Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Study Design
2.2. Measurement of Biochemical Variables
2.3. DNA Extraction, Bisulfite Reaction, and Pyrosequencing for Methylation Analysis
2.4. Statistical Analysis
3. Results
3.1. Tumor LINE-1 Methylation Level and General Clinical and Pathological Characteristic Data of the Participants
3.2. Pathological and Oncological Data, Tumor LINE-1 Methylation and Colorectal Cancer Patient Survival
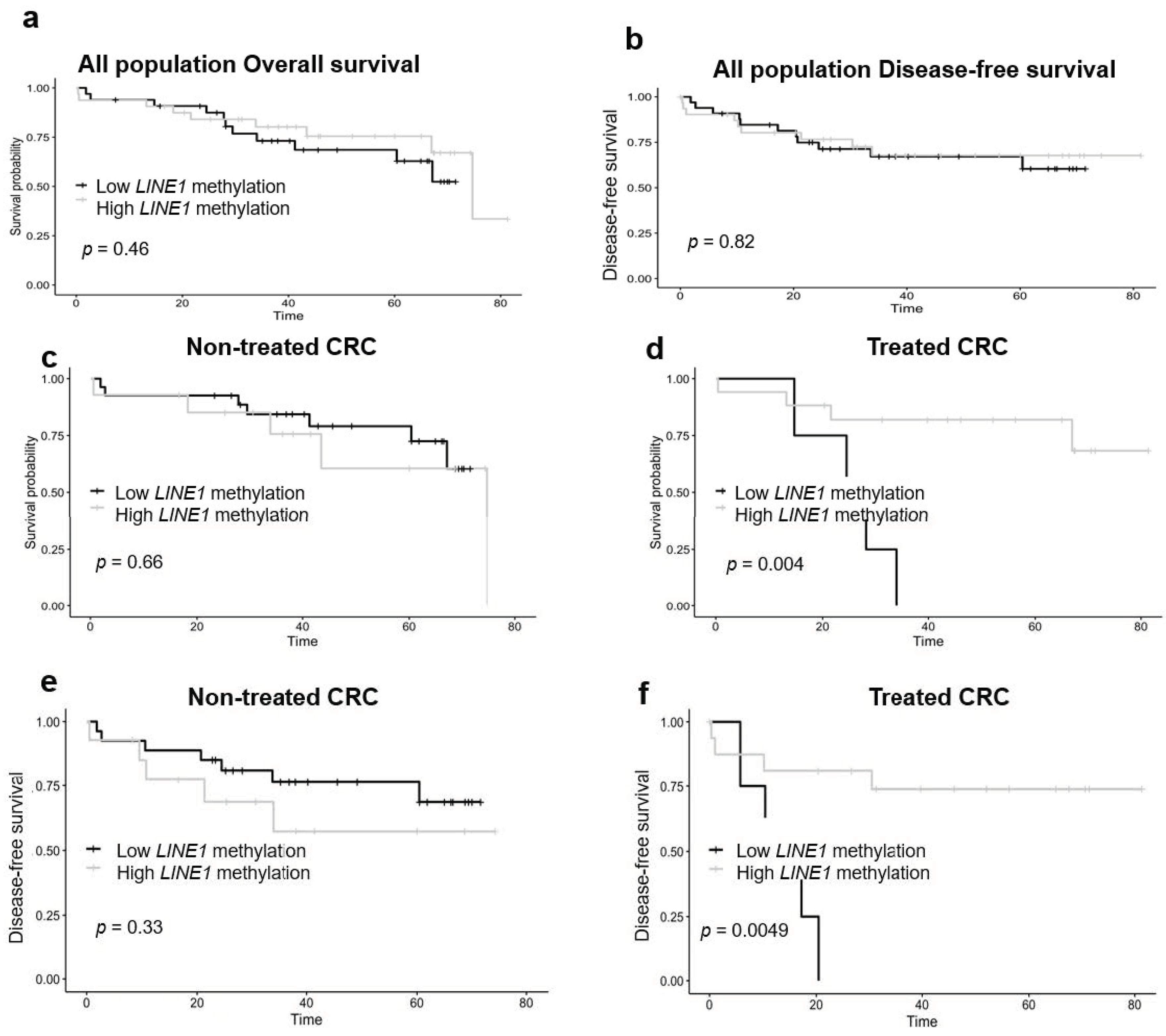
3.3. Tumor LINE-1 Methylation and Colorectal Cancer Patient Survival in Strata of Neoadjuvant Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- International Agency for Research on Cancer (IARC). GLOBOCAN 2018: Latest global cancer data. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar]
- Bray, F.; Me, J.F.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Marí-Alexandre, J.; Diaz-Lagares, A.; Villalba, M.; Juan, O.; Crujeiras, A.B.; Calvo, A.; Sandoval, J. Translating cancer epigenomics into the clinic: Focus on lung cancer. Transl. Res. 2017, 189, 76–92. [Google Scholar] [CrossRef] [PubMed]
- Boughanem, H.; Cabrera-Mulero, A.; Hernández-Alonso, P.; Bandera-Merchán, B.; Tinahones, A.; Tinahones, F.J.; Morcillo, S.; Macias-Gonzalez, M. The Expression/Methylation Profile of Adipogenic and Inflammatory Transcription Factors in Adipose Tissue Are Linked to Obesity-Related Colorectal Cancer. Cancers 2019, 11, 1629. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Williams, M.; Cheng, Y.Y.; Leung, W.K. Roles of Methylated DNA Biomarkers in Patients with Colorectal Cancer. Dis. Markers 2019, 2019, 1–8. [Google Scholar] [CrossRef]
- Dong, L.; Ren, H. Blood-based DNA Methylation Biomarkers for Early Detection of Colorectal Cancer. J. Proteom. Bioinform. 2018, 11, 120–126. [Google Scholar] [CrossRef]
- Diaz-Lagares, A.; Crujeiras, A.B.; Lopez-Serra, P.; Soler, M.; Setien, F.; Goyal, A.; Sandoval, J.; Hashimoto, Y.; Martinez-Cardús, A.; Gomez, A.; et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E7535–E7544. [Google Scholar] [CrossRef]
- Clemente-Postigo, M.; Muñoz-Garach, A.; Serrano, M.; Garrido-Sánchez, L.; Bernal-López, M.R.; Fernández-García, D.; Moreno-Santos, I.; Garriga, N.; Castellano, D.; Camargo, A.; et al. Serum 25-Hydroxyvitamin D and Adipose Tissue Vitamin D Receptor Gene Expression: Relationship With Obesity and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2015, 100. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Weber, B.; Kimhi, S.; Howard, G.; Eden, A.; Lyko, F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 2010, 29, 5775–5784. [Google Scholar] [CrossRef]
