Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging
Abstract
:1. Introduction
2. Methods
2.1. Study Population
2.2. Imaging Protocols and Data Preparation
Cardiac Magnetic Resonance Imaging
2.3. Experimental Setting
2.4. Data Preprocessing
2.5. Statistical Analysis of Convolutional Neural Network Performance
3. Results
3.1. Clinical Characteristics of Study Participants
3.2. Cardiac Magnetic Resonance Imaging-Based Diagnostic Ability of the Convolutional Neural Network
3.3. The effect of Imaging Protocol on Diagnostic Accuracy
3.4. The Effect of the Deep Learning Technique on Diagnostic Accuracy
3.5. The Effect of Prediction Protocol on Diagnostic Accuracy
4. Discussion
4.1. Cardiac Magnetic Resonance Imaging for the Diagnosis of Cardiac Amyloidosis
4.2. Role and Contribution of AI for CA Diagnosis
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations and Acronyms
| CA | cardiac amyloidosis |
| CMR | cardiac magnetic resonance |
| HFpEF | heart failure with preserved ejection fraction |
| HF | heart failure |
| AL | amyloid light chains |
| LV | left ventricle |
| HF | heart failure |
| CNN | convolutional neural network |
| ECG | electrocardiogram |
| EMB | endomyocardial biopsy |
| LGE | late gadolinium enhancement |
| MOLLI | modified look-locker inversion recovery |
| ECV | extracellular volume |
| CV | cross-validation |
| ROC AUC | varea under the receiver-operating characteristic curve |
| NT-proBNP | N-terminal prohormone of brain natriuretic peptide |
| XAI | explainable artificial intelligence |
References
- Palladini, G.; Campana, C.; Klersy, C.; Balduini, A.; Vadacca, G.; Perfetti, V.; Perlini, S.; Obici, L.; Ascari, E.; D’Eril, G.M.; et al. Serum N-Terminal Pro–Brain Natriuretic Peptide Is a Sensitive Marker of Myocardial Dysfunction in AL Amyloidosis. Circulation 2003, 107, 2440–2445. [Google Scholar] [CrossRef] [PubMed]
- Falk, R.H.; Alexander, K.M.; Liao, R.; Dorbala, S. AL (Light-Chain) Cardiac Amyloidosis: A Review of Diagnosis and Therapy. J. Am. Coll. Cardiol. 2016, 68, 1323–1341. [Google Scholar] [CrossRef]
- Tanskanen, M.; Peuralinna, T.; Polvikoski, T. Senile systemic amyloidosis affects 25% of the very aged and associates with genetic variation in alpha2-macroglobulin and tau: A population-based autopsy study. Ann. Med. 2008, 40, 232–239. [Google Scholar] [CrossRef]
- González-López, E.; Gallego-Delgado, M.; Guzzo-Merello, G.; Moral, F.J.D.H.-D.; Cobo-Marcos, M.; Robles, C.; Bornstein, B.; Salas, C.; Lara-Pezzi, E.; Alonso-Pulpon, L.; et al. Wild-type transthyretin amyloidosis as a cause of heart failure with preserved ejection fraction. Eur. Heart J. 2015, 36, 2585–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scully, P.R.; Treibel, T.; Fontana, M.; Hartman, N.; Lloyd, G.; Pugliese, F.; Sabharwal, N.; Newton, J.; Kelion, A.; Mullen, M.; et al. 1 A multi-centre study of cardiac amyloidosis in tavi patients. Heart 2018, 104, A15. [Google Scholar] [CrossRef]
- Cavalcante, J.L.; Rijal, S.; Abdelkarim, I.; Althouse, A.D.; Sharbaugh, M.S.; Fridman, Y.; Soman, P.; Forman, D.E.; Schindler, J.T.; Gleason, T.G.; et al. Cardiac amyloidosis is prevalent in older patients with aortic stenosis and carries worse prognosis. J. Cardiovasc. Magn. Reson. 2017, 19, 98. [Google Scholar] [CrossRef] [Green Version]
