Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans
Abstract
:1. Introduction
2. Material and Methods
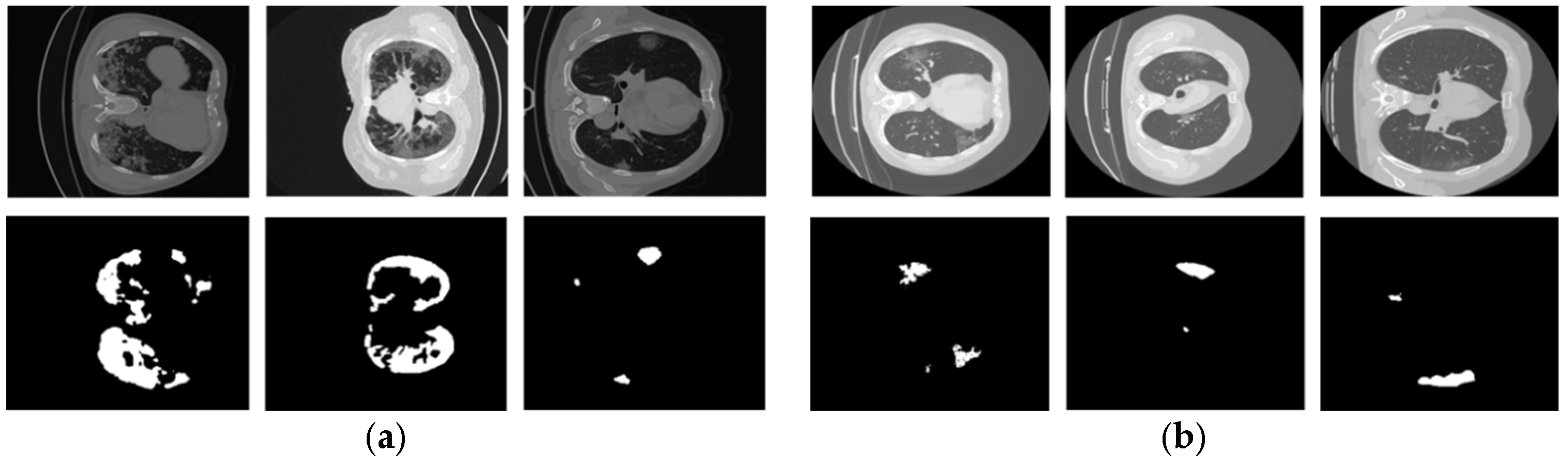
2.1. Datasets
2.2. Method
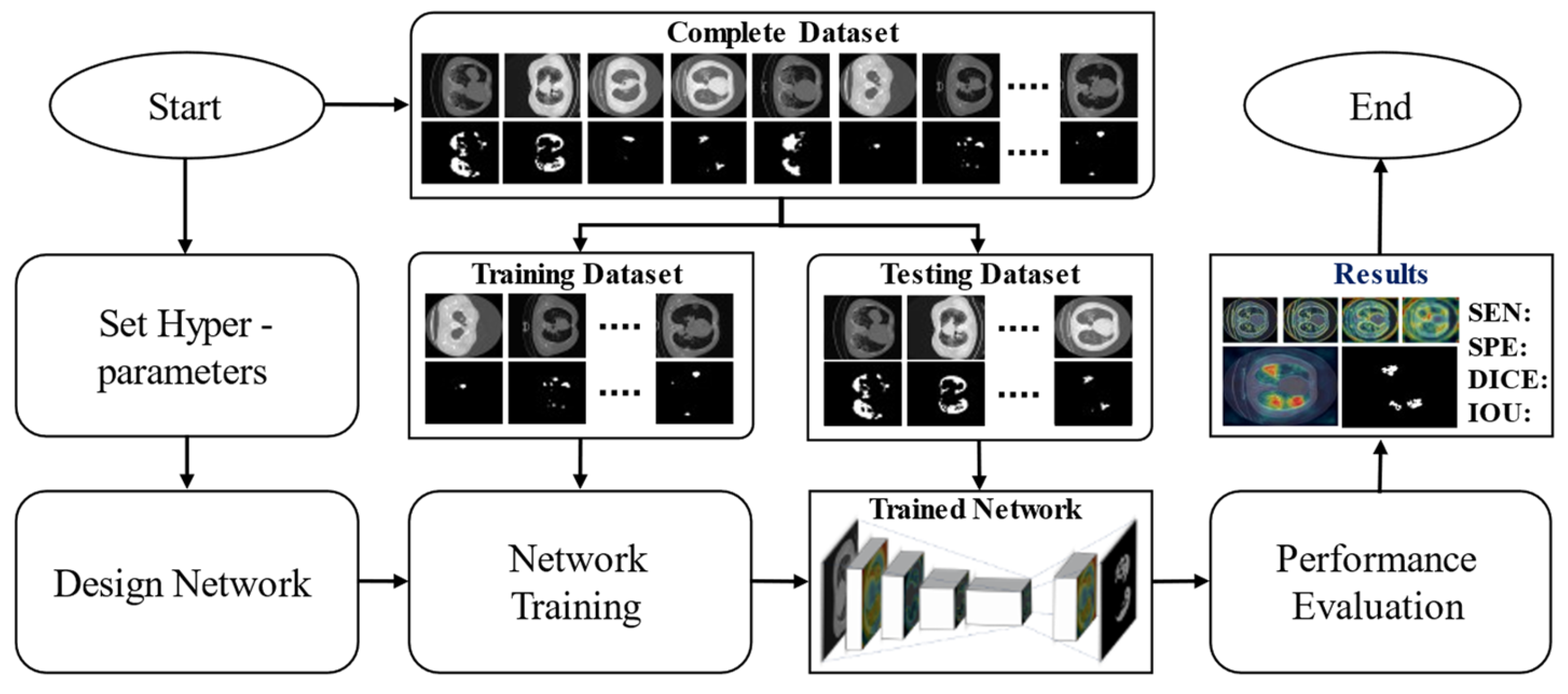
2.2.1. Overview of the Proposed Method
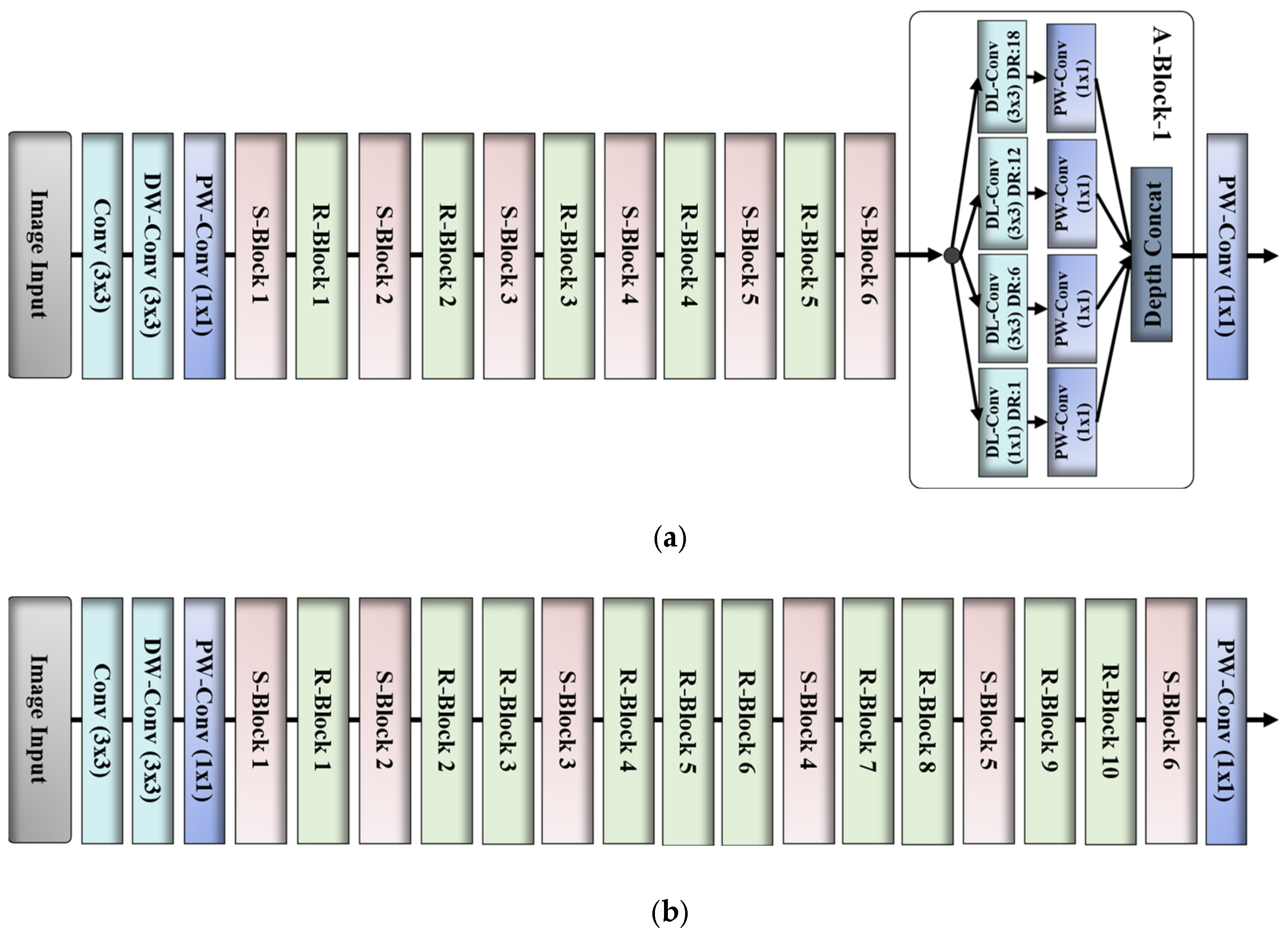
2.2.2. Network Design
2.2.3. Loss Function and Network Training
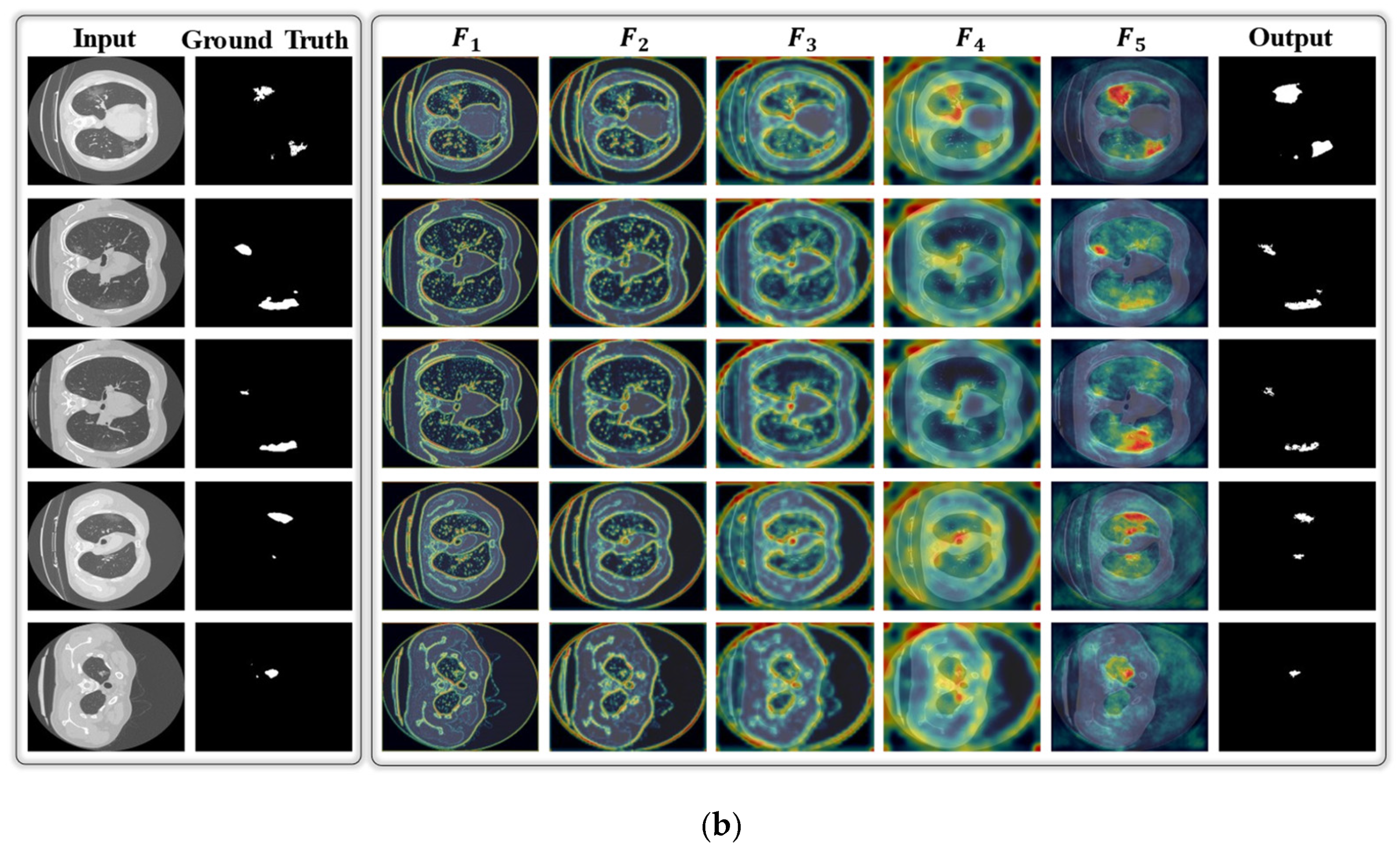
3. Results
3.1. Experimental Setup
| Algorithm 1: Training procedure of the proposed DAL-Net | |
| Input:: a total of training data samples, : input image, : corresponding ground-truth mask Output: Learned parameters, Parameters: Learnable parameters, ; initial learning rate, ; maximum epoch, ; mini-batch size, ; | |
| 1 | Initialize parameters (Pre-trained weights of MobileNetV2 model that was trained on large ImageNet dataset) |
| 2 | //Continue the training procedure |
| 3 | fordo //Loop for number of epochs |
| 4 | Randomly divide the whole dataset into mini-batches of size : |
| 5 | fordo //Loop for number of iterations |
| 6 | obtain:// presents our model |
| 7 | update://loss function Equation (5) |
| 8 | end |
| 9 | end |
| 10 | //Training stop and finally we obtain learned weights, |
3.2. Results
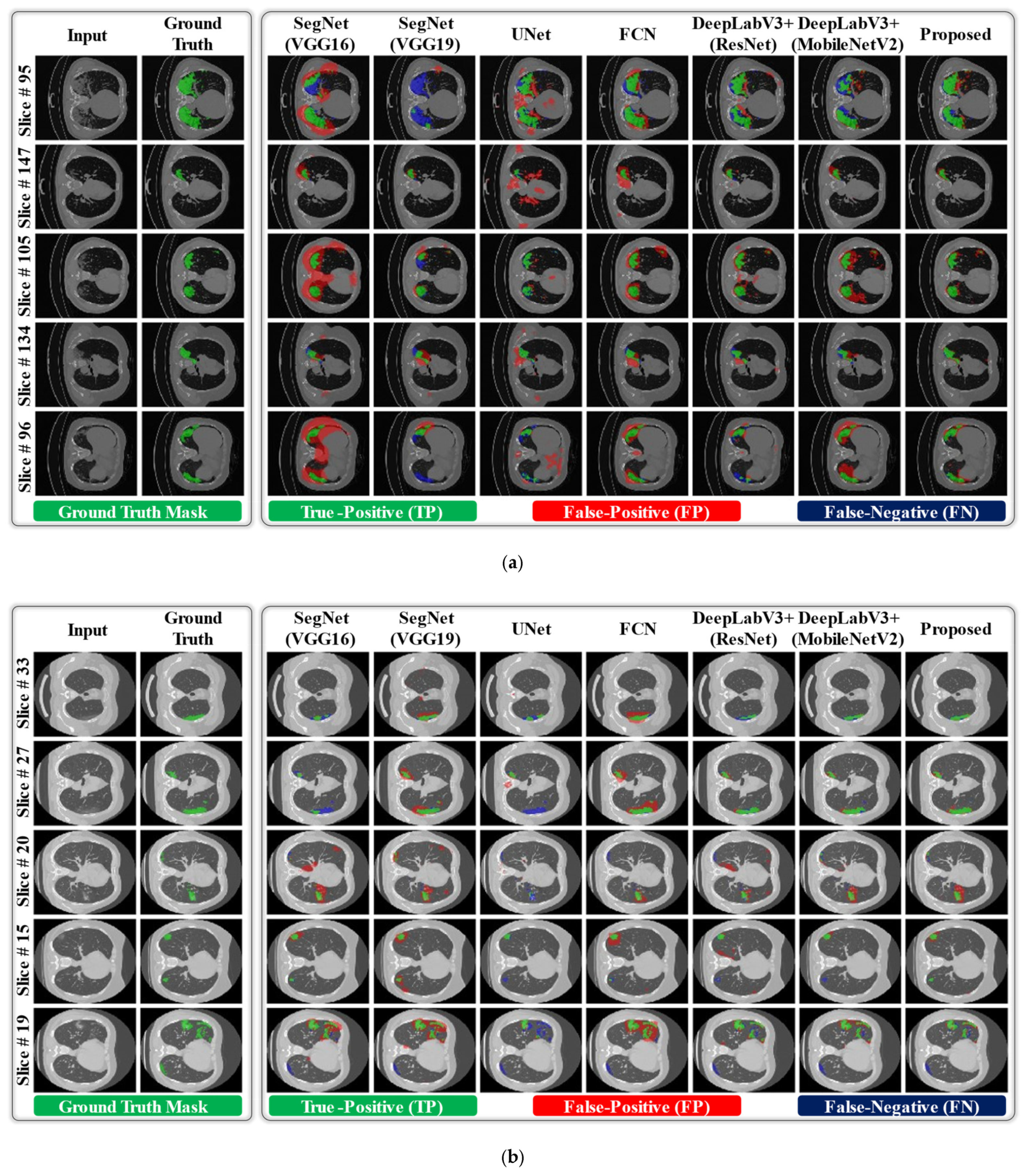
3.3. Comparisons with the State-of-the-Art Methods
4. Discussion
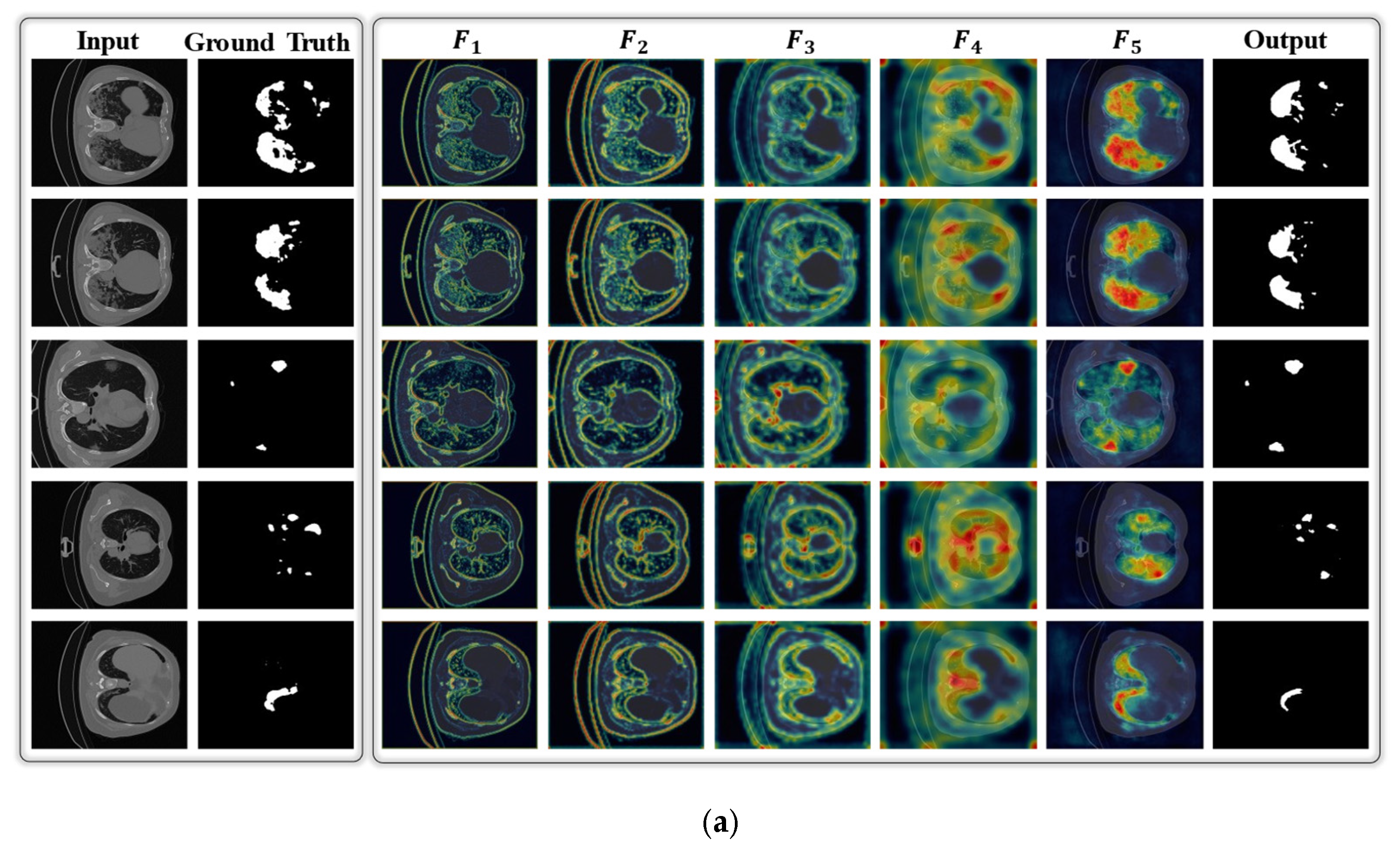
4.1. Principal Findings
4.2. Limitations and Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization, WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 29 March 2021).
