Role of Sex on the Genetic Susceptibility to Childhood Asthma in Latinos and African Americans
Abstract
:1. Introduction
2. Results
2.1. Study Populations
2.2. Significance Threshold Estimation
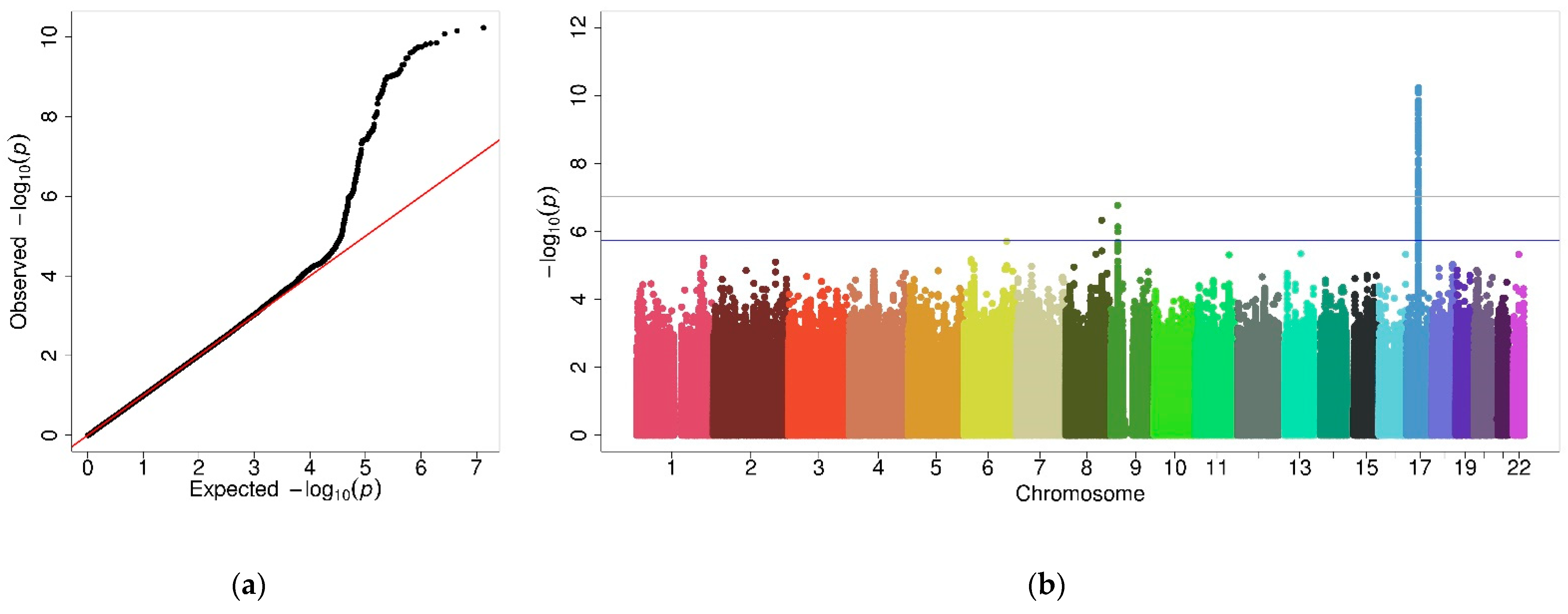
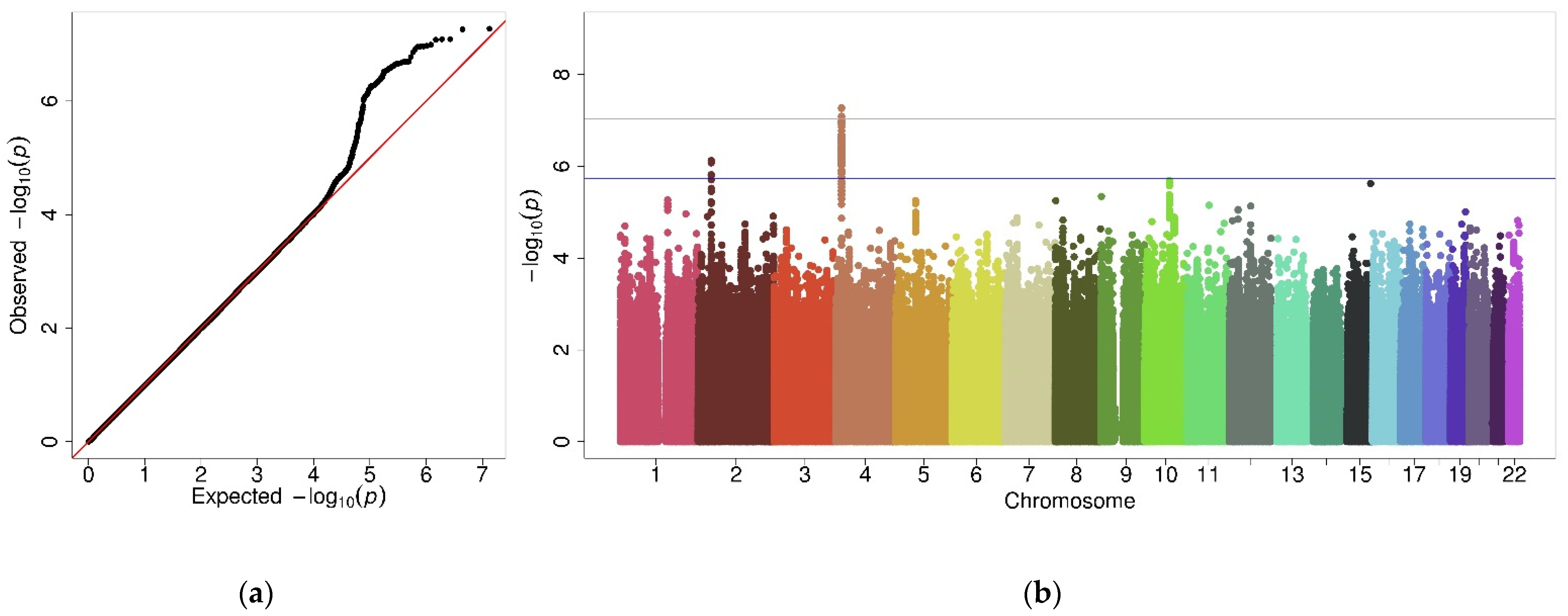
2.3. Interaction GWAS
2.4. Stratified GWAS in Females
2.5. Stratified GWAS in Males
2.6. PQTL Analyses and Enrichment Analysis
2.7. Validation of Variants Previously Reported Having a Sex-Specific Role in Asthma or an Interaction Effect
3. Discussion
4. Materials and Methods
4.1. Study Sample
4.2. Genotyping, Imputation of Genetic Variants and Ancestry Assessment
4.3. Significance Threshold Estimation
4.4. Statistical Analysis
4.5. In Silico Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, V.M. Why are sex and gender important to basic physiology and translational and individualized medicine? Am. J. Physiol. Heart Circ. Physiol. 2014, 306. [Google Scholar] [CrossRef] [Green Version]
- Mersha, T.B.; Martin, L.J.; Biagini Myers, J.M.; Kovacic, M.B.; He, H.; Lindsey, M.; Sivaprasad, U.; Chen, W.; Khurana Hershey, G.K. Genomic architecture of asthma differs by sex. Genomics 2015, 106, 15–22. [Google Scholar] [CrossRef]
- Mauvais-Jarvis, F.; Bairey Merz, N.; Barnes, P.J.; Brinton, R.D.; Carrero, J.J.; DeMeo, D.L.; De Vries, G.J.; Epperson, C.N.; Govindan, R.; Klein, S.L.; et al. Sex and gender: Modifiers of health, disease, and medicine. Lancet 2020, 396, 565–582. [Google Scholar] [CrossRef]
- Weiss, L.A.; Pan, L.; Abney, M.; Ober, C. The sex-specific genetic architecture of quantitative traits in humans. Nat. Genet. 2006, 38, 218–222. [Google Scholar] [CrossRef] [PubMed]
- GINA Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. 2019. Available online: http://www.ginasthma.org (accessed on 1 March 2021).
