Lung Segmentation with Lightweight Convolutional Attention Residual U-Net
Abstract
1. Introduction
- A custom Lightweight U-Net model is proposed by combining the strengths of residual paths, CBAM, and ASPP with LeakyReLU activations for feature extraction to handle the various channel and spatial perspectives of CXR images and predict segmentation accurately.
- The effectiveness of the model was examined on three different popular datasets (JSRT, SZ, and MC), where it outperformed all other SOTA models.
- A random chest X-ray dataset from Kaggle was used for external validation, demonstrating the model’s effectiveness and robustness.
- The complexity of the model was analyzed, demonstrating that the proposed model is lighter than all other SOTA models and also more efficient while maintaining high performance.
2. Literature Review
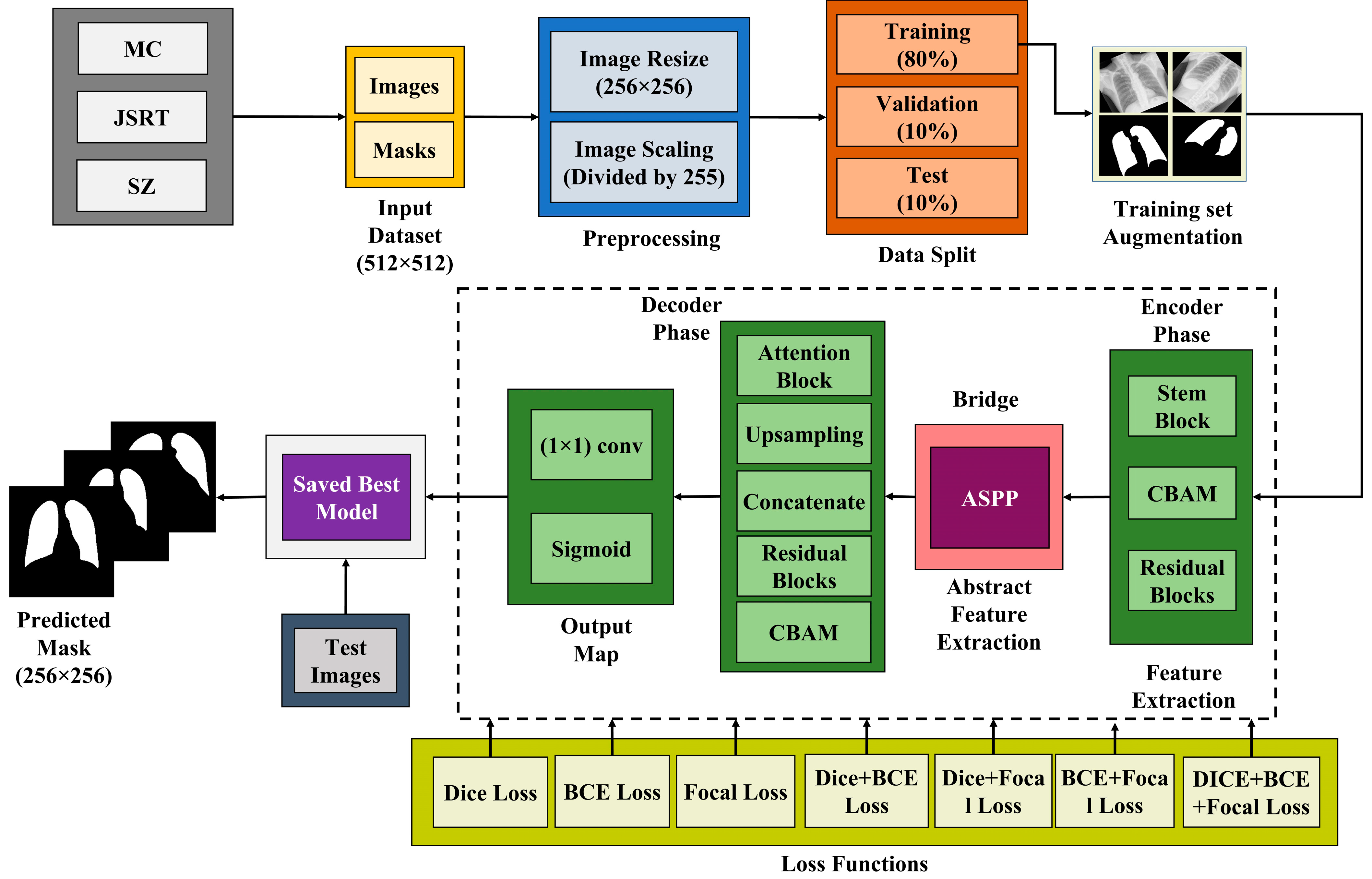
3. Proposed Method
| Algorithm 1: Lung Segmentation using Lightweight Residual U-Net |