- Kim, J.H.; Marks, F.; Clemens, J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021, 27, 205–211. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Ng, M.-Y.; Lee, E.Y.P.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.M.; Lo, C.S.-Y.; Leung, B.; Khong, P.-L.; et al. Imaging profile of the COVID19 infection: Radiologic findings and literature review. Radiol. Cardiothorac. Imaging 2020, 2, 200034. [Google Scholar]
- Kim, K.M.; Heo, T.-Y.; Kim, A.; Kim, J.; Han, K.J.; Yun, J.; Min, J.K. Development of a fundus image-based deep learning diagnostic tool for various retinal diseases. J. Pers. Med. 2021, 11, 321. [Google Scholar] [CrossRef] [PubMed]
- de Jong, D.J.; Veldhuis, W.B.; Wessels, F.J.; de Vos, B.; Moeskops, P.; Kok, M. Towards personalised contrast injection: Artificial-intelligence-derived body composition and liver enhancement in computed tomography. J. Pers. Med. 2021, 11, 159. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Owais, M.; Yoon, H.S.; Mahmood, T.; Haider, A.; Sultan, H.; Park, K.R. Light-weighted ensemble network with multilevel activation visualization for robust diagnosis of COVID19 pneumonia from large-scale chest radiographic database. Appl. Soft Comput. 2021, 108, 107490. [Google Scholar] [CrossRef]