- Toskala, E.; Kennedy, D.W. Asthma risk factors. Int. Forum Allergy Rhinol. 2015, 5, S11–S16. [Google Scholar] [CrossRef]
- Flores, C.; Ma, S.-F.; Pino-Yanes, M.; Wade, M.S.; Pérez-Méndez, L.; Kittles, R.A.; Wang, D.; Papaiahgari, S.; Ford, J.G.; Kumar, R.; et al. African ancestry is associated with asthma risk in African Americans. PLoS ONE 2012, 7, e26807. [Google Scholar] [CrossRef] [Green Version]
- Pino-Yanes, M.; Thakur, N.; Gignoux, C.R.; Galanter, J.M.; Roth, L.A.; Eng, C.; Nishimura, K.K.; Oh, S.S.; Vora, H.; Huntsman, S.; et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J. Allergy Clin. Immunol. 2015, 135, 228–235. [Google Scholar] [CrossRef] [Green Version]
- Vergara, C.; Murray, T.; Rafaels, N.; Lewis, R.; Campbell, M.; Foster, C.; Gao, L.; Faruque, M.; Oliveira, R.R.; Carvalho, E.; et al. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet. Epidemiol. 2013, 37, 393–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergara, C.; Caraballo, L.; Mercado, D.; Jimenez, S.; Rojas, W.; Rafaels, N.; Hand, T.; Campbell, M.; Tsai, Y.J.; Gao, L.; et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Hum. Genet. 2009, 125, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Van Bever, H.P. Determinants in early life for asthma development. Allergy Asthma Clin. Immunol. 2009, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Skobeloff, E.M.; Spivey, W.H.; Clair, S.S.S.; Schoffstall, J.M. The Influence of Age and Sex on Asthma Admissions. JAMA J. Am. Med. Assoc. 1992, 268, 3437–3440. [Google Scholar] [CrossRef]
- Almqvist, C.; Worm, M.; Leynaert, B. Impact of gender on asthma in childhood and adolescence: A GA2LEN review. Allergy 2007, 63, 47–57. [Google Scholar] [CrossRef] [PubMed]
- National Health Interview Survey (NHIS) Data Lifetime Asthma Prevalence Percents by Age, United States: National Health Interview Survey. Available online: https://www.cdc.gov/asthma/nhis/2019/table2-1.htm (accessed on 1 March 2021).
- Shah, R.; Newcomb, D.C. Sex Bias in Asthma Prevalence and Pathogenesis. Front. Immunol. 2018, 9, 2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.-Y.; Forno, E.; Celedón, J.C. Sex Steroid Hormones and Asthma in a Nationwide Study of U.S. Adults. Am. J. Respir. Crit. Care Med. 2020, 201. [Google Scholar] [CrossRef]
- Patel, R.; Solatikia, F.; Zhang, H.; Wolde, A.; Kadalayil, L.; Karmaus, W.; Ewart, S.; Arathimos, R.; Relton, C.; Ring, S.; et al. Sex-specific associations of asthma acquisition with changes in DNA methylation during adolescence. Clin. Exp. Allergy 2020, 11, cea.13776. [Google Scholar] [CrossRef]
- Gautam, Y.; Afanador, Y.; Abebe, T.; López, J.E.; Mersha, T.B. Genome-wide analysis revealed sex-specific gene expression in asthmatics. Hum. Mol. Genet. 2019, 28, 2600–2614. [Google Scholar] [CrossRef]
- Loisel, D.A.; Tan, Z.; Tisler, C.J.; Evans, M.D.; Gangnon, R.E.; Jackson, D.J.; Gern, J.E.; Lemanske, R.F.; Ober, C. IFNG genotype and sex interact to influence the risk of childhood asthma. J. Allergy Clin. Immunol. 2011, 128, 524–531. [Google Scholar] [CrossRef] [Green Version]