- Wechalekar, A.D.; Schönland, S.; Kastritis, E.; Gillmore, J.D.; Dimopoulos, M.; Lane, T.; Foli, A.; Foard, D.; Milani, P.; Rannigan, L.; et al. A European collaborative study of treatment outcomes in 346 patients with cardiac stage III AL amyloidosis. Blood 2013, 121, 3420–3427. [Google Scholar] [CrossRef] [Green Version]
- Gillmore, J.D.; Maurer, M.S.; Falk, R.H.; Merlini, G.; Damy, T.; Dispenzieri, A.; Wechalekar, A.D.; Berk, J.L.; Quarta, C.C.; Grogan, M.; et al. Nonbiopsy Diagnosis of Cardiac Transthyretin Amyloidosis. Circulation 2016, 133, 2404–2412. [Google Scholar] [CrossRef] [PubMed]
- Bonderman, D.; Agis, H.; Kain, R.; Mascherbauer, J. Amyloid in the heart: An under-recognized threat at the interface of cardiology, haematology, and pathology. Eur. Heart J. Cardiovasc. Imaging 2016, 17, 978–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonderman, D.; Pölzl, G.; Ablasser, K.; Agis, H.; Aschauer, S.; Auer-Grumbach, M.; Binder, C.; Dörler, J.; Duca, F.; Ebner, C.; et al. Diagnosis and treatment of cardiac amyloidosis: An interdisciplinary consensus statement. Wien. Klin. Wochenschr. 2020, 132, 742–761. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.M.; Barkhausen, J.; Flamm, S.D.; Kim, R.J.; Nagel, E. Standardized cardiovascular magnetic resonance imaging (CMR) protocols, society for cardiovascular magnetic resonance: Board of trustees task force on standardized protocols. J. Cardiovasc. Magn. Reson. 2008, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellman, P.; Wilson, J.R.; Xue, H.; Ugander, M.; Arai, A.E. Extracellular volume fraction mapping in the myocardium, part 1: Evaluation of an automated method. J. Cardiovasc. Magn. Reson. 2012, 14, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Ruopp, M.D.; Perkins, N.J.; Whitcomb, B.W.; Schisterman, E.F. Youden Index and Optimal Cut-Point Estimated from Observations Affected by a Lower Limit of Detection. Biom. J. 2008, 50, 419–430. [Google Scholar] [CrossRef] [Green Version]
- Sutskever, I.; Martens, J.; Dahl, G.; Hinton, G. On the importance of initialization and momentum in deep learning. In Proceedings of the 30th International Conference on Machine Learning. Proceedings of Machine Learning Research, Atlanta, GA, USA, 16–21 June 2013; Sanjoy, D., David, M., Eds.; PMLR: Atlanta, GA, USA, 2013; pp. 1139–1147. [Google Scholar]
- Yao, Y.; Rosasco, L.; Caponnetto, A. On Early Stopping in Gradient Descent Learning. Constr. Approx. 2007, 26, 289–315. [Google Scholar] [CrossRef]
- Fontana, M.; Chung, R.; Hawkins, P.N.; Moon, J.C. Cardiovascular magnetic resonance for amyloidosis. Heart Fail. Rev. 2014, 20, 133–144. [Google Scholar] [CrossRef]
- Chacko, L.; Martone, R.; Cappelli, F.; Fontana, M. Cardiac Amyloidosis: Updates in Imaging. Curr. Cardiol. Rep. 2019, 21, 108. [Google Scholar] [CrossRef] [Green Version]
- Fontana, M.; Corovic, A.; Scully, P.; Moon, J.C. Myocardial Amyloidosis: The Exemplar Interstitial Disease. JACC Cardiovasc. Imaging 2019, 12, 2345–2356. [Google Scholar] [CrossRef]