- Lee, K.-S.; Kim, J.Y.; Jeon, E.-T.; Choi, W.S.; Kim, N.H.; Lee, K.Y. Evaluation of scalability and degree of fine-tuning of deep convolutional neural networks for COVID-19 screening on chest X-ray images using explainable deep-learning algorithm. J. Pers. Med. 2020, 10, 213. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, H.; Loew, M.H.; Ko, H. COVID-19 CT image synthesis with a conditional generative adversarial network. IEEE J. Biomed. Health Inform. 2021, 25, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhong, Y.; Deng, Y.; Tang, X.; Li, X. CoSinGAN: Learning COVID-19 infection segmentation from a single radiological image. Diagnostics 2020, 10, 901. [Google Scholar] [CrossRef]
- Fan, D.P.; Zhou, T.; Ji, G.P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-net: Automatic COVID-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Y.; An, X.; Ge, C.; Yu, Z.; Chen, J.; Zhu, Q.; Dong, G.; He, J.; He, Z.; et al. Towards data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation. Med. Phys. 2021, 48, 1197–1210. [Google Scholar] [CrossRef]
- Oulefki, A.; Agaian, S.; Trongtirakul, T.; Laouar, A.K. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit. 2021, 114, 107747. [Google Scholar] [CrossRef]
- El-bana, S.; Al-Kabbany, A.; Sharkas, M. A multi-task pipeline with specialized streams for classification and segmentation of infection manifestations in COVID-19 scans. PeerJ Comput. Sci. 2020, 6, e303. [Google Scholar] [CrossRef]