- Speek, M. Antisense Promoter of Human L1 Retrotransposon Drives Transcription of Adjacent Cellular Genes. Mol. Cell. Biol. 2001, 21, 1973–1985. [Google Scholar] [CrossRef] [PubMed]
- Kerachian, M.A.; Kerachian, M. Long interspersed nucleotide element-1 (LINE-1) methylation in colorectal cancer. Clin. Chim. Acta 2019, 488, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Mulero, A.; Crujeiras, A.B.; Izquierdo, A.G.; Torres, E.; Ayers, D.; Casanueva, F.F.; Tinahones, F.J.; Morcillo, S.; Macias-Gonzalez, M. Novel SFRP2 DNA Methylation Profile Following Neoadjuvant Therapy in Colorectal Cancer Patients with Different Grades of BMI. J. Clin. Med. 2019, 8, 1041. [Google Scholar] [CrossRef] [PubMed]
- Sunami, E.; De Maat, M.; Vu, A.; Turner, R.R.; Hoon, D.S. LINE-1 Hypomethylation during Primary Colon Cancer Progression. PLoS ONE 2011, 6, e18884. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Sunami, E.; Yamamoto, Y.; Hata, K.; Okada, S.; Murono, K.; Yasuda, K.; Otani, K.; Nishikawa, T.; Tanaka, T.; et al. LINE-1 hypomethylation status of circulating cell-free DNA in plasma as a biomarker for colorectal cancer. Oncotarget 2017, 8, 11906–11916. [Google Scholar] [CrossRef] [PubMed]
- Swets, M.; Zaalberg, A.; Boot, A.; Van Wezel, T.; Frouws, M.A.; Bastiaannet, E.; Gelderblom, H.; Van De Velde, C.; Kuppen, P.J.K. Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer. Int. J. Mol. Sci. 2016, 18, 36. [Google Scholar] [CrossRef]
- Chen, D.; Wen, X.; Song, Y.S.; Rhee, Y.-Y.; Lee, T.H.; Cho, N.-Y.; Han, S.-W.; Kim, T.; Kang, G.H. Associations and prognostic implications of Eastern Cooperative Oncology Group performance status and tumoral LINE-1 methylation status in stage III colon cancer patients. Clin. Epigenet. 2016, 8, 36. [Google Scholar] [CrossRef]
- Ye, D.; Jiang, D.; Li, Y.; Jin, M.; Chen, K. The role of LINE-1 methylation in predicting survival among colorectal cancer patients: A meta-analysis. Int. J. Clin. Oncol. 2017, 14, 2253–2757. [Google Scholar] [CrossRef]
- Kuan, T.-C.; Lin, P.-C.; Yang, S.-H.; Lin, C.-C.; Lan, Y.-T.; Lin, H.-H.; Liang, W.-Y.; Chen, W.-S.; Lin, J.-K.; Jiang, J.-K.; et al. Impact of LINE-1 hypomethylation on the clinicopathological and molecular features of colorectal cancer patients. PLoS ONE 2018, 13, e0197681. [Google Scholar] [CrossRef]