- Maceira, A.M.; Joshi, J.; Prasad, S.K.; Moon, J.; Perugini, E.; Harding, I.; Sheppard, M.; Poole-Wilson, P.A.; Hawkins, P.N.; Pennell, D. Cardiovascular Magnetic Resonance in Cardiac Amyloidosis. Circulation 2005, 111, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Banypersad, S.M.; Fontana, M.; Maestrini, V.; Sado, D.M.; Captur, G.; Petrie, A.; Piechnik, S.K.; Whelan, C.J.; Herrey, A.S.; Gillmore, J.D.; et al. T1 mapping and survival in systemic light-chain amyloidosis. Eur. Heart J. 2015, 36, 244–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, B.A.; Tang, W.W.; Rodriguez, E.R.; Tan, C.; Flamm, S.D.; Taylor, D.O.; Starling, R.C.; Desai, M.Y. Delayed Hyper-Enhancement Magnetic Resonance Imaging Provides Incremental Diagnostic and Prognostic Utility in Suspected Cardiac Amyloidosis. JACC Cardiovasc. Imaging 2009, 2, 1369–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatti, S.; Watts, E.; Syed, F.; Vallurupalli, S.; Pandey, T.; Jambekar, K.; Mazur, W.; Hakeem, A. Clinical and prognostic utility of cardiovascular magnetic resonance imaging in myeloma patients with suspected cardiac amyloidosis. Eur. Heart J. Cardiovasc. Imaging 2016, 17, 970–977. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.X.; Petersen, S.E.; Sanghvi, M.M.; Lu, G.M.; Zhang, L.J. Cardiovascular magnetic resonance imaging for amyloidosis: The state-of-the-art. Trends Cardiovasc. Med. 2018, 29, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Mahara, K.; Beussink-Nelson, L.; Ikura, H.; Katsumata, Y.; Endo, J.; Gaggin, H.K.; Shah, S.J.; Itabashi, Y.; MacRae, C.A.; et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat. Commun. 2021, 12, 2726. [Google Scholar] [CrossRef]
- Agibetov, A.; Seirer, B.; Dachs, T.-M.; Koschutnik, M.; Dalos, D.; Rettl, R.; Duca, F.; Schrutka, L.; Agis, H.; Kain, R.; et al. Machine Learning Enables Prediction of Cardiac Amyloidosis by Routine Laboratory Parameters: A Proof-of-Concept Study. J. Clin. Med. 2020, 9, 1334. [Google Scholar] [CrossRef]
- Martini, N.; Aimo, A.; Barison, A.; Della Latta, D.; Vergaro, G.; Aquaro, G.D.; Ripoli, A.; Emdin, M.; Chiappino, D. Deep learning to diagnose cardiac amyloidosis from cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2020, 22, 84. [Google Scholar] [CrossRef]
- Slomka, P.J.; Dey, D.; Sitek, A.; Motwani, M.; Berman, D.S.; Germano, G. Cardiac imaging: Working towards fully-automated machine analysis & interpretation. Expert Rev. Med. Devices 2017, 14, 197–212. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Martin-Isla, C.; Campello, V.M.; Izquierdo, C.; Raisi-Estabragh, Z.; Baeßler, B.; Petersen, S.E.; Lekadir, K. Image-Based Cardiac Diagnosis With Machine Learning: A Review. Front. Cardiovasc. Med. 2020, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Ploeg, T.; Austin, P.C.; Steyerberg, E.W. Modern modelling techniques are data hungry: A simulation study for predicting dichotomous endpoints. BMC Med. Res. Methodol. 2014, 14, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perfetto, F.; Bergesio, F.; Emdin, M.; Cappelli, F. Troponins in cardiac amyloidosis: Multipurpose markers. Nat. Rev. Cardiol. 2014, 11, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajpurkar, P.; Irvin, J.; Bagul, A. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. Med. Imaging Deep. Learn. 2017. Available online: https://arxiv.org/abs/1712.06957 (accessed on 28 September 2021).