- Zheng, B.; Liu, Y.; Zhu, Y.; Yu, F.; Jiang, T.; Yang, D.; Xu, T. MSD-Net: Multi-scale discriminative network for COVID-19 lung infection segmentation on CT. IEEE Access 2020, 8, 185786–185795. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Chang, V.; Hawash, H.; Chakrabortty, R.K.; Ryan, M. FSS-2019-nCov: A deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection. Knowl.-Based Syst. 2021, 212, 106647. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, D.; Venkatesan, A.; Mahesh, V.G.; Raj, A.N.J. An integrated feature frame work for automated segmentation of COVID-19 infection from lung CT images. Int. J. Imaging Syst. Technol. 2021, 31, 28–46. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Canu, S.; Ruan, S. Automatic COVID-19 CT segmentation using U-Net integrated spatial and channel attention mechanism. Int. J. Imaging Syst. Technol. 2021, 31, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Jun, M.; Cheng, G.; Yixin, W.; Xingle, A.; Jiantao, G.; Ziqi, Y.; Minqing, Z.; Xin, L.; Xueyuan, D.; Shucheng, C.; et al. COVID-19 CT lung and infection segmentation dataset (Version 1.0) [Data set]. Zenodo 2020. [Google Scholar] [CrossRef]
- Morozov, S.P.; Andreychenko, A.E.; Pavlov, N.A.; Vladzymyrskyy, A.V.; Ledikhova, N.V.; Gombolevskiy, V.A.; Blokhin, I.A.; Gelezhe, P.B.; Gonchar, A.V.; Chernina, V.Y. Mosmeddata: Chest ct scans with COVID-19 related findings dataset. arXiv 2020, arXiv:2005.06465. [Google Scholar]
- Dongguk Light-Weighted Segmentation Model for Effective Diagnosis of COVID-19 Infection. Available online: http://dm.dgu.edu/link.html (accessed on 1 March 2021).