- Myers, R.A.; Scott, N.M.; Gauderman, W.J.; Qiu, W.; Mathias, R.A.; Romieu, I.; Levin, A.M.; Pino-Yanes, M.; Graves, P.E.; Villarreal, A.B.; et al. Genome-wide interaction studies reveal sex-specific asthma risk alleles. Hum. Mol. Genet. 2014, 23, 5251–5259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James Gauderman, W.; Kim, A.; Conti, D.V.; Morrison, J.; Thomas, D.C.; Vora, H.; Lewinger, J.P. A Unified Model for the Analysis of Gene-Environment Interaction. Am. J. Epidemiol. 2019, 188, 760–767. [Google Scholar] [CrossRef]
- Nishimura, K.K.; Galanter, J.M.; Roth, L.A.; Oh, S.S.; Thakur, N.; Nguyen, E.A.; Thyne, S.; Farber, H.J.; Serebrisky, D.; Kumar, R.; et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am. J. Respir. Crit. Care Med. 2013, 188, 309–318. [Google Scholar] [CrossRef]
- Fadista, J.; Manning, A.K.; Florez, J.C.; Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 2016, 24, 1202–1205. [Google Scholar] [CrossRef] [Green Version]
- Kanai, M.; Tanaka, T.; Okada, Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J. Hum. Genet. 2016, 61, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Luis, E.; Espuela-Ortiz, A.; Lorenzo-Diaz, F.; Keys, K.L.; Mak, A.C.Y.; Eng, C.; Huntsman, S.; Villar, J.; Rodriguez-Santana, J.R.; Burchard, E.G.; et al. Genome-wide association study reveals a novel locus for asthma with severe exacerbations in diverse populations. Pediatr. Allergy Immunol. 2021, 32, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Soranzo, N.; Rivadeneira, F.; Chinappen-Horsley, U.; Malkina, I.; Richards, J.B.; Hammond, N.; Stolk, L.; Nica, A.; Inouye, M.; Hofman, A.; et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009, 5, e1000445. [Google Scholar] [CrossRef]
- Sovio, U.; Bennett, A.J.; Millwood, L.Y.; Molitor, J.; O’Reilly, P.F.; Timpson, N.J.; Kaakinen, M.; Laitinen, J.; Haukka, J.; Pillas, D.; et al. Genetic determinants of height growth assessed longitudinally from infancy to adulthood in the northern finland birth cohort 1966. PLoS Genet. 2009, 5, 1000409. [Google Scholar] [CrossRef]
- Carty, C.L.; Johnson, N.A.; Hutter, C.M.; Reiner, A.P.; Peters, U.; Tang, H.; Kooperberg, C. Genome-wide association study of body height in African Americans: The Women’s Health Initiative SNP Health Association Resource (SHARe). Hum. Mol. Genet. 2012, 21, 711–720. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, M.; Yaghootkar, H.; Mook-Kanamori, D.O.; Sovio, U.; Taal, H.R.; Hennig, B.J.; Bradfield, J.P.; St Pourcain, B.; Evans, D.M.; Charoen, P.; et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013, 45, 76–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, K.; Ogasawara, M. The Role of Histamine in the Pathophysiology of Asthma and the Clinical Efficacy of Antihistamines in Asthma Therapy. Int. J. Mol. Sci. 2019, 20, 1733. [Google Scholar] [CrossRef] [Green Version]
- Billcliff, P.G.; Lowe, M. Inositol lipid phosphatases in membrane trafficking and human disease. Biochem. J. 2014, 461, 159–175. [Google Scholar] [CrossRef]