| Non-Amyloidosis Related HF (n = 420) | Amyloidosis (n = 82) | p-Value (Adjusted) | |
|---|---|---|---|
| Clinical parameters, median (IQR) | |||
| Age, years | 66.0 (50.0–75.0) | 75.0 (68.0–82.5) | <0.001 |
| Male sex, no. (%) | 188 (44.9) | 52 (65.8) | 0.003 |
| Height, cm | 169.5 (162.0–178.0) | 167.0 (162.5–172.0) | 0.578 |
| Weight, kg | 76.0 (65.8–88.2) | 75.0 (65.0–79.5) | 0.596 |
| Body mass index, kg/m2 | 27.0 (23.9–31.0) | 25.5 (23.9–28.7) | 0.066 |
| Laboratory parameters, median (IQR) | |||
| NT-proBNP, pg/mL | 452.0 (143.9–1380.0) | 3002.0 (1282.5–7453.0) | <0.001 |
| Serum creatinine, mg/dL | 0.9 (0.8–1.1) | 1.2 (1.0–1.6) | <0.001 |
| Estimated GFR, mL/min/1.73 m2 | 78.0 (55.0–106.0) | 50.0 (38.8–60.5) | <0.001 |
| C-Reactive Protein, mg/dL | 0.3 (0.1–0.7) | 0.3 (0.2–0.7) | 0.340 |
| Troponin T, mg/L | 17.0 (7.0–29.0) | 79.0 (64.0–122.0) | <0.001 |
| NYHA functional class, no. (%) | <0.001 | ||
| I | 167 (40.3) | 10 (12.8) | |
| II | 125 (30.2) | 24 (30.8) | |
| III | 107 (25.8) | 40 (51.3) | |
| IV | 13 (3.1) | 2 (2.6) | |
| Missing data | 2 (0.5) | 2 (2.6) | |