- Heaton, J. Artificial Intelligence for Humans; Deep learning and neural networks; Heaton Research Inc.: St. Louis, MO, USA, 2015; Volume 3. [Google Scholar]
- Majid, M.; Owais, M.; Anwar, S.M. Visual saliency based redundancy allocation in HEVC compatible multiple description video coding. Multimed. Tools App. 2018, 77, 20955–20977. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18-23 June 2018; pp. 4510–4520. [Google Scholar]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the IEEE Symposium on Computational Intelligence and Bioinformatics and Computational Biology, Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Ni, J.; Wu, J.; Tong, J.; Chen, Z.; Zhao, J. GC-Net: Global context network for medical image segmentation. Comput. Meth. Programs Biomed. 2020, 190, 105121. [Google Scholar] [CrossRef]
- Li, X.; Yu, L.; Chen, H.; Fu, C.W.; Xing, L.; Heng, P.A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Oda, H.; Zhou, X.; Shimizu, N.; Yang, Y.; Hayashi, Y.; Oda, M.; Fujiwara, M.; Misawa, K.; Mori, K. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput. Med. Imaging Graph. 2018, 66, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Li, X.-L. Preconditioned stochastic gradient descent. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1454–1466. [Google Scholar] [CrossRef] [Green Version]
- Prabowo, D.A.; Herwanto, G.B. Duplicate question detection in question answer website using convolutional neural network. In Proceedings of the International Conference on Science and Technology-Computer, Yogyakarta, Indonesia, 30–31 July 2019; pp. 1–6. [Google Scholar]
- Kandel, I.; Castelli, M. The effect of batch size on the generalizability of the convolutional neural networks on a histopathology dataset. ICT Express 2020, 6, 312–315. [Google Scholar] [CrossRef]
- Mahmood, T.; Owais, M.; Noh, K.J.; Yoon, H.S.; Koo, J.H.; Haider, A.; Sultan, H.; Park, K.R. Accurate Segmentation of Nuclear Regions with Multi-Organ Histopathology Images Using Artificial Intelligence for Cancer Diagnosis in Personalized Medicine. J. Pers. Med. 2021, 11, 515. [Google Scholar] [CrossRef]
- Suh, Y.J.; Jung, J.; Cho, B.J. Automated breast cancer detection in digital mammograms of various densities via deep learning. J. Pers. Med. 2020, 10, 211. [Google Scholar] [CrossRef]
- Qiu, Y.; Liu, Y.; Li, S.; Xu, J. Miniseg: An extremely minimum network for efficient COVID-19 segmentation. arXiv 2020, arXiv:2004.09750. [Google Scholar]
- Johnson, R.; Zhang, T. Accelerating stochastic gradient descent using predictive variance reduction. Adv. Neural. Inf. Process. Syst. 2013, 26, 315–323. [Google Scholar]
