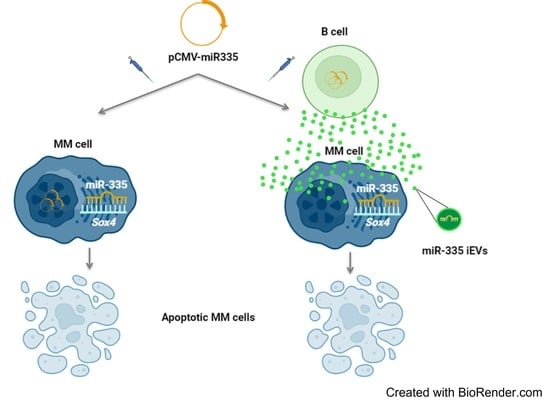
miR-335-laden B Cell-Derived Extracellular Vesicles Promote SOX4-Dependent Apoptosis in Human Multiple Myeloma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. B Cell Purification
2.2. Cells Culture
2.3. Plasmid Construct and Cells Transfection
2.4. miR-335 Analysis
2.5. IEV Isolation and Quantification
2.6. B Cell-Derived IEVS and U266B1 Cells
2.7. RT-qPCR
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
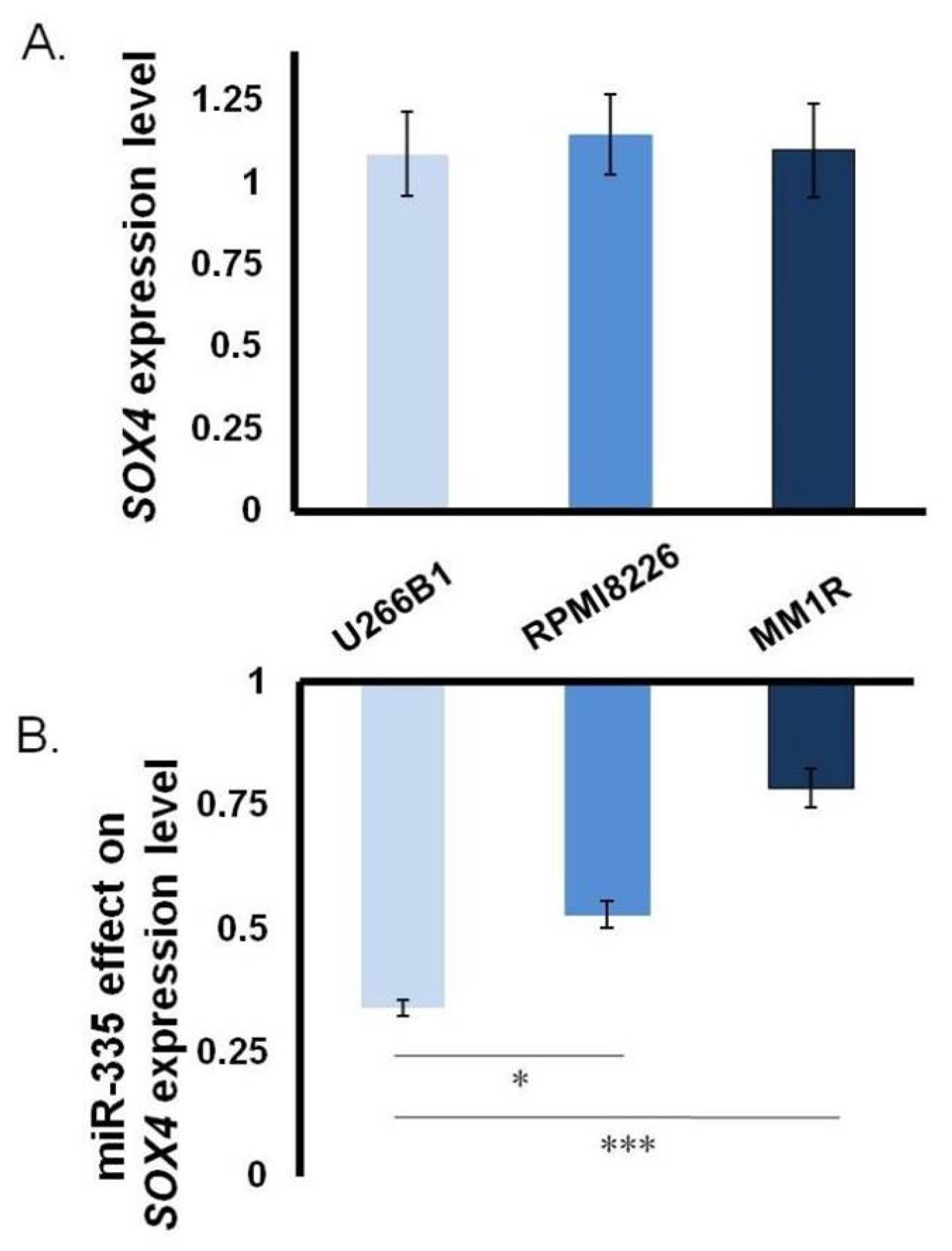
3.1. Close Correlation between miR-335 and SOX4 in MM
3.2. Direct Effect of miR-335 Effect on Tumor Cells
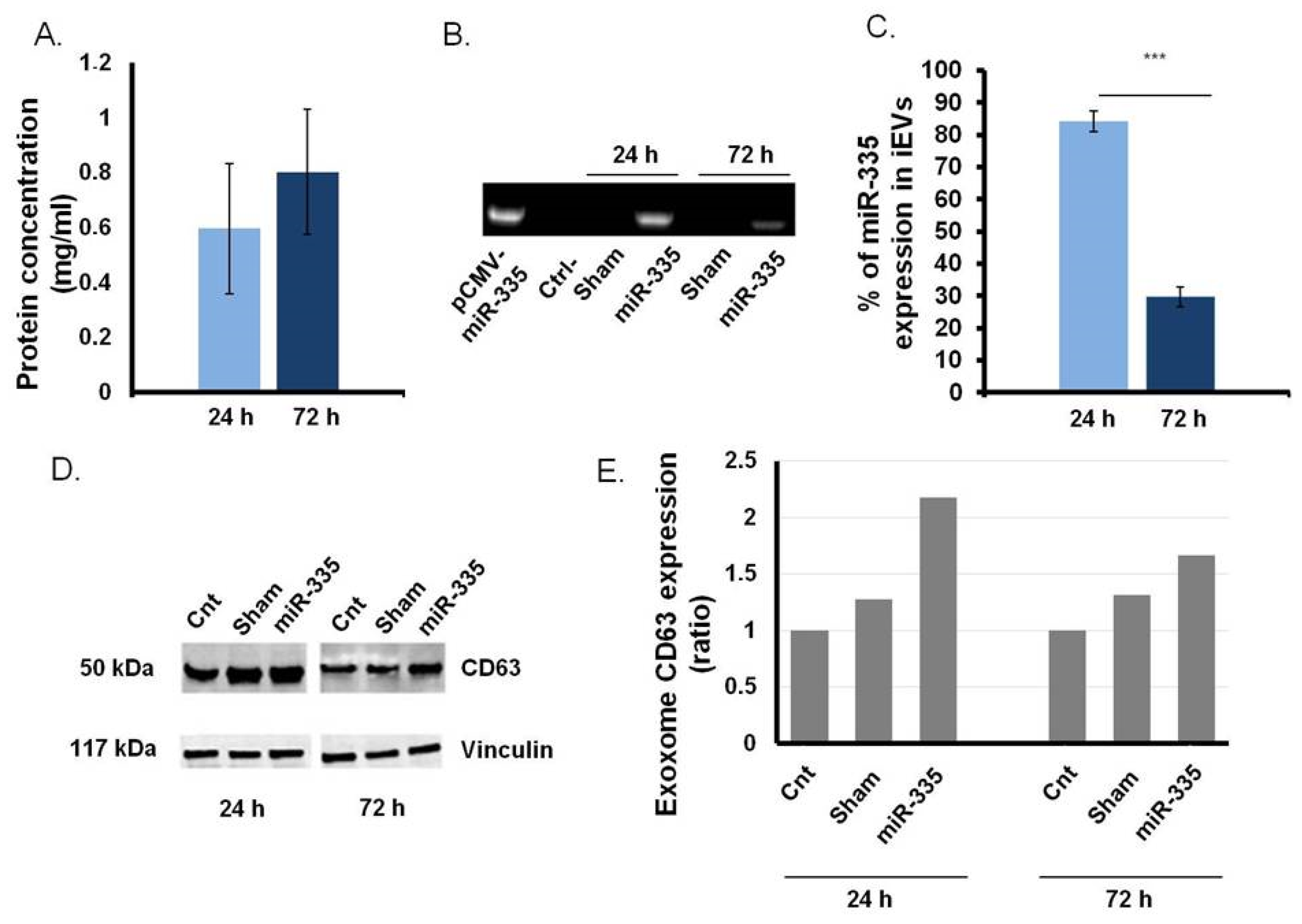
3.3. B Cells as Efficient Bioreactor for iEVs Production
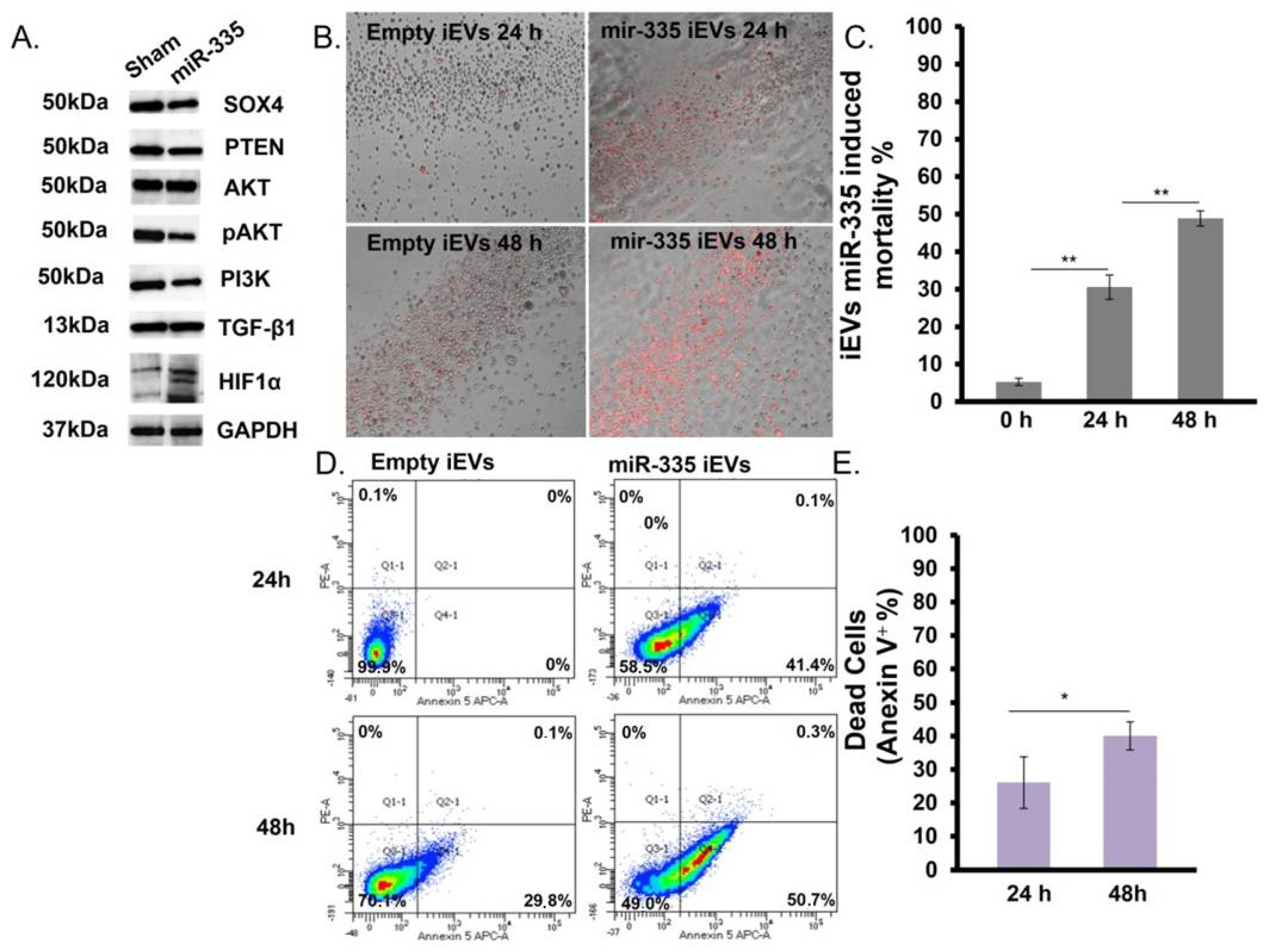
3.4. IEVs Effect on U266B1 MM Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Storti, P.; Bolzoni, M.; Donofrio, G.; Airoldi, I.; Guasco, D.; Toscani, D.; Martella, E.; Lazzaretti, M.; Mancini, C.; Agnelli, L.; et al. Hypoxia-Inducible Factor (HIF)-1α Suppression in Myeloma Cells Blocks Tumoral Growth in Vivo Inhibiting Angiogenesis and Bone Destruction. Leukemia 2013, 27, 1697–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacca, A.; Ribatti, D.; Roncali, L.; Ranieri, G.; Serio, G.; Silvestris, F.; Dammacco, F. Bone Marrow Angiogenesis and Progression in Multiple Myeloma. Br. J. Haematol. 1994, 87, 503–508. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple Myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef] [PubMed]
- Kehrer, M.; Koob, S.; Strauss, A.; Wirtz, D.; Schmolders, J. Multiples Myelom–Aktuelle Standards in Diagnostik und Therapie. Z. Orthop. Unf. 2017, 155, 575–586. [Google Scholar] [CrossRef]
- Dong, C.; Wilhelm, D.; Koopman, P. Sox Genes and Cancer. Cytogenet. Genome Res. 2004, 105, 442–447. [Google Scholar] [CrossRef]