| Medical history, no. (%) | |||
| Hypertension | 304 (72.7) | 50 (63.3) | 0.236 |
| Atrial fibrillation | 127 (30.8) | 36 (46.2) | 0.031 |
| Coronary artery disease | 110 (26.6) | 20 (25.3) | 0.917 |
| Myocardial infarction | 42 (10.2) | 4 (5.1) | 0.339 |
| Percutaneous coronary intervention | 57 (13.7) | 8 (10.1) | 0.591 |
| Coronary artery bypass grafting | 22 (5.3) | 5 (6.3) | 0.829 |
| Diabetes mellitus type II | 77 (18.4) | 13 (16.5) | 0.829 |
| Treatment, no. (%) * | |||
| Oral anticoagulants | 134 (32.4) | 41 (52.6) | 0.003 |
| Diuretic agent | 153 (37.0) | 50 (64.1) | <0.001 |
| Mineralocorticoid-receptor antagonist | 99 (24.0) | 28 (35.9) | 0.084 |
| ACE inhibitor or ARB | 229 (55.4) | 36 (46.2) | 0.278 |
| Beta-blocker | 232 (56.2) | 36 (46.2) | 0.245 |
| Calcium channel antagonist | 66 (16.0) | 8 (10.3) | 0.478 |
| Statin | 159 (38.4) | 23 (29.5) | 0.278 |
| Cardiac magnetic resonance imaging parameters, median (IQR) | |||
| Myocardial native T1 time, ms | 1050.9 (998.1–1103.8) | 1107.6 (1074.5–1140.7) | <0.001 |