- Zhaobin, W.; Wang, E.; Zhu, Y. Image segmentation evaluation: A survey of methods. Artif. Intell. Rev. 2020, 53, 5637–5674. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Xu, Z.; Cao, Y.; Jin, C.; Shao, G.; Liu, X.; Zhou, J.; Shi, H.; Feng, J. GASNet: Weakly-supervised Framework for COVID-19 Lesion Segmentation. arXiv 2020, arXiv:2010.09456. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Yao, Q.; Xiao, L.; Liu, P.; Zhou, S.K. Label-Free Segmentation of COVID-19 Lesions in Lung CT. IEEE Trans. Med. Imaging 2021, 40, 2808–2819. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Cui, H.; Sun, C.; Meng, Z.; Wei, L.; Su, R. Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images. Expert Syst. Appl. 2021, 176, 114848. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Hope, M.D.; Raptis, C.A.; Shah, A.; Hammer, M.M.; Henry, T.S. A role for CT in COVID-19? What data really tell us so far. Lancet 2020, 395, 1189–1190. [Google Scholar] [CrossRef]
- Zhang, J.; Meng, G.; Li, W.; Shi, B.; Dong, H.; Su, Z.; Huang, Q.; Gao, P. Relationship of chest CT score with clinical characteristics of 108 patients hospitalized with COVID-19 in Wuhan, China. Resp. Res. 2020, 21, 180. [Google Scholar] [CrossRef] [PubMed]
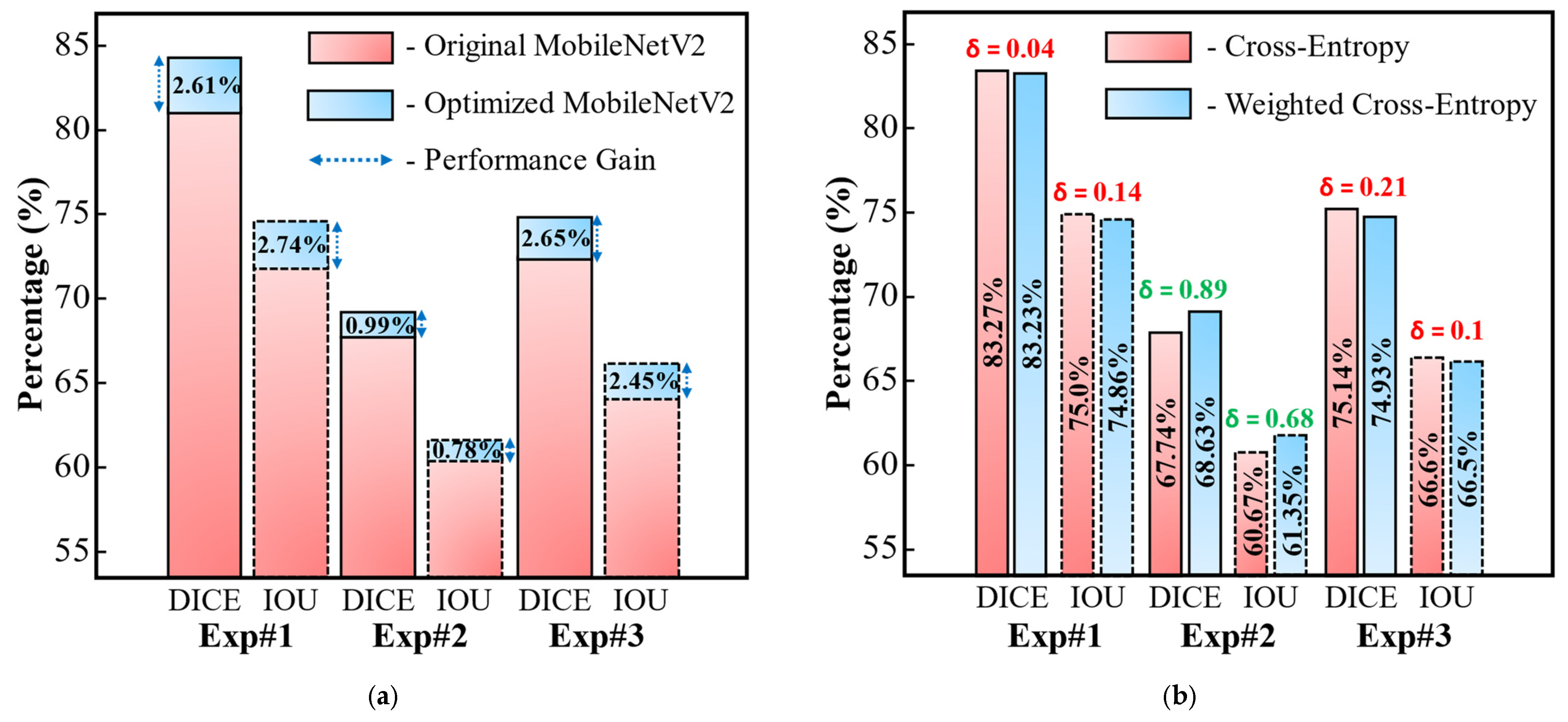

| Experiment# | #Fold | SEN | SPE | PPV | DICE | IOU | AUC | |
|---|---|---|---|---|---|---|---|---|
| Same-Dataset | Exp#1 (COVID-19-CT-Seg) | 1 | 87.75 | 99.02 | 81.22 | 86.26 | 78.21 | 98.83 |
| 2 | 88.98 | 98.67 | 70.9 | 78.13 | 69.2 | 97.87 | ||
| 3 | 93.01 | 99.06 | 74.57 | 81.93 | 73.22 | 99.06 | ||
| 4 | 96.24 | 99.61 | 74.54 | 82.41 | 73.88 | 99.66 | ||
| 5 | 89.97 | 99.54 | 82.2 | 87.42 | 79.8 | 98.79 | ||
| Avg. ± Std | 91.19 ± 3.43 | 99.18 ± 0.39 | 76.69 ± 4.84 | 83.23 ± 3.71 | 74.86 ± 4.22 | 98.84 ± 0.65 | ||
| Exp#2 (MosMed) | 1 | 87.76 | 99.2 | 59.22 | 65.05 | 58.59 | 97.55 | |
| 2 | 87.8 | 99.65 | 65.13 | 72.43 | 64.35 | 98.49 | ||
| 3 | 93.98 | 99.43 | 60.79 | 67.42 | 60.36 | 99.06 | ||
| 4 | 87.54 | 99.61 | 64.97 | 72.21 | 64.16 | 98.49 | ||
| 5 | 90.15 | 99.17 | 59.9 | 66.03 | 59.27 | 98.74 | ||
| Avg. ± Std | 89.45 ± 2.75 | 99.41 ± 0.22 | 62.00 ± 2.84 | 68.63 ± 3.47 | 61.35 ± 2.73 | 98.47 ± 0.56 | ||
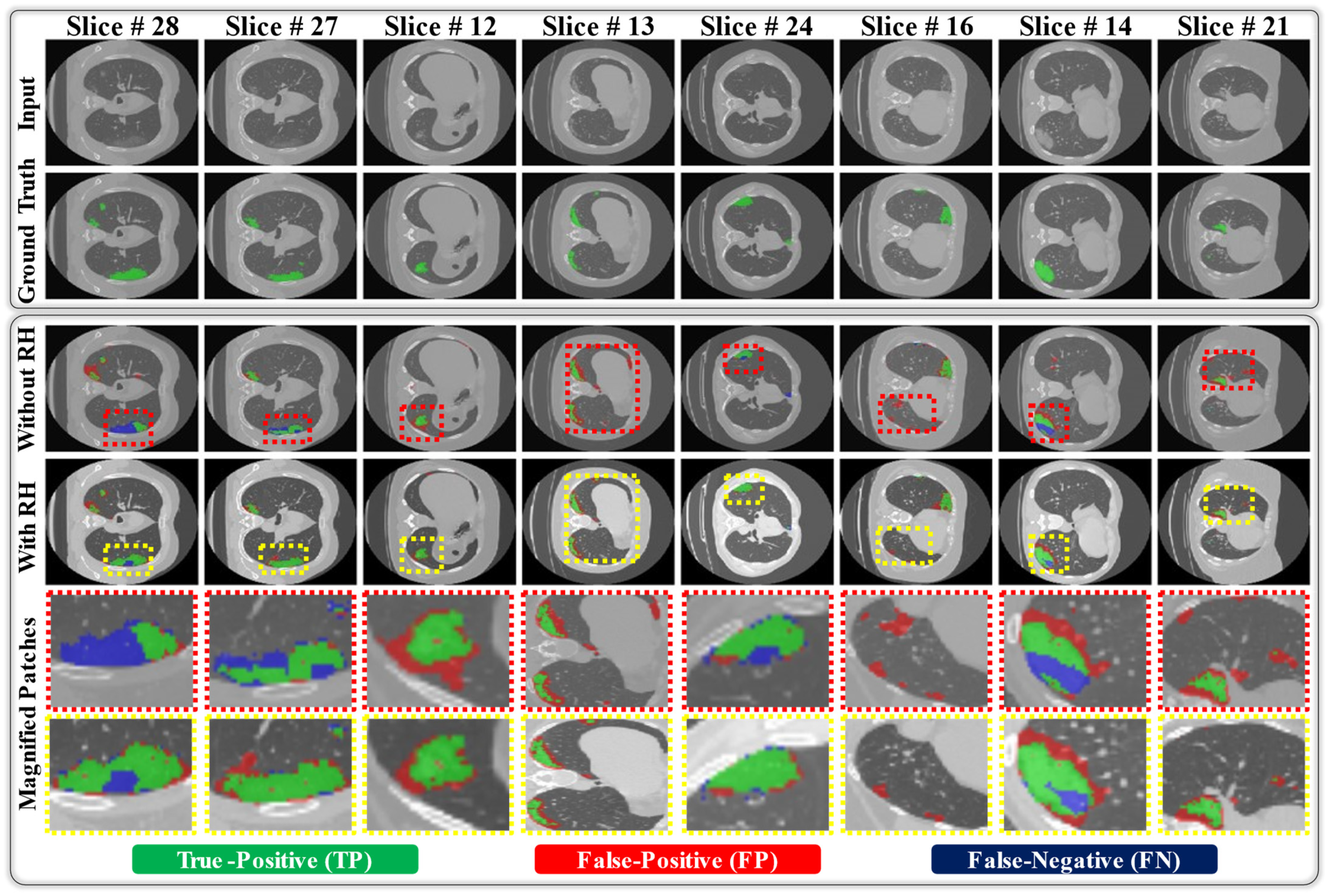
| Cross-Dataset | Exp#3 (Without RH) | 1 | 71.88 | 99.63 | 63.7 | 69.76 | 62.18 | 95.43 |
| 2 | 37.72 | 99.53 | 70.34 | 69.44 | 61.76 | 79.49 | ||
| Avg. ± Std | 54.8 ± 24.15 | 99.58 ± 0.07 | 67.02 ± 4.7 | 69.6 ± 0.23 | 61.97 ± 0.3 | 87.46 ± 11.27 | ||
| Exp#3 (With RH) | 1 | 76.42 | 99.69 | 66.01 | 72.5 | 64.41 | 96.09 | |
| 2 | 69.97 | 99.28 | 72.67 | 77.36 | 68.58 | 94.93 | ||
| Avg. ± Std | 73.2 ± 4.56 | 99.49 ± 0.29 | 69.34 ± 4.71 | 74.93 ± 3.44 | 66.5 ± 2.95 | 95.51 ± 0.82 | ||
| Experiment# | A- Block 1 | A- Block 2 | #Par. (M) | Std | Std | Std | Std | Std | Std | |
|---|---|---|---|---|---|---|---|---|---|---|
| Same-Dataset | Exp#1 (COVID-19-CT-Seg) | x | x | 5.67 | 92.493.51 | 98.191.3 | 68.95.26 | 75.975.85 | 67.335.37 | 98.620.64 |
| x | ✓ | 6.13 | 91.384.56 | 98.990.63 | 74.956.86 | 81.495.49 | 73.095.95 | 98.51.01 | ||
