| DD | 7 | 111 | 0.019 | 0.38 | 0.14–0.90 | |||
| SLC6A3 | rs27072 | CC + CT | 43 | 149 | 1.452 | 0.377 | 2.45 | 0.55–22.65 |
| TT | 2 | 17 | 0.228 | 0.41 | 0.04–1.83 | |||
| DRD1 | rs686 | CC + CT | 39 | 137 | 0.438 | 0.653 | 1.38 | 0.51–4.34 |
| TT | 6 | 29 | 0.508 | 0.73 | 0.23–1.96 |
| Informative Allelic Pattern | Genotype Carriers | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|
| PD1 | Control | ||||
| ACE_rs4646994:I; BDNF_rs6265:A; DRD2_rs1076560:A | 25.6% | 0.006% | 2.68 × 10−7 | 55.17 | 6.80–447.57 |
| BDNF_rs6265:A; DRD2_rs1076560:A | 28.2% | 2.5% | 3.28 × 10−6 | 15.42 | 4.58–51.86 |
| BDNF_rs6265:A; DBH_rs1611115:T | 43.2% | 12.4% | 1.89 × 10−5 | 5.36 | 2.51–11.44 |
| BDNF_rs6265:G; DBH_rs1611115:T | 72.7% | 37.3% | 2.63 × 10−5 | 4.49 | 2.15–9.37 |
| Informative Allelic Pattern | Genotype Carriers | Fi (p) | OR | CI95% | |
|---|---|---|---|---|---|
| PD1 | Control | ||||
| BDNF_rs6265:G; DBH_rs1611115:C,C | 22.3% | 60.8% | 5.73 × 10−6 | 0.19 | 0.09–0.41 |
| BDNF_rs6265:G,G; DRD2_rs6275:C | 27.3% | 64.6% | 9.77 × 10−6 | 0.21 | 0.10–0.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedosova, A.; Titova, N.; Kokaeva, Z.; Shipilova, N.; Katunina, E.; Klimov, E. Genetic Markers as Risk Factors for the Development of Impulsive-Compulsive Behaviors in Patients with Parkinson’s Disease Receiving Dopaminergic Therapy. J. Pers. Med. 2021, 11, 1321. https://doi.org/10.3390/jpm11121321
Fedosova A, Titova N, Kokaeva Z, Shipilova N, Katunina E, Klimov E. Genetic Markers as Risk Factors for the Development of Impulsive-Compulsive Behaviors in Patients with Parkinson’s Disease Receiving Dopaminergic Therapy. Journal of Personalized Medicine. 2021; 11(12):1321. https://doi.org/10.3390/jpm11121321
Chicago/Turabian StyleFedosova, Anna, Nataliya Titova, Zarema Kokaeva, Natalia Shipilova, Elena Katunina, and Eugene Klimov. 2021. "Genetic Markers as Risk Factors for the Development of Impulsive-Compulsive Behaviors in Patients with Parkinson’s Disease Receiving Dopaminergic Therapy" Journal of Personalized Medicine 11, no. 12: 1321. https://doi.org/10.3390/jpm11121321
APA StyleFedosova, A., Titova, N., Kokaeva, Z., Shipilova, N., Katunina, E., & Klimov, E. (2021). Genetic Markers as Risk Factors for the Development of Impulsive-Compulsive Behaviors in Patients with Parkinson’s Disease Receiving Dopaminergic Therapy. Journal of Personalized Medicine, 11(12), 1321. https://doi.org/10.3390/jpm11121321