| Extracellular volume, % | 33.5 (28.8–38.3) | 46.7 (40.6–52.8) | <0.001 |
| Imaging Protocol | Feature Extraction | From Scratch | Fine-Tuning | ||||
|---|---|---|---|---|---|---|---|
| ROC AUC | Se (Sp) | ROC AUC | Se (Sp) | ROC AUC | Se (Sp) | ||
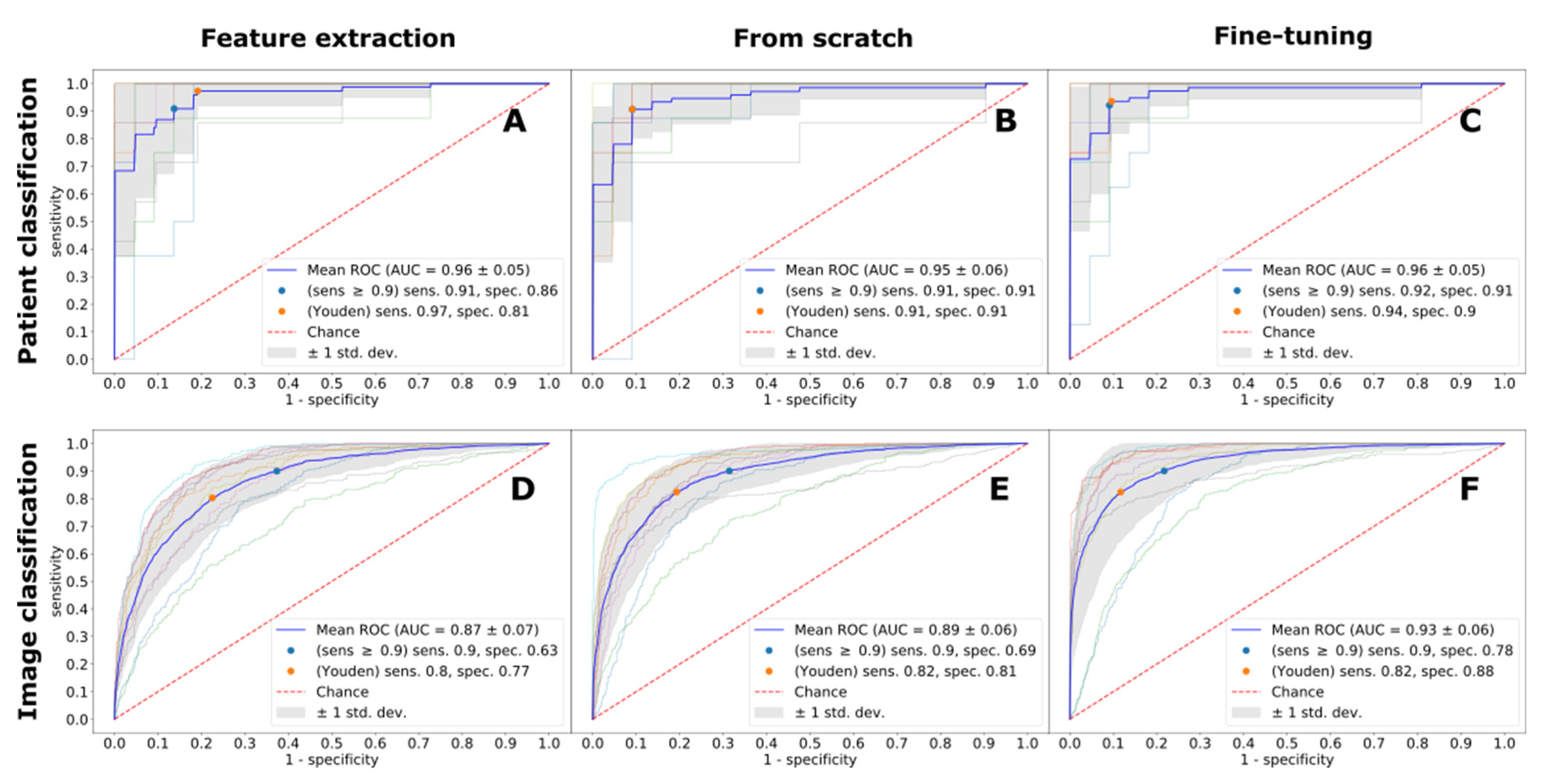
| LGE | Patient | 0.96 | 0.97 (0.81) | 0.95 | 0.91 (0.91) | 0.96 | 0.94 (0.9) |
| Image | 0.87 | 0.8 (0.77) | 0.89 | 0.82 (0.81) | 0.93 | 0.82 (0.88) | |
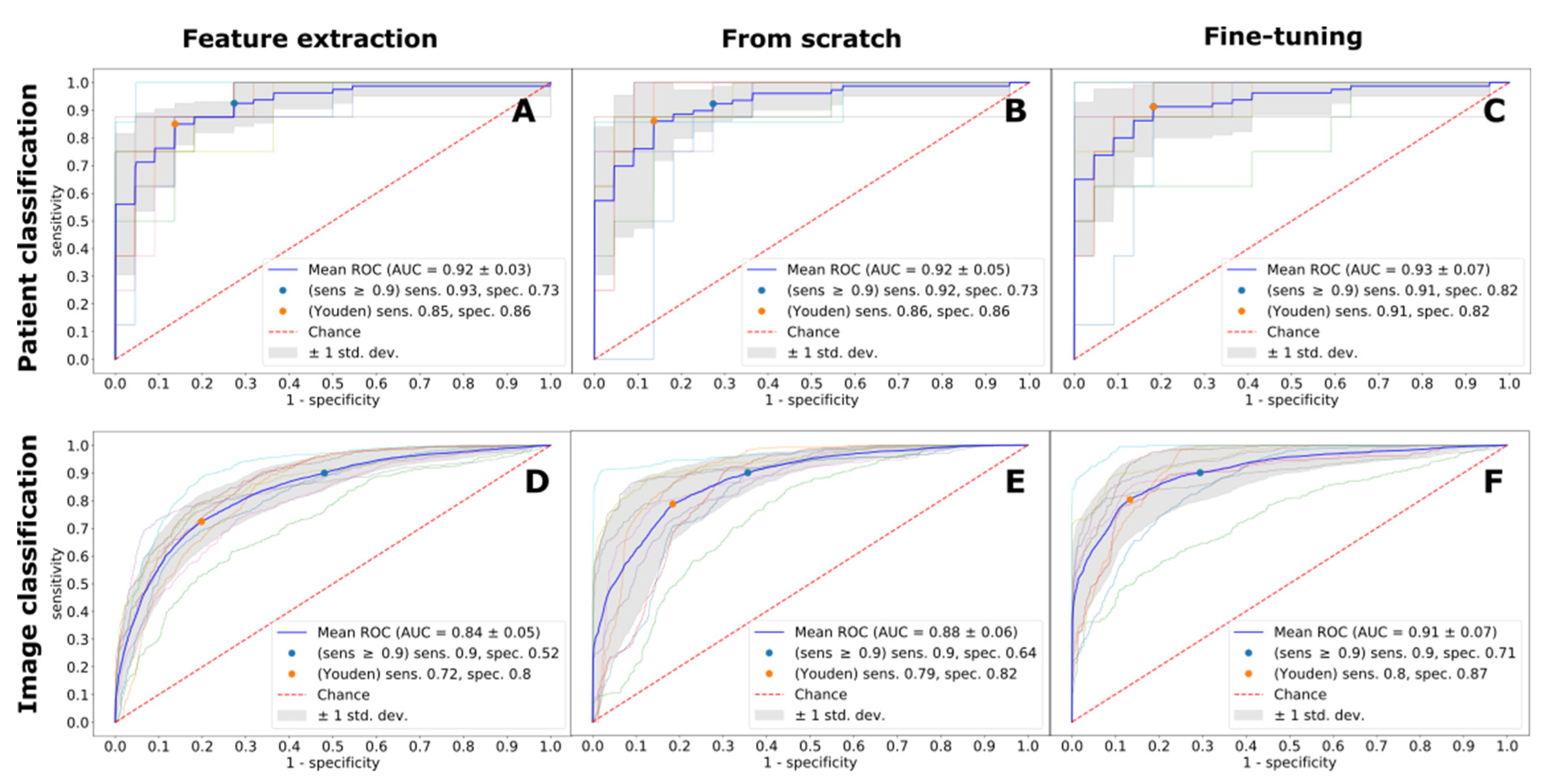
| MOLLI | Patient | 0.92 | 0.85 (0.86) | 0.92 | 0.86 (0.86) | 0.93 | 0.91 (0.82) |
| Image | 0.84 | 0.72 (0.8) | 0.88 | 0.79 (0.82) | 0.91 | 0.8 (0.87) | |
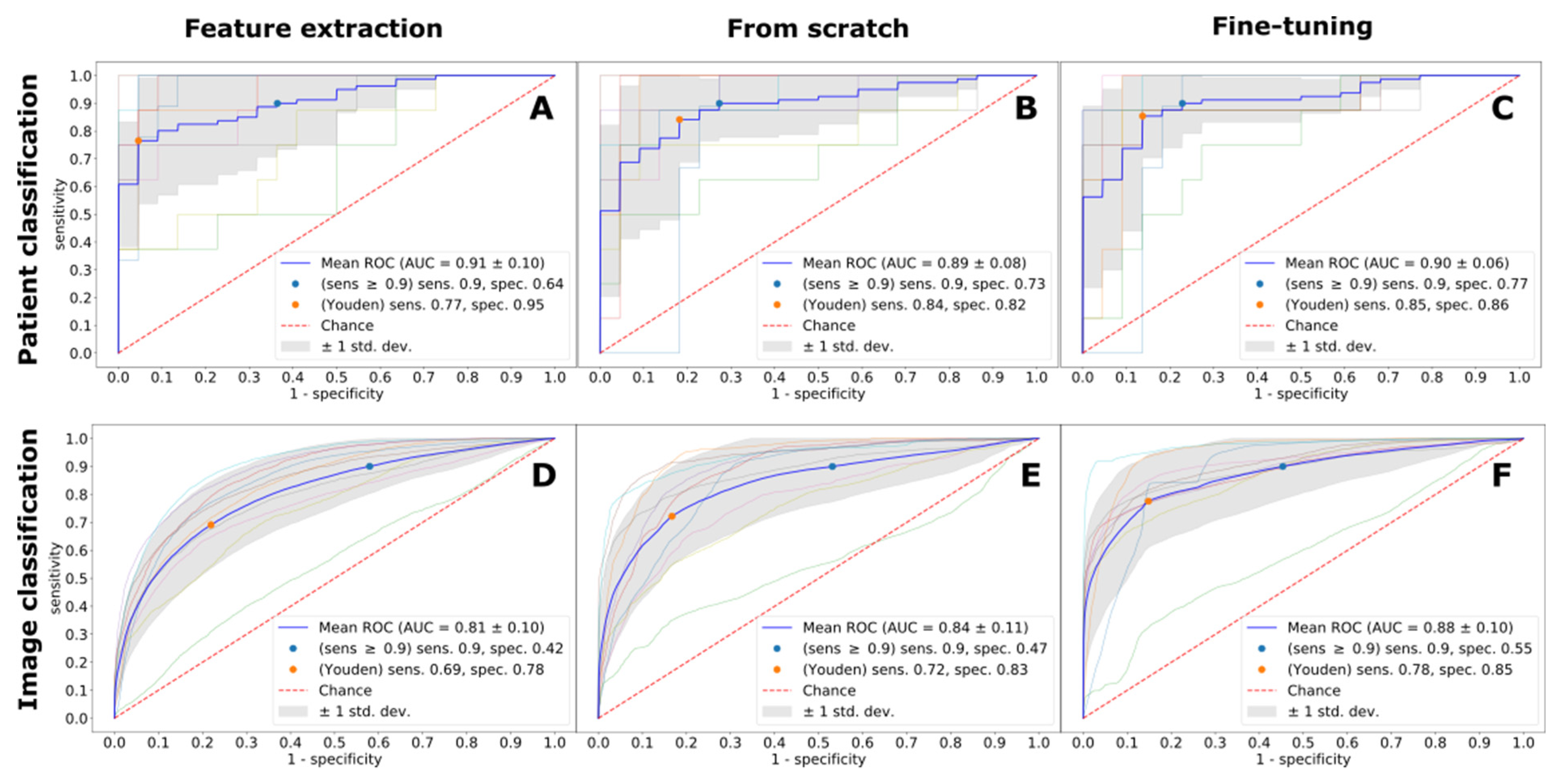
| CINE | Patient | 0.91 | 0.77 (0.95) | 0.89 | 0.84 (0.82) | 0.90 | 0.85 (0.86) |
| Image | 0.81 | 0.69 (0.78) | 0.84 | 0.72 (0.83) | 0.88 | 0.78 (0.85) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agibetov, A.; Kammerlander, A.; Duca, F.; Nitsche, C.; Koschutnik, M.; Donà, C.; Dachs, T.-M.; Rettl, R.; Stria, A.; Schrutka, L.; et al. Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging. J. Pers. Med. 2021, 11, 1268. https://doi.org/10.3390/jpm11121268
Agibetov A, Kammerlander A, Duca F, Nitsche C, Koschutnik M, Donà C, Dachs T-M, Rettl R, Stria A, Schrutka L, et al. Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging. Journal of Personalized Medicine. 2021; 11(12):1268. https://doi.org/10.3390/jpm11121268
Chicago/Turabian StyleAgibetov, Asan, Andreas Kammerlander, Franz Duca, Christian Nitsche, Matthias Koschutnik, Carolina Donà, Theresa-Marie Dachs, René Rettl, Alessa Stria, Lore Schrutka, and et al. 2021. "Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging" Journal of Personalized Medicine 11, no. 12: 1268. https://doi.org/10.3390/jpm11121268
APA StyleAgibetov, A., Kammerlander, A., Duca, F., Nitsche, C., Koschutnik, M., Donà, C., Dachs, T.-M., Rettl, R., Stria, A., Schrutka, L., Binder, C., Kastner, J., Agis, H., Kain, R., Auer-Grumbach, M., Samwald, M., Hengstenberg, C., Dorffner, G., Mascherbauer, J., & Bonderman, D. (2021). Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging. Journal of Personalized Medicine, 11(12), 1268. https://doi.org/10.3390/jpm11121268