- Albores, S.J.; Adsay, N.V.; Crawford, J.M. WHO classification of Tumours. In Digestive System Tumours, 5th ed.; WHO Classification of Tumours Editorial Board: Lyon, France, 2010. [Google Scholar]
- Bairaktari, E.T.; Seferiadis, K.I.; Elisaf, M.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nowak, J.A.; Qian, Z.R.; Cao, Y.; Song, M.; Masugi, Y.; Shi, Y.; Da Silva, A.; Gu, M.; Li, W.; et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget 2016, 7, 55098–55109. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Yamauchi, M.; Nishihara, R.; Lochhead, P.; Qian, Z.R.; Kuchiba, A.; Kim, S.A.; Mima, K.; Sukawa, Y.; Jung, S.; et al. Tumor LINE-1 Methylation Level and Microsatellite Instability in Relation to Colorectal Cancer Prognosis. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Murata, A.; Baba, Y.; Watanabe, M.; Shigaki, H.; Miyake, K.; Ishimoto, T.; Iwatsuki, M.; Iwagami, S.; Sakamoto, Y.; Miyamoto, Y.; et al. Methylation levels of LINE-1 in primary lesion and matched metastatic lesions of colorectal cancer. Br. J. Cancer 2013, 109, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, S.; Liu, Y.; Zhang, C.; Li, H.; Lai, B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med. 2019, 9, 361–373. [Google Scholar] [CrossRef] [PubMed]
| Variables | Low LINE-1 Methylation (%) | High LINE-1 Methylation (%) | p |
|---|---|---|---|
| n | 34 | 33 | |
| Age (years) | 66.29 ± 10.75 | 67.27 ± 10.19 | 0.703 |
| Sex (male/female) | 24/10 | 21/12 | 0.544 |
| BMI (kg/m2) | 26.87 ± 4.37 | 27.05 ± 3.46 | 0.853 |
| Glucose (mg/dL) | 117.51 ± 34.94 | 124.63 ± 52.16 | 0.517 |
| Insulin (µUI/mL) | 5.54 ± 3.71 | 6.60 ± 5.57 | 0.380 |
| HOMA-IR | 1.68 ± 1.37 | 2.06 ± 2.08 | 0.383 |
| Total cholesterol (mg/dL) | 166.58 ± 41.02 | 177.27 ± 36.58 | 0.268 |
| Triglycerides (mg/dL) | 167.51 ± 97.85 | 162.42 ± 66.97 | 0.806 |
| LDL (mg/dL) | 99.11 ± 32.84 | 107.09 ± 29.64 | 0.978 |
| HDL (mg/dL) | 40.33 ± 11.44 | 40.24 ± 15.37 | 0.304 |
| CEA (mg/dL) | 7.27 ± 11.27 | 4.42 ± 9.62 | 0.297 |
| CA19.9 (U/mL) | 21.78 ± 28.24 | 21.93 ± 34.72 | 0.984 |
| Variables | Overall Survival | Disease-Free Survival | ||
|---|---|---|---|---|
| Univariate HR (95% CI) | Multivariate HR (95% CI) | Univariate HR (95% CI) | Multivariate HR (95% CI) | |
| Location (rectum vs. colon) | 1.28 (0.57–2.90) | 1.09 (0.29–4.18) | 1.1 (0.48–2.50) | 1.44 (0.37–5.4) |
| Stage (I + II vs. III + IV) | 1.60 (0.62–3.84) | 1.11 (0.09–13.72) | 2.3 (0.97–5.4) | 2.18 (0.20–24.0) |