- Vervoort, S.J.; van Boxtel, R.; Coffer, P.J. The Role of SRY-Related HMG Box Transcription Factor 4 (SOX4) in Tumorigenesis and Metastasis: Friend or Foe? Oncogene 2013, 32, 3397–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, R.; Pan, S.; Qi, S.; Lin, X.; Cheng, S. Epigenetic Repression of MicroRNA-129-2 Leads to Overexpression of SOX4 in Gastric Cancer. Biochem. Biophys. Res. Commun. 2010, 394, 1047–1052. [Google Scholar] [CrossRef]
- Hur, W.; Rhim, H.; Jung, C.K.; Kim, J.D.; Bae, S.H.; Jang, J.W.; Yang, J.M.; Oh, S.-T.; Kim, D.G.; Wang, H.J.; et al. SOX4 Overexpression Regulates the P53-Mediated Apoptosis in Hepatocellular Carcinoma: Clinical Implication and Functional Analysis in Vitro. Carcinogenesis 2010, 31, 1298–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, J.; Kim, H.; Li, Z.; Moreno, C. SOX4 Regulates Invasion of Bladder Cancer Cells via Repression of WNT5a. Int. J. Oncol. 2019, 55, 359–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, C.S. The Sex-Determining Region Y-Box 4 and Homeobox C6 Transcriptional Networks in Prostate Cancer Progression. Am. J. Pathol. 2010, 176, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Madan, A.; Yoon, J.-G.; Fang, X.; Yan, X.; Kim, T.-K.; Hwang, D.; Hood, L.; Foltz, G. Massively Parallel Signature Sequencing and Bioinformatics Analysis Identifies Up-Regulation of TGFBI and SOX4 in Human Glioblastoma. PLoS ONE 2010, 5, e10210. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Sigvardsson, M. The Roles of Transcription Factors in B Lymphocyte Commitment, Development, and Transformation. J. Leukoc. Biol. 2004, 75, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.S.; Otsuyama, K.; Shamsasenjan, K.; Asaoku, H.; Kawano, M.M. CD56 Expression in Human Myeloma Cells Derived from the Neurogenic Gene Expression: Possible Role of the SRY-HMG Box Gene, SOX4. Int. J. Hematol. 