Restoration of Normal NF1 Function with Antisense Morpholino Treatment of Recurrent Pathogenic Patient-Specific Variant c.1466A>G; p.Y489C
Abstract
1. Introduction
2. Materials and Methods
2.1. Mutant Cell Line Generation and Characterization
2.2. Cell Line Characterization
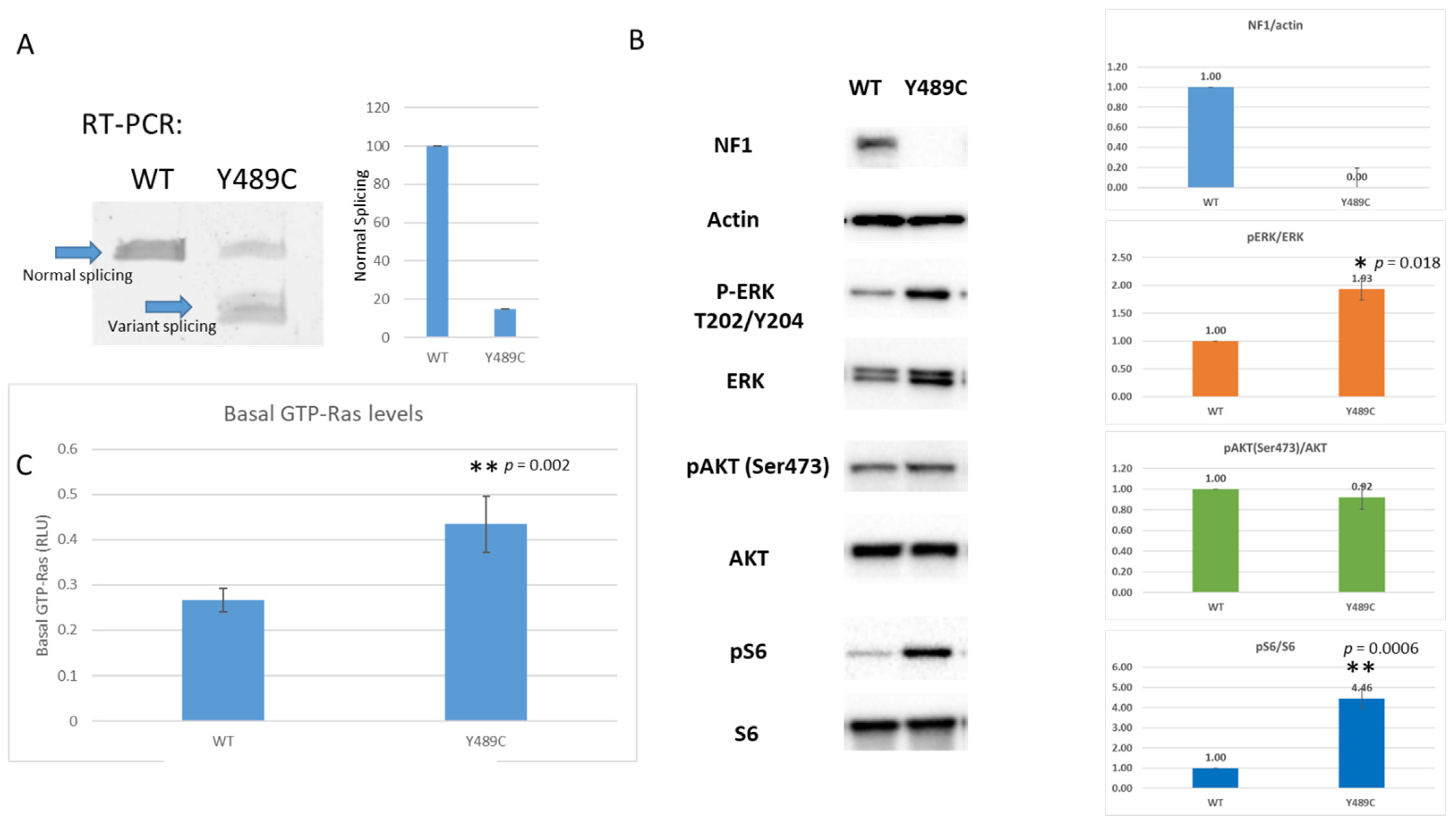
2.2.1. Western Blotting
2.2.2. RAS-G-LISA Assay
2.3. PMO Design and Treatment
2.4. PMO Efficiency Assay
2.5. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutmann, D.; Collins, F.S. The neurofibromatosis type 1 gene and its protein product, neurofibromin. Neuron 1993, 10, 335–343. [Google Scholar] [CrossRef]
- Messiaen, L.; Wimmer, K. Mutational Spectrum. In Neurofibromatoses; Kaufmann, D., Ed.; Karger: Basel, Switzerland, 2008; Volume 16, pp. 63–77. [Google Scholar]
- Pros, E.; Gómez, C.; Martín, T.; Fábregas, P.; Serra, E.; Lázaro, C. Nature and mRNA effect of 282 differentNF1point mutations: Focus on splicing alterations. Hum. Mutat. 2008, 29, E173–E193. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, L.M.; Callens, T.; Mortier, G.; Beysen, D.; Vandenbroucke, I.; Van Roy, N.; Speleman, F.; De Paepe, A. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 2000, 15, 541–555. [Google Scholar] [CrossRef]
- Rojnueangnit, K.; Xie, J.; Gomes, A.; Sharp, A.; Callens, T.; Chen, Y.; Liu, Y.; Cochran, M.; Abbott, M.; Atkin, J.; et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype–Phenotype Correlation. Hum. Mutat. 2015, 36, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, L.M.; Callens, T.; Roux, K.J.; Mortier, G.R.; De Paepe, A.; Abramowicz, M.; Perick-Vance, M.A.; Vance, J.M.; Wallace, M.R. Exon 10b of the NF1 gene represented a mutational hotspot and harbors a recurrent missense mutation y489c associated with aberrant splicing. Genet. Med. 1999, 1, 248–253. [Google Scholar] [CrossRef][Green Version]
- Wallis, D.; Li, K.; Lui, H.; Hu, K.; Chen, M.-J.; Li, J.; Kang, J.; Das, S.; Korf, B.R.; Kesterson, R.A. Neurofibromin (NF1 ) genetic variant structure-function analyses using a full-length mouse cDNA. Hum. Mutat. 2018, 39, 816–821. [Google Scholar] [CrossRef]
- Long, A.; Liu, H.; Liu, J.; Daniel, M.; Bedwell, D.M.; Korf, B.; Kesterson, R.A.; Wallis, D. Analysis of patient-specific NF1 variants leads to functional insights for Ras signaling that can impact personalized medicine. Hum. Mutat. 2021. [Google Scholar] [CrossRef]
- El-Beshlawy, A.; Mostafa, A.; Youssry, I.; Gabr, H.; Mansour, I.M.; El-Tablawy MAziz, M.; Hussein, I.R. Correction of aberrant pre-mRNA splicing by antisense oligonucleotides in beta-thalassemia Egyptian patients with IVSI-110 mutation. J. Pediatr. Hematol. Oncol. 2008, 30, 281–284. [Google Scholar] [CrossRef]
- Akpulat, U.; Wang, H.; Becker, K.; Contreras, A.; Partridge, T.A.; Novak, J.S.; Cirak, S. Shorter Phosphorodiamidate Morpholino Splice-Switching Oligonucleotides May Increase Exon-Skipping Efficacy in DMD. Mol. Ther.-Nucleic Acids 2018, 13, 534–542. [Google Scholar] [CrossRef]