| ✓ | x | 6.2 | 89.622.58 | 98.60.84 | 70.335.35 | 77.255.19 | 68.65.02 | 98.110.54 | ||
| ✓ | ✓ | 6.65 | 91.193.43 | 99.180.39 | 76.694.84 | 83.233.71 | 74.864.22 | 98.840.65 | ||
| Exp#2 (MosMed) | x | x | 5.67 | 89.71.65 | 98.980.39 | 57.751.81 | 62.892.71 | 57.091.91 | 98.50.24 | |
| x | ✓ | 6.13 | 90.614.39 | 99.250.4 | 61.024.43 | 67.115.51 | 60.294.23 | 98.60.66 | ||
| ✓ | x | 6.2 | 90.144.67 | 98.840.51 | 57.322.56 | 62.123.76 | 56.582.64 | 98.120.75 | ||
| ✓ | ✓ | 6.65 | 89.452.75 | 99.410.22 | 62.002.84 | 68.633.47 | 61.352.73 | 98.470.56 | ||
| Cross-Dataset | Exp#3 (With RH) | x | x | 5.67 | 72.053.96 | 99.090.86 | 65.070.33 | 71.040.17 | 63.010.43 | 94.012.04 |
| x | ✓ | 6.13 | 79.488.3 | 98.721.44 | 64.884.00 | 71.063.95 | 63.03.64 | 94.832.23 | ||
| ✓ | x | 6.2 | 76.953.9 | 98.990.72 | 63.33.1 | 69.483.54 | 61.822.51 | 94.722.45 | ||
| ✓ | ✓ | 6.65 | 73.24.56 | 99.490.29 | 69.344.71 | 74.933.44 | 66.52.95 | 95.510.82 | ||
| Experiment# | #Fold | SEN | SPE | PPV | DICE | IOU | AUC | |
|---|---|---|---|---|---|---|---|---|
| Mixed-Datasets | Exp#1 (COVID-19-CT Seg) Exp#2 (MosMed) | 1 | 83.5 | 99.32 | 74.72 | 80.9 | 72.17 | 97.35 |
| 2 | 88.28 | 98.63 | 65.86 | 73.02 | 64.53 | 96.89 | ||
| 3 | 94.61 | 99.17 | 71.38 | 79.25 | 70.46 | 99.22 | ||
| 4 | 95.56 | 99.6 | 72.21 | 80.23 | 71.56 | 99.24 | ||
| 5 | 87.03 | 99.53 | 78.56 | 84.38 | 76.08 | 97.58 | ||
| Avg.Std | 89.85.15 | 99.250.39 | 72.554.67 | 79.564.13 | 70.964.17 | 98.061.1 | ||
| Experiment# | Models | Std | Std | Std | Std | Std | Std |
|---|---|---|---|---|---|---|---|
| Exp#1 (COVID-19-CT-Seg) | SegNet (VGG16) [44] | 93.524.49 | 97.81.63 | 66.585.96 | 73.356.78 | 64.976.11 | 98.730.69 |
| SegNet (VGG19) [44] | 89.295.56 | 98.450.95 | 69.646.52 | 76.316.18 | 67.785.97 | 98.550.54 | |
| U-Net (E.D 4) [45] | 85.797.04 | 93.766.72 | 51.0715.82 | 73.7815.27 | 62.768.38 | 96.831.25 | |
| FCN (USF 32) [46] | 91.915.55 | 97.771.25 | 65.514.74 | 72.235.27 | 63.854.63 | 98.410.9 | |
| DeepLabV3+(ResNet) [22] | 87.375.63 | 98.930.89 | 74.437.18 | 81.645.73 | 71.936.2 | 97.411.3 | |
| DeepLabV3+(MobileNetV2) [29] | 90.622.83 | 98.860.79 | 73.885.98 | 80.625.63 | 72.125.74 | 98.490.97 | |
| DAL-Net (Proposed) | 91.193.43 | 99.180.39 | 76.694.84 | 83.233.71 | 74.864.22 | 98.840.65 | |
| Exp#2 (MosMed) | SegNet (VGG16) [44] | 90.122.62 | 97.990.71 | 54.20.79 | 57.171.5 | 53.151.11 | 98.350.47 |
| SegNet (VGG19) [44] | 92.322.84 | 98.360.61 | 55.190.99 | 58.91.76 | 54.321.27 | 98.780.3 | |
| U-Net (E.D 4) [45] | 89.674.76 | 96.791.9 | 53.181.63 | 55.053.24 | 51.542.46 | 98.270.43 | |
| FCN (USF 32) [46] | 89.942.66 | 98.70.39 | 56.151.18 | 60.461.88 | 55.411.3 | 98.130.42 | |
| DeepLabV3+(ResNet) [22] | 85.633.1 | 99.450.23 | 62.432.84 | 68.983.42 | 61.622.66 | 97.70.78 | |
| DeepLabV3+(MobileNetV2) [29] | 88.282.81 | 99.370.24 | 61.242.44 | 67.643.11 | 60.572.37 | 98.150.9 | |