2010, 91, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Blobe, G.C. Role of Transforming Growth Factor-β in Hematologic Malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Li, L.; Piontek, K.; Sakaguchi, M.; Selaru, F.M. Exosome MiR-335 as a Novel Therapeutic Strategy in Hepatocellular Carcinoma: Wang et Al. Hepatology 2018, 67, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Scharer, C.D.; McCabe, C.D.; Ali-Seyed, M.; Berger, M.F.; Bulyk, M.L.; Moreno, C.S. Genome-Wide Promoter Analysis of the SOX4 Transcriptional Network in Prostate Cancer Cells. Cancer Res. 2009, 69, 709–717. [Google Scholar] [CrossRef] [Green Version]
- Almanza, G.; Rodvold, J.J.; Tsui, B.; Jepsen, K.; Carter, H.; Zanetti, M. Extracellular Vesicles Produced in B Cells Deliver Tumor Suppressor MiR-335 to Breast Cancer Cells Disrupting Oncogenic Programming In Vitro and In Vivo. Sci. Rep. 2018, 8, 17581. [Google Scholar] [CrossRef] [PubMed]
- Burkova, E.E.; Grigor’eva, A.E.; Bulgakov, D.V.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Sedykh, S.E.; Nevinsky, G.A. Extra Purified Exosomes from Human Placenta Contain an Unpredictable Small Number of Different Major Proteins. Int. J. Mol. Sci. 2019, 20, 2434. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A Genome-Wide Transcriptomic Analysis of Protein-Coding Genes in Human Blood Cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.A.; Parker, J.S.; Silva, G.O.; Hoadley, K.A.; Perou, C.M.; Gatza, M.L. Amplification of SOX4 Promotes PI3K/Akt Signaling in Human Breast Cancer. Breast Cancer Res. Treat. 2017, 162, 439–450. [Google Scholar] [CrossRef]