- Pros, E.; Rodríguez, J.F.; Canet, B.; Benito, L.; Sánchez, A.; Benavides, A.; Ramos, F.J.; López-Ariztegui, M.A.; Capella, G.; Blanco, I.; et al. Antisense therapeutics for neurofibromatosis type 1 caused by deep intronic mutations. Hum. Mutat. 2009, 30, 454–462. [Google Scholar] [CrossRef]
- Popplewell, L.J.; Trollet, C.; Dickson, G.; Graham, I.R. Design of Phosphorodiamidate Morpholino Oligomers (PMOs) for the Induction of Exon Skipping of the Human DMD Gene. Mol. Ther. 2009, 17, 554–561. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Gianino, S.M.; Gutmann, D.H. Neurofibromatosis-1 regulation of neural stem cell proliferation and multilineage differentiation operates through distinct RAS effector pathways. Genes Dev. 2015, 29, 1677–1682. [Google Scholar] [CrossRef]
- Rajalingam, K.; Schreck, R.; Rapp, U.R.; Albert, Š. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2007, 1773, 1177–1195. [Google Scholar] [CrossRef]
- Koczkowska, M.; Callens, T.; Chen, Y.; Gomes, A.; Hicks, A.D.; Sharp, A.; Johns, E.; Uhas, K.A.; Armstrong, L.; Bosanko, K.A.; et al. Clinical spectrum of individuals with pathogenic NF1 missense variants affecting p.Met1149, p.Arg1276, and p.Lys1423: Genotype–phenotype study in neurofibromatosis type 1. Hum. Mutat. 2020, 41, 299–315. [Google Scholar] [CrossRef]
- Koczkowska, M.; Callens, T.; Gomes, A.; Sharp, A.; Chen, Y.; Hicks, A.D.; Aylsworth, A.S.; Azizi, A.A.; Basel, D.G.; Bellus, G.; et al. Expanding the clinical phenotype of individuals with a 3-bp in-frame deletion of the NF1 gene (c.2970_2972del): An update of genotype-phenotype correlation. Genet. Med. 2019, 21, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Chen, Y.; Callens, T.; Gomes, A.; Sharp, A.; Johnson, S.; Hsiao, M.-C.; Chen, Z.; Balasubramanian, M.; Barnett, C.P.; et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844–848. Am. J. Hum. Genet. 2018, 102, 69–87. [Google Scholar] [CrossRef]
- Upadhyaya, M.; Huson, S.M.; Davies, M.; Thomas, N.; Chuzhanova, N.; Giovannini, S.; Evans, D.G.; Howard, E.; Kerr, B.; Griffiths, S.; et al. An Absence of Cutaneous Neurofibromas Associated with a 3-bp Inframe Deletion in Exon 17 of the NF1 Gene (c.2970-2972 delAAT): Evidence of a Clinically Significant NF1 Genotype-Phenotype Correlation. Am. J. Hum. Genet. 2007, 80, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Trevisson, E.; Morbidoni, V.; Forzan, M.; Daolio, C.; Fumini, V.; Parrozzani, R.; Cassina, M.; Midena, E.; Salviati, L.; Clementi, M. The Arg1038Gly missense variant in the NF1 gene causes a mild phenotype without neurofibromas. Mol. Genet. Genom. Med. 2019, 7, e616. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, K.; Pyronneau, A.; Chao, M.; Bennett, M.V.L.; Zukin, R.S. Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in fragile X mice. Proc. Natl. Acad. Sci. USA 2016, 113, E6290–E6297. [Google Scholar] [CrossRef]