| DAL-Net (Proposed) | 89.452.75 | 99.410.22 | 62.002.84 | 68.633.47 | 61.352.73 | 98.470.56 | |
| Exp#3 (Cross-Dataset (with RH)) | SegNet (VGG16) [44] | 72.6513.41 | 97.283.22 | 58.533.39 | 62.814.72 | 56.474.23 | 94.430.93 |
| SegNet (VGG19) [44] | 66.8214.8 | 99.020.44 | 61.263.14 | 66.242.46 | 59.351.56 | 95.12.57 | |
| U-Net (E.D 4) [45] | 55.0721.99 | 97.882.64 | 58.923.42 | 62.223.03 | 56.222.9 | 91.961.26 | |
| FCN (USF 32) [46] | 78.158.28 | 98.731.05 | 61.821.89 | 67.862.43 | 60.481.47 | 95.012.36 | |
| DeepLabV3+(ResNet) [22] | 66.923.1 | 99.480.32 | 68.315.19 | 73.414.68 | 65.213.88 | 94.090.08 | |
| DeepLabV3+(MobileNetV2) [29] | 73.261.48 | 99.170.79 | 66.20.79 | 72.280.79 | 64.050.92 | 95.150.71 | |
| DAL-Net (Proposed) | 73.24.56 | 99.490.29 | 69.344.71 | 74.933.44 | 66.52.95 | 95.510.82 |
| Experiment# | Models | SEN | SPE | PPV | DICE | IOU | AUC |
|---|---|---|---|---|---|---|---|
| Exp#1 (COVID-19-CT-Seg) | CoSinGAN+3D U-Net [12] | – | – | – | 61.5 | – | – |
| Inf-Net [13] | 69.46 | 99.02 | – | 63.38 | 64.62 | – | |
| 3D nnU-Net [14,48] | – | – | – | 67.3 | – | – | |
| Label-Free [49] | 66.2 | – | – | 69.8 | – | – | |
| CoSinGAN+2D U-Net [12] | – | – | – | 71.3 | – | – | |
| Miniseg [41] | 85.06 | 99.05 | – | 76.27 | 84.49 | – | |
| GASNet [47] | 84.6 | 99.2 | – | 76.7 | – | – | |
| DAL-Net (Proposed) | 91.19 | 99.18 | 76.69 | 83.23 | 74.86 | 98.84 | |
| Exp#2 (MosMed) | Inf-Net [13] | 62.93 | 93.45 | – | 56.39 | 74.32 | – |
| Miniseg [41] | 79.62 | 97.71 | – | 64.84 | 78.33 | – | |
| DAL-Net (Proposed) | 89.45 | 99.41 | 62.00 | 68.63 | 61.35 | 98.47 | |
| Exp#3 (Fold 1) (Cross-Dataset) | CoSinGAN+3D U-Net [12] | – | – | – | 44.9 | – | – |
| CoSinGAN+2D U-Net [12] | – | – | – | 47.4 | – | – | |
| 3D nnU-Net [14,48] | – | – | – | 58.8 | – | – | |
| GASNet [47] | 60.4 | 99.8 | – | 58.9 | – | – | |
| AFD-DA [50] | 75.17 | 99.74 | – | 59.04 | – | – | |
| DASC-Net [50] | 72.44 | 99.78 | – | 60.66 | – | – | |
| DAL-Net (Proposed) | 76.42 | 99.69 | 66.01 | 72.5 | 64.41 | 96.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owais, M.; Baek, N.R.; Park, K.R. Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans. J. Pers. Med. 2021, 11, 1008. https://doi.org/10.3390/jpm11101008
Owais M, Baek NR, Park KR. Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans. Journal of Personalized Medicine. 2021; 11(10):1008. https://doi.org/10.3390/jpm11101008
Chicago/Turabian StyleOwais, Muhammad, Na Rae Baek, and Kang Ryoung Park. 2021. "Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans" Journal of Personalized Medicine 11, no. 10: 1008. https://doi.org/10.3390/jpm11101008
APA StyleOwais, M., Baek, N. R., & Park, K. R. (2021). Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans. Journal of Personalized Medicine, 11(10), 1008. https://doi.org/10.3390/jpm11101008