- Ramezani-Rad, P.; Geng, H.; Hurtz, C.; Chan, L.N.; Chen, Z.; Jumaa, H.; Melnick, A.; Paietta, E.; Carroll, W.L.; Willman, C.L.; et al. SOX4 Enables Oncogenic Survival Signals in Acute Lymphoblastic Leukemia. Blood 2013, 121, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, P.P.; Castillo, S.D.; Blanco, S.; Sanz-Garcia, M.; Largo, C.; Alvarez, S.; Yokota, J.; Gonzalez-Neira, A.; Benitez, J.; Clevers, H.C.; et al. The SRY-HMG Box Gene, SOX4, Is a Target of Gene Amplification at Chromosome 6p in Lung Cancer. Hum. Mol. Genet. 2009, 18, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Almanza, G.; Anufreichik, V.; Rodvold, J.J.; Chiu, K.T.; DeLaney, A.; Akers, J.C.; Chen, C.C.; Zanetti, M. Synthesis and Delivery of Short, Noncoding RNA by B Lymphocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 20182–20187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urashima, M.; Ogata, A.; Chauhan, D.; Hatziyanni, M.; Vidriales, M.; Dedera, D.; Schlossman, R.; Anderson, K. Transforming Growth Factor-Beta1: Differential Effects on Multiple Myeloma versus Normal B Cells. Blood 1996, 87, 1928–1938. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.; Miao, L.; Lin, Y.; Huang, X.; Ge, X.; Moosa, S.L.; Liu, B.; Ren, M.; Zhou, Q.; Liang, H.; et al. Regulation Mechanism of Oxidative Stress Induced by High Glucose through PI3K/Akt/Nrf2 Pathway in Juvenile Blunt Snout Bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2017, 70, 66–75. [Google Scholar] [CrossRef]
- Kabir, T.; Leigh, R.; Tasena, H.; Mellone, M.; Coletta, R.; Parkinson, E.; Prime, S.; Thomas, G.; Paterson, I.; Zhou, D.; et al. A MiR-335/COX-2/PTEN Axis Regulates the Secretory Phenotype of Senescent Cancer-Associated Fibroblasts. Aging 2016, 8, 1608–1635. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zeng, F.; Wu, J.-Y.; Li, H.-Y.; Fan, J.-J.; Mai, L.; Zhang, J.; Ma, D.-M.; Li, Y.; Song, F. MiR-335 Inhibits Migration of Breast Cancer Cells through Targeting Oncoprotein c-Met. Tumor Biol. 2015, 36, 2875–2883. [Google Scholar] [CrossRef]