- Matthews, S.P.; McMillan, S.J.; Colbert, J.D.; Lawrence, R.A.; Watts, C. Cystatin F Ensures Eosinophil Survival by Regulating Granule Biogenesis. Immunity 2016, 44, 795. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, M.F.; Kabesch, M.; Liang, L.; Dixon, A.L.; Strachan, D.; Heath, S.; Depner, M.; Von Berg, A.; Bufe, A.; Rietschel, E.; et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007, 448, 470–473. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Pino-Yanes, M.; Flores, C. Genomic predictors of asthma phenotypes and treatment response. Front. Pediatr. 2019, 7, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Stein, M.M.; Thompson, E.E.; Schoettler, N.; Helling, B.A.; Magnaye, K.M.; Stanhope, C.; Igartua, C.; Morin, A.; Washington, C.; Nicolae, D.; et al. A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle. J. Allergy Clin. Immunol. 2018, 142, 749–764.e3. [Google Scholar] [CrossRef] [Green Version]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, T.R.; Shihab, H.A.; Hemani, G.; Min, J.L.; Woodward, G.; Lyttleton, O.; Zheng, J.; Duggirala, A.; McArdle, W.L.; Ho, K.; et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016, 17, 61. [Google Scholar] [CrossRef] [Green Version]
- Ober, C.; McKennan, C.G.; Magnaye, K.M.; Altman, M.C.; Washington, C.; Stanhope, C.; Naughton, K.A.; Rosasco, M.G.; Bacharier, L.B.; Billheimer, D.; et al. Expression quantitative trait locus fine mapping of the 17q12–21 asthma locus in African American children: A genetic association and gene expression study. Lancet Respir. Med. 2020, 8, 482–492. [Google Scholar] [CrossRef]
- Byun, J.; Schwartz, A.G.; Lusk, C.; Wenzlaff, A.S.; De Andrade, M.; Mandal, D.; Gaba, C.; Yang, P.; You, M.; Kupert, E.Y.; et al. Genome-wide association study of familial lung cancer. Carcinogenesis 2018, 39, 1135–1140. [Google Scholar] [CrossRef]
- Paulissen, G.; Rocks, N.; Gueders, M.M.; Crahay, C.; Quesada-Calvo, F.; Bekaert, S.; Hacha, J.; El Hour, M.; Foidart, J.-M.; Noel, A.; et al. Role of ADAM and ADAMTS metalloproteinases in airway diseases. Respir. Res. 2009, 10, 127. [Google Scholar] [CrossRef] [Green Version]
- Hunninghake, G.M.; Soto-Quirós, M.E.; Avila, L.; Kim, H.P.; Lasky-Su, J.; Rafaels, N.; Ruczinski, I.; Beaty, T.H.; Mathias, R.A.; Barnes, K.C.; et al. TSLP Polymorphisms are Associated with Asthma in a Sex-Specific Fashion. Allergy 2010, 65, 1566–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, J.C.; Thompson, C.B. The central effectors of cell death in the immune system. Annu. Rev. Immunol 1999, 17, 781–828. [Google Scholar] [CrossRef] [PubMed]
- Yung, J.A.; Fuseini, H.; Newcomb, D.C. Hormones, sex, and asthma. Ann. Allergy, Asthma Immunol. 2018, 120, 488–494. [Google Scholar] [CrossRef] [Green Version]
- Skol, A.D.; Scott, L.J.; Abecasis, G.R.; Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006, 38, 209–213. [Google Scholar] [CrossRef]
- Dimas, A.S.; Nica, A.C.; Montgomery, S.B.; Stranger, B.E.; Raj, T.; Buil, A.; Giger, T.; Lappalainen, T.; Gutierrez-Arcelus, M.; Consortium, M.; et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012, 22, 2368. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- Altshuler, D.L.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Collins, F.S.; De La Vega, F.M.; Donnelly, P.; Egholm, M.; et al. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [CrossRef] [Green Version]