- Leier, A.; Bedwell, D.M.; Chen, A.T.; Dickson, G.; Keeling, K.M.; Kesterson, R.A.; Korf, B.R.; Lago, T.T.M.; Müller, U.F.; Popplewell, L.; et al. Mutation-Directed Therapeutics for Neurofibromatosis Type I. Mol. Ther.-Nucleic Acids 2020, 20, 739–753. [Google Scholar] [CrossRef]
- Anastasaki, C.; Woo, A.S.; Messiaen, L.M.; Gutmann, D.H. Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum. Mol. Genet. 2015, 24, 3518–3528. [Google Scholar] [CrossRef]
- Li, K.; Turner, A.N.; Chen, M.; Brosius, S.N.; Schoeb, T.R.; Messiaen, L.M.; Bedwell, D.M.; Zinn, K.R.; Anastasaki, C.; Gutmann, D.H.; et al. Mice with missense and nonsense NF1 mutations display divergent phenotypes compared with human neurofibromatosis type I. Dis. Model. Mech. 2016, 9, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Toonen, J.A.; Anastasaki, C.; Smithson, L.J.; Gianino, S.M.; Li, K.; Kesterson, R.A.; Gutmann, D.H. NF1 germline mutation differentially dictates optic glioma formation and growth in neurofibromatosis-1. Hum. Mol. Genet. 2016, 25, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Carnes, R.M.; Kesterson, R.A.; Korf, B.R.; Mobley, J.A.; Wallis, D. Affinity Purification of NF1 Protein-Protein Interactors Identifies Keratins and Neurofibromin Itself as Binding Partners. Genes 2019, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Sherekar, M.; Han, S.W.; Ghirlando, R.; Messing, S.; Drew, M.; Rabara, D.; Waybright, T.; Juneja, P.; O’Neill, H.; Stanley, C.B.; et al. Biochemical and structural analyses reveal that the tumor suppressor neurofibromin (NF1) forms a high-affinity dimer. J. Biol. Chem. 2020, 295, 1105–1119. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awad, E.K.; Moore, M.; Liu, H.; Ciszewski, L.; Lambert, L.; Korf, B.R.; Popplewell, L.; Kesterson, R.A.; Wallis, D. Restoration of Normal NF1 Function with Antisense Morpholino Treatment of Recurrent Pathogenic Patient-Specific Variant c.1466A>G; p.Y489C. J. Pers. Med. 2021, 11, 1320. https://doi.org/10.3390/jpm11121320
Awad EK, Moore M, Liu H, Ciszewski L, Lambert L, Korf BR, Popplewell L, Kesterson RA, Wallis D. Restoration of Normal NF1 Function with Antisense Morpholino Treatment of Recurrent Pathogenic Patient-Specific Variant c.1466A>G; p.Y489C. Journal of Personalized Medicine. 2021; 11(12):1320. https://doi.org/10.3390/jpm11121320
Chicago/Turabian StyleAwad, Elias K., Marc Moore, Hui Liu, Lukasz Ciszewski, Laura Lambert, Bruce R. Korf, Linda Popplewell, Robert A. Kesterson, and Deeann Wallis. 2021. "Restoration of Normal NF1 Function with Antisense Morpholino Treatment of Recurrent Pathogenic Patient-Specific Variant c.1466A>G; p.Y489C" Journal of Personalized Medicine 11, no. 12: 1320. https://doi.org/10.3390/jpm11121320
APA StyleAwad, E. K., Moore, M., Liu, H., Ciszewski, L., Lambert, L., Korf, B. R., Popplewell, L., Kesterson, R. A., & Wallis, D. (2021). Restoration of Normal NF1 Function with Antisense Morpholino Treatment of Recurrent Pathogenic Patient-Specific Variant c.1466A>G; p.Y489C. Journal of Personalized Medicine, 11(12), 1320. https://doi.org/10.3390/jpm11121320