- Carmeliet, P.; Jain, R.K. Angiogenesis in Cancer and Other Diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardi, E.; Almanza, G.; Kowal, K.; Valvasori, M.; Agostini, F.; Vicinanza, C.; Da Ros, F.; Durante, C.; Marangon, M.; Michieli, M.; et al. miR-335-laden B Cell-Derived Extracellular Vesicles Promote SOX4-Dependent Apoptosis in Human Multiple Myeloma Cells. J. Pers. Med. 2021, 11, 1240. https://doi.org/10.3390/jpm11121240
Lombardi E, Almanza G, Kowal K, Valvasori M, Agostini F, Vicinanza C, Da Ros F, Durante C, Marangon M, Michieli M, et al. miR-335-laden B Cell-Derived Extracellular Vesicles Promote SOX4-Dependent Apoptosis in Human Multiple Myeloma Cells. Journal of Personalized Medicine. 2021; 11(12):1240. https://doi.org/10.3390/jpm11121240
Chicago/Turabian StyleLombardi, Elisabetta, Gonzalo Almanza, Kinga Kowal, Marco Valvasori, Francesco Agostini, Carla Vicinanza, Francesco Da Ros, Cristina Durante, Miriam Marangon, Mariagrazia Michieli, and et al. 2021. "miR-335-laden B Cell-Derived Extracellular Vesicles Promote SOX4-Dependent Apoptosis in Human Multiple Myeloma Cells" Journal of Personalized Medicine 11, no. 12: 1240. https://doi.org/10.3390/jpm11121240
APA StyleLombardi, E., Almanza, G., Kowal, K., Valvasori, M., Agostini, F., Vicinanza, C., Da Ros, F., Durante, C., Marangon, M., Michieli, M., Rupolo, M., Mazzucato, M., & Zanetti, M. (2021). miR-335-laden B Cell-Derived Extracellular Vesicles Promote SOX4-Dependent Apoptosis in Human Multiple Myeloma Cells. Journal of Personalized Medicine, 11(12), 1240. https://doi.org/10.3390/jpm11121240