- Delaneau, O.; Coulonges, C.; Zagury, J.-F. Shape-IT: New rapid and accurate algorithm for haplotype inference. BMC Bioinform. 2008, 9, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Pino-Yanes, M.; Gignoux, C.R.; Galanter, J.M.; Levin, A.M.; Campbell, C.D.; Eng, C.; Huntsman, S.; Nishimura, K.K.; Gourraud, P.A.; Mohajeri, K.; et al. Genome-wide association study and admixture mapping reveal new loci associated with total IgE levels in Latinos. J. Allergy Clin. Immunol. 2015, 135, 1502–1510. [Google Scholar] [CrossRef] [Green Version]
- Duggal, P.; Gillanders, E.M.; Holmes, T.N.; Bailey-Wilson, J.E. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genom. 2008, 9, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011, 88, 586–598. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Huang, D.; Wang, J.; Zhao, K.; Zhou, Y.; Guo, Z.; Zhai, S.; Xu, H.; Cui, H.; Yao, H.; et al. QTLbase: An integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020, 48, D983–D991. [Google Scholar] [CrossRef] [Green Version]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Kamat, M.A.; Blackshaw, J.A.; Young, R.; Surendran, P.; Burgess, S.; Danesh, J.; Butterworth, A.S.; Staley, J.R. PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019, 35, 4851–4853. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| GALA II (n = 4291) | SAGE (n = 1730) | |||||
|---|---|---|---|---|---|---|
| Cases (n = 2263) | Controls (n = 2028) | p-Value | Cases (n = 1108) | Controls (n = 622) | p-Value | |
| Characteristic | Overall (n = 6021) | |||||
| Sex (% female) | 1031 (45.6) | 1137 (56.1) | 7.8 × 10−12 | 549 (49.5) | 358 (57.6) | 1.6 × 10−3 |
| Age (years) | 12.7 ± 3.3 | 13.9 ± 3.6 | 7.0 × 10−28 | 14.0 ± 3.6 | 15.8 ± 3.8 | 2.5 × 10−20 |
| Height (cm) a | 151 ± 14.0 | 155.5 ± 13.1 | 3.4 × 10−18 | 157.9 ± 14.6 | 162 ± 13.9 | 1.2 × 10−6 |
| African ancestry (%) | 16.2 ± 13.1 | 14.1 ± 12.0 | 4.4 × 10−8 | 78.9 ± 11.4 | 78.1 ± 11.4 | 0.06 |
| European ancestry (%) | 54.6 ± 18.8 | 52.0 ± 20.9 | 1.4 × 10−3 | 21.1 ± 11.4 | 21.9 ± 11.4 | 0.06 |
| Native American ancestry (%) | 29.2 ± 25.0 | 33.9 ± 27.8 | 4.8 × 10−6 | NA b | NA b | NA b |
| Characteristic | Female (n = 3075) | |||||
| Age (years) | 13.3 ± 3.6 | 14.1 ± 3.6 | 1.2 × 10−8 | 14.4 ± 3.8 | 16.0 ± 3.8 | 8.4 × 10−10 |
| Height (cm) a | 150.5 ± 11.9 | 153.2 ± 10.3 | 7.9 × 10−5 | 156.9 ± 11.1 | 159.7 ± 10.8 | 4.2 × 10−4 |
| African ancestry (%) | 16.5 ± 13.1 | 14.0 ± 12.1 | 5.9 × 10−7 | 78.5 ± 12.4 | 78.3 ± 11.0 | 0.08 |
| European ancestry (%) | 54.9 ± 18.7 | 51.4 ± 21.1 | 9.9 × 10−4 | 21.5 ± 12.4 | 21.7 ± 11.0 | 0.08 |
| Native American ancestry (%) | 28.6 ± 25.0 | 35.0 ± 28.0 | 4.9 × 10−6 | NA b | NA b | NA b |
| Characteristic | Male (n = 2946) | |||||
| Age (years) | 12.3 ± 3.1 | 13.6 ± 3.6 | 4.1 × 10−18 | 13.5 ± 3.4 | 15.4 ± 3.9 | 1.5 × 10−10 |
| Height (cm) a | 151.3 ± 15.6 | 158.5 ± 15.4 | 6.8 × 10−17 | 158.9 ± 17.4 | 165 ± 16.6 | 1.1 × 10−4 |
| African ancestry (%) | 16.0 ± 13.0 | 14.4 ± 12.0 | 8.6 × 10−3 | 79.2 ± 10.3 | 77.8 ± 12.0 | 0.33 |
| European ancestry (%) | 54.3 ± 18.8 | 52.7 ± 20.7 | 0.27 | 20.8 ± 10.3 | 22.2 ± 12.0 | 0.33 |
| Native American ancestry (%) | 29.7 ± 24.9 | 32.9 ± 27.4 | 0.06 | NA b | NA b | NA b |