| Lymph node (Negative vs. positive) | 1.41 (0.58–3.43) | 0.68 (0.07–6.44) | 2.1 (0.86–4.9) | 0.62 (0.07–5.7) |
| Vascular invasion (Negative vs. positive) | 1.84 (0.71–4.79) | 1.94 (0.58–6.45) | 2.1 (0.82–5.5) | 2.2 (0.67–7.4) |
| Metastasis (Negative vs. positive) | 5.22 (2.11–12.86) * | 8.65 (1.54–45.31) * | 8.9 (3.4–24.0) * | 8.99 (1.74–46.4) * |
| Neoadjuvant therapy (Negative vs. positive) | 1.12 (0.48–2.64) | 0.85 (0.20–3.70) | 1.4 (0.6–3.3) | 0.78 (0.18–3.3) |
| LINE-1 methylation (Low vs. high) | 0.71 (0.28–1.76) | 0.95 (0.28–3.20) | 0.9 (0.37–2.2) | 1.29 (0.42–3.9) |
| Variables | LINE-1 Methylation Levels (Continuous) β (SD) | LINE-1 Methylation Levels (Low vs. High) OR (95% CI) |
|---|---|---|
| Location (rectum vs. colon) | 3.26 (1.44) * | 1.19 (0.91–1.56) |
| Stage (I + II vs. III + IV) | 4.36 (3.60) | 1.92 (0.97–3.77) |
| Lymph node (Negative vs. positive) | −2.27 (3.53) | 0.55 (0.28–1.06) |
| Vascular invasion (Negative vs. positive) | −1.31 (1.65) | 1.01 (0.74–1.38) |
| Metastasis (Negative vs. positive) | −6.54 (2.61) * | 0.55 (0.33–0.89) * |
| Neoadjuvant treatment (Negative vs. positive) | 8.65 (1.55) * | 1.91 (1.43–2.56) * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boughanem, H.; Martin-Nuñez, G.M.; Torres, E.; Arranz-Salas, I.; Alcaide, J.; Morcillo, S.; Tinahones, F.J.; Crujeiras, A.B.; Macias-Gonzalez, M. Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival. J. Pers. Med. 2020, 10, 219. https://doi.org/10.3390/jpm10040219
Boughanem H, Martin-Nuñez GM, Torres E, Arranz-Salas I, Alcaide J, Morcillo S, Tinahones FJ, Crujeiras AB, Macias-Gonzalez M. Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival. Journal of Personalized Medicine. 2020; 10(4):219. https://doi.org/10.3390/jpm10040219
Chicago/Turabian StyleBoughanem, Hatim, Gracia María Martin-Nuñez, Esperanza Torres, Isabel Arranz-Salas, Julia Alcaide, Sonsoles Morcillo, Francisco J Tinahones, Ana B Crujeiras, and Manuel Macias-Gonzalez. 2020. "Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival" Journal of Personalized Medicine 10, no. 4: 219. https://doi.org/10.3390/jpm10040219
APA StyleBoughanem, H., Martin-Nuñez, G. M., Torres, E., Arranz-Salas, I., Alcaide, J., Morcillo, S., Tinahones, F. J., Crujeiras, A. B., & Macias-Gonzalez, M. (2020). Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival. Journal of Personalized Medicine, 10(4), 219. https://doi.org/10.3390/jpm10040219