| rsID | Chr:pos a | A1/A2 | Closest Gene | OR (95% CI) Interaction | p-Value Interaction | OR (95% CI) Female | p-Value Female | OR (95% CI) Male | p-Value Male |
|---|---|---|---|---|---|---|---|---|---|
| rs146406602 | 2:139218399 | CA7/C b | AC114763.1 | 0.56 (0.45–0.70) | 1.76 × 10−7 | 0.76 (0.63–0.92) | 6.09 × 10−4 | 1.39 (1.19–1.64) | 6.42 × 10−5 |
| rs73021932 | 6:158513756 | T/C | SYNJ2 | 0.55 (0.29–1.08) | 1.72 × 10−6 | 0.70 (0.51–0.98) | 1.48 × 10−5 | 1.26 (0.91–1.73) | 2.48 × 10−2 |
| rs12539647 | 7:46170582 | C/T | AC023669.2 | 0.70 (0.60–0.81) | 1.53 × 10−6 | 0.82 (0.74–0.91) | 3.10 × 10−4 | 1.18 (1.05–1.32) | 6.01 × 10−3 |
| rs6036804 | 20:24433071 | C/A | GAPDHP53 | 0.63 (0.39–1.01) | 8.49 × 10−7 | 0.86 (0.77–0.95) | 5.72 × 10−3 | 1.36 (0.90–2.04) | 2.43 × 10−5 |
| Analysis | rsID | Chr:pos a | A1/A2 | Closest Gene | OR (95% CI) Female | p-Value Female | OR (95% CI) Male | p-Value Male | OR (95% CI) Interaction | p-Value Interaction |
|---|---|---|---|---|---|---|---|---|---|---|
| Female | rs72683967 | 8:117337397 | G/A | LINC00536 | 0.66 (0.49–0.89) | 4.70 × 10−7 | 1.05 (0.89–1.23) | 6.18 × 10−1 | 0.62 (0.40–0.97) | 1.43 × 10−4 |
| rs4400476 | 9:22862535 | T/G | AL391117.1 | 0.78 (0.60–1.00) | 1.71 × 10−7 | 1.05 (0.91–1.21) | 5.54 × 10−1 | 0.70 (0.60–0.82) | 1.08 × 10−5 | |
| rs907092 | 17:37922259 | A/G | IKZF3 | 0.69 (0.54–0.89) | 5.85 × 10−11 | 0.83 (0.67–1.02) | 4.51 × 10−4 | 0.82 (0.69–0.97) | 2.72 × 10−2 | |
| Male | rs10207164 | 2:42031062 | C/T | LDHAP3 | 1.15 (1.01–1.33) | 4.89 × 10−2 | 1.45 (1.25–1.68) | 7.50 × 10−7 | 0.79 (0.64–0.96) | 1.89 × 10−2 |
| rs4593128 | 4:18221770 | G/T | LCORL | 0.92 (0.78–1.07) | 2.95 × 10−1 | 0.64 (0.54–0.75) | 5.31 × 10−8 | 1.36 (1.09–1.70) | 7.38 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espuela-Ortiz, A.; Herrera-Luis, E.; Lorenzo-Díaz, F.; Hu, D.; Eng, C.; Villar, J.; Rodriguez-Santana, J.R.; Burchard, E.G.; Pino-Yanes, M. Role of Sex on the Genetic Susceptibility to Childhood Asthma in Latinos and African Americans. J. Pers. Med. 2021, 11, 1140. https://doi.org/10.3390/jpm11111140
Espuela-Ortiz A, Herrera-Luis E, Lorenzo-Díaz F, Hu D, Eng C, Villar J, Rodriguez-Santana JR, Burchard EG, Pino-Yanes M. Role of Sex on the Genetic Susceptibility to Childhood Asthma in Latinos and African Americans. Journal of Personalized Medicine. 2021; 11(11):1140. https://doi.org/10.3390/jpm11111140
Chicago/Turabian StyleEspuela-Ortiz, Antonio, Esther Herrera-Luis, Fabián Lorenzo-Díaz, Donglei Hu, Celeste Eng, Jesús Villar, Jose R. Rodriguez-Santana, Esteban G. Burchard, and María Pino-Yanes. 2021. "Role of Sex on the Genetic Susceptibility to Childhood Asthma in Latinos and African Americans" Journal of Personalized Medicine 11, no. 11: 1140. https://doi.org/10.3390/jpm11111140
APA StyleEspuela-Ortiz, A., Herrera-Luis, E., Lorenzo-Díaz, F., Hu, D., Eng, C., Villar, J., Rodriguez-Santana, J. R., Burchard, E. G., & Pino-Yanes, M. (2021). Role of Sex on the Genetic Susceptibility to Childhood Asthma in Latinos and African Americans. Journal of Personalized Medicine, 11(11), 1140. https://doi.org/10.3390/jpm11111140