| 1: Input: CXR images, mask images. 2: Preprocess the data by resizing the images to 256 × 256 pixels and normalizing the pixel values between 0 to 1 by dividing each pixel by 255. 3: Split the images into training, validation, and testing sets. 4: Augment the training set using the different techniques mentioned in the study. 5: Select Dice loss as the primary loss function with the Adam optimizer, train the lightweight segmentation model, and save the best-performing model based on validation loss. 6: Output: Predict the results on the test set. |
3.1. Dataset Description
3.2. Data Preprocessing and Augmentation
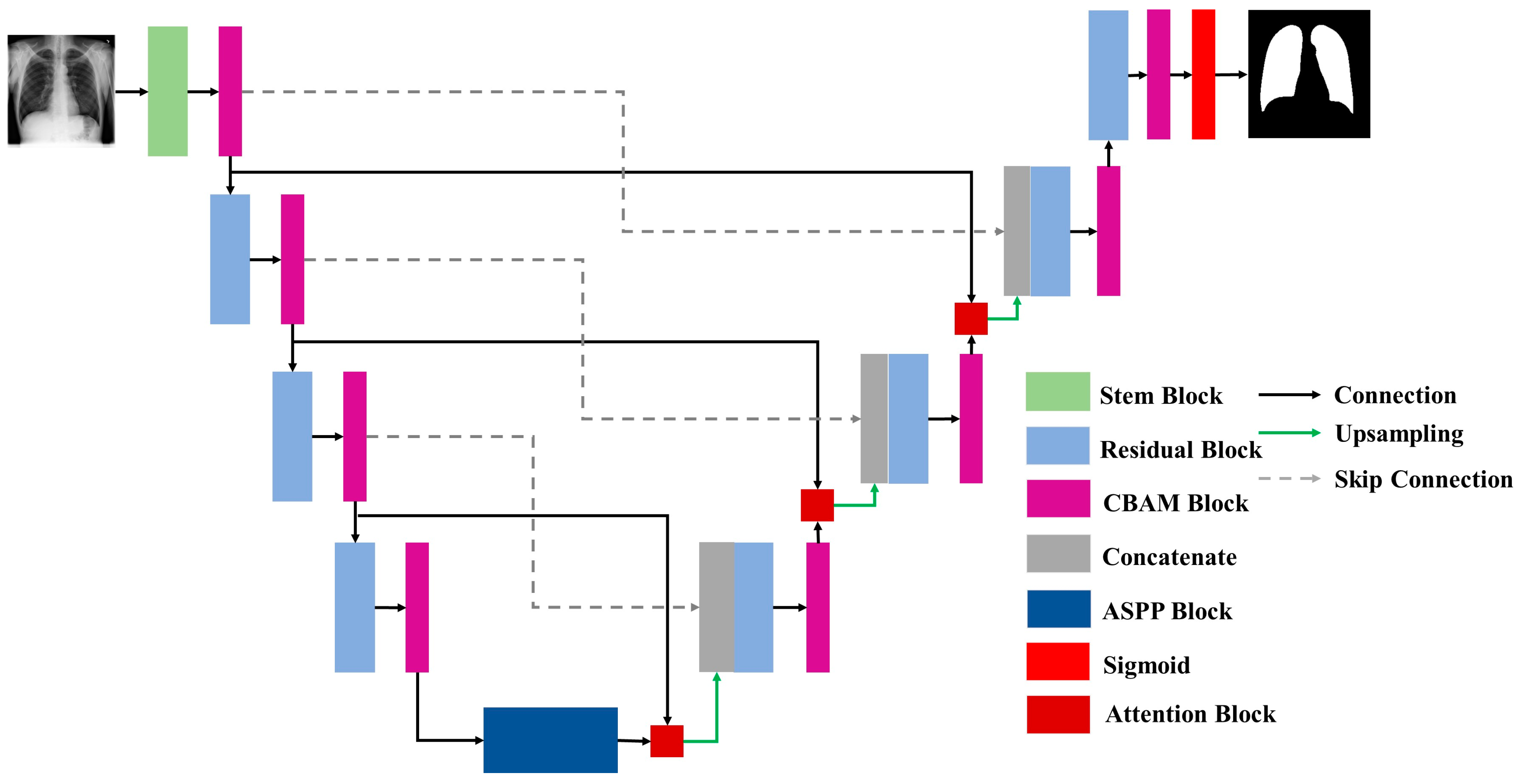
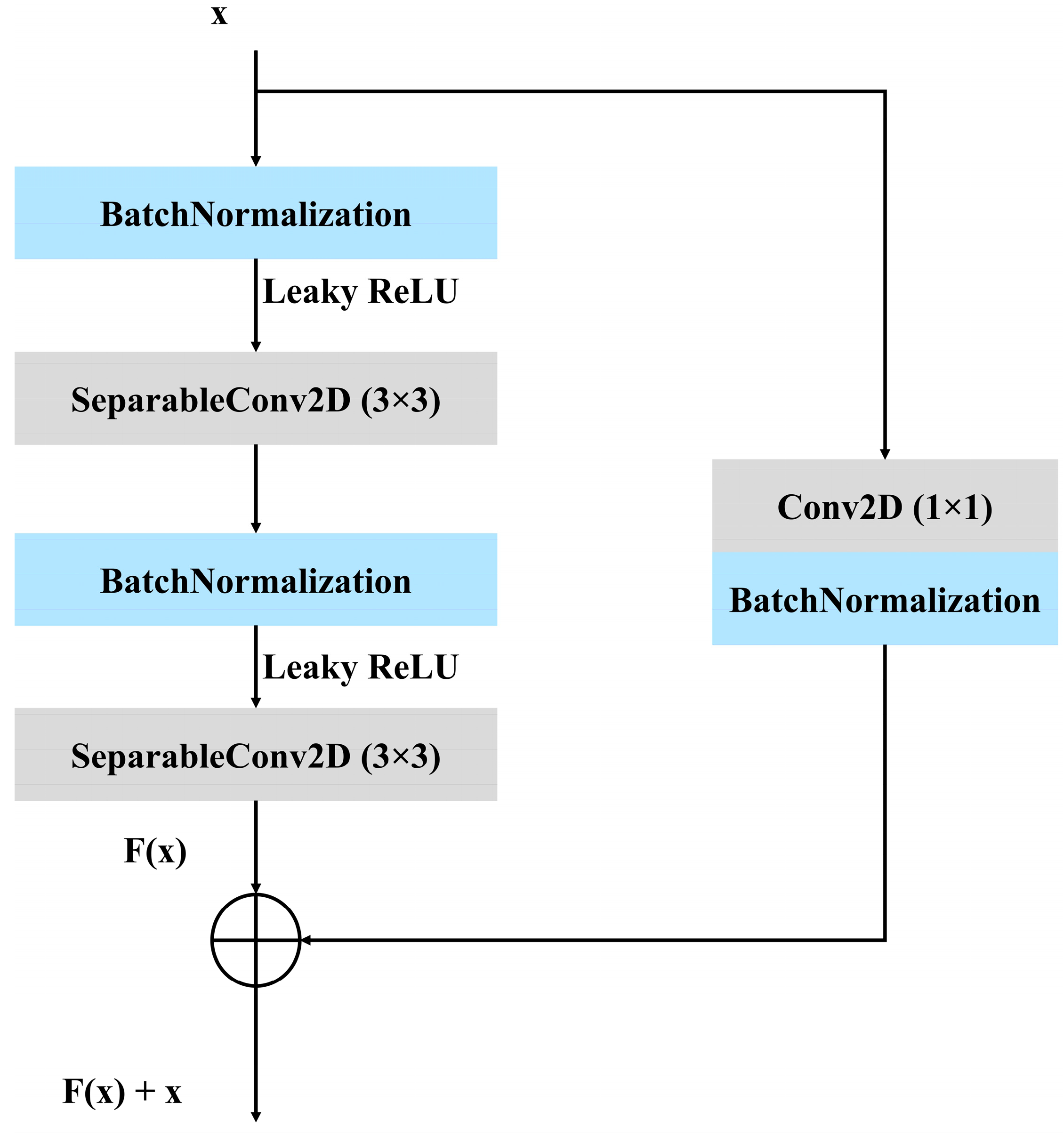
3.3. Proposed Architecture
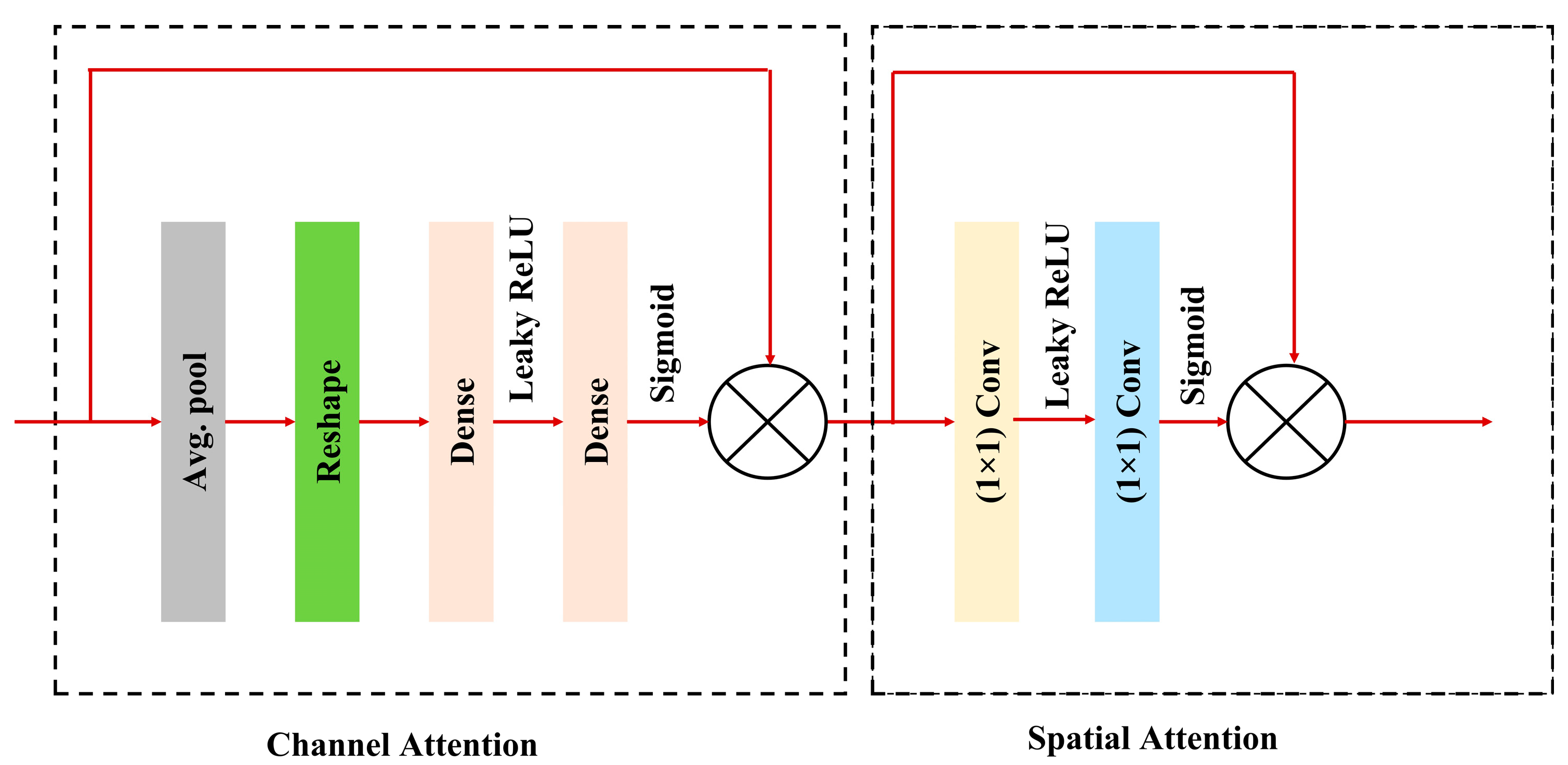
3.3.1. Convolutional Block Attention Module (CBAM)
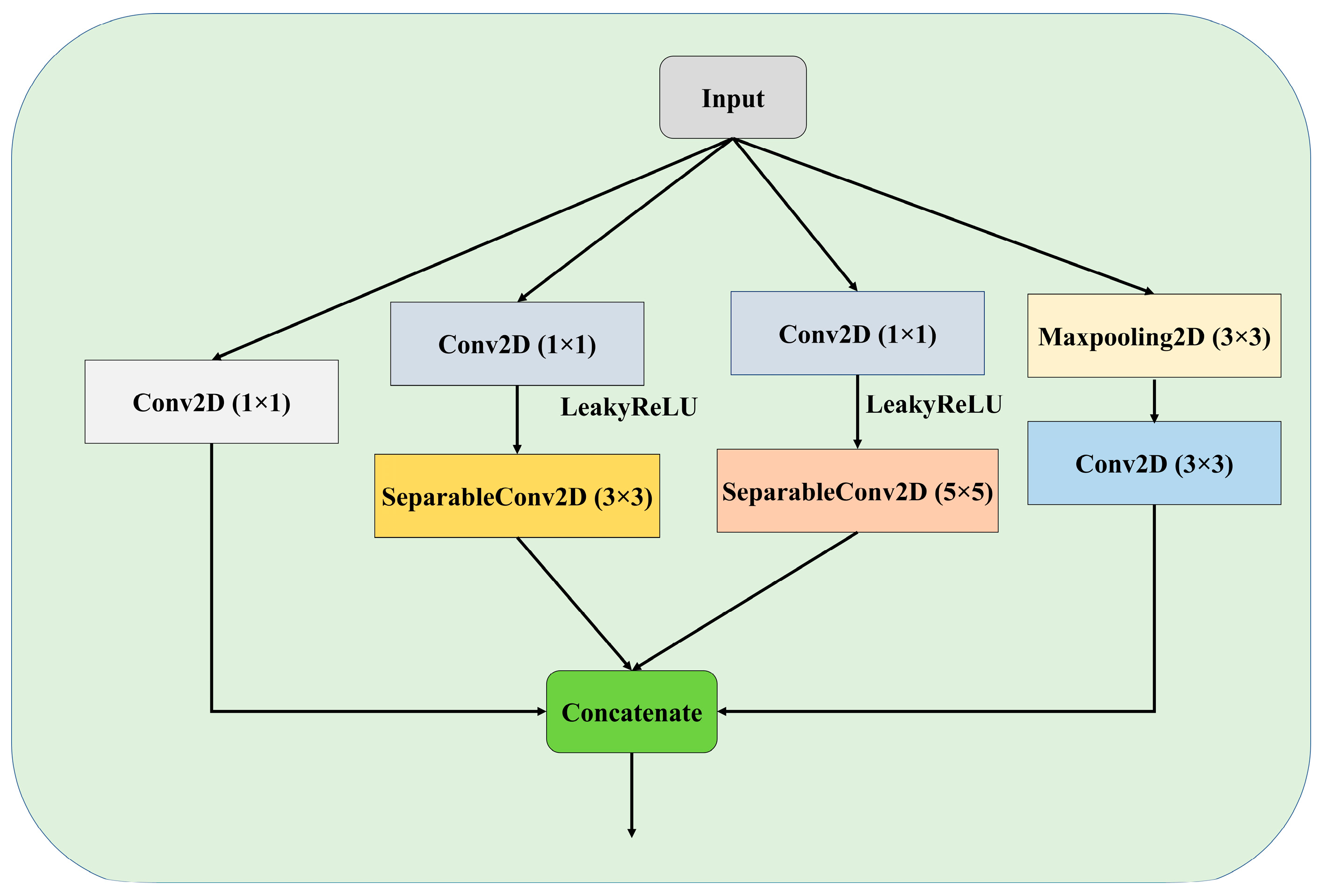
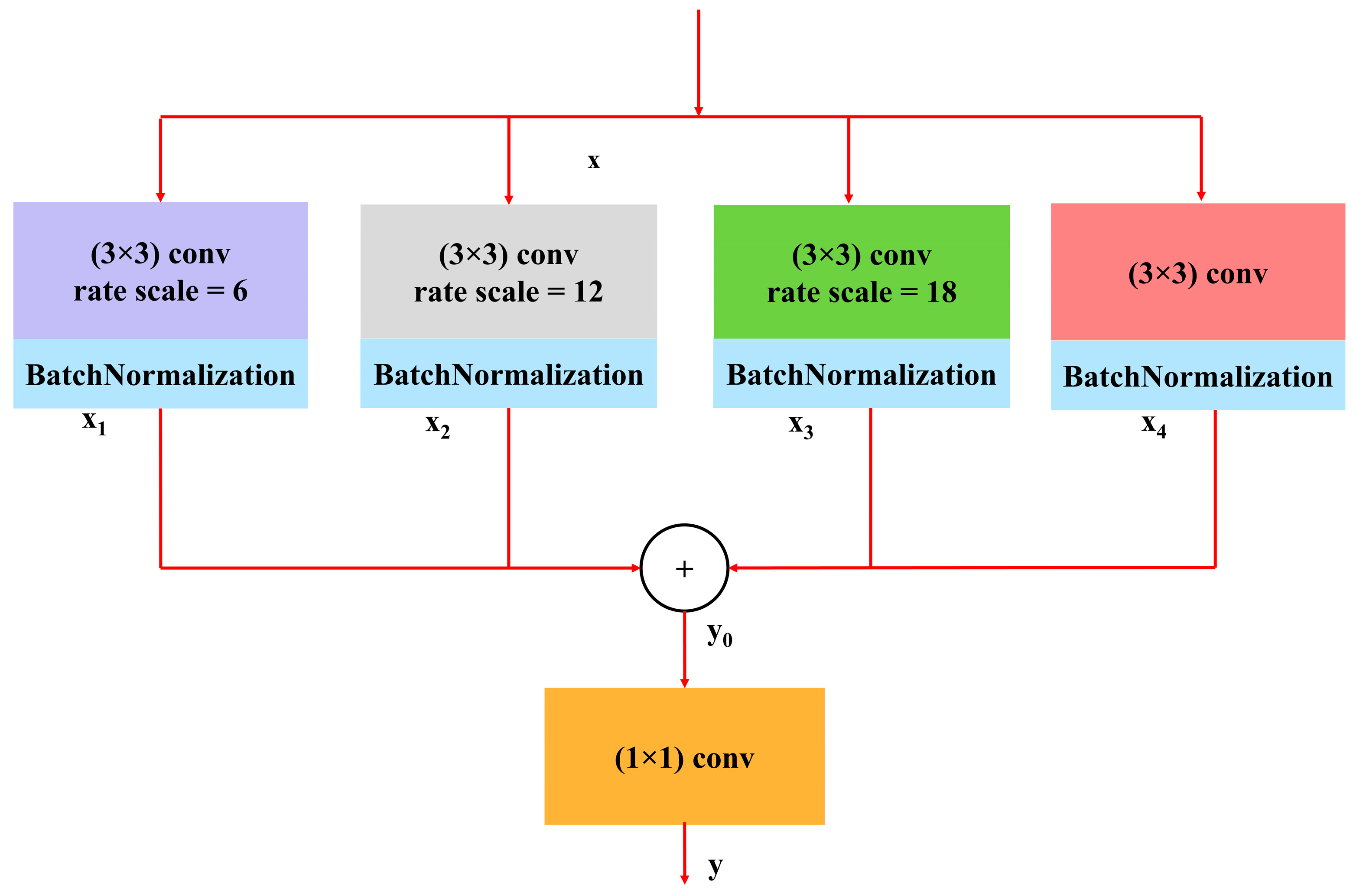
3.3.2. Atrous Spatial Pyramidal Pooling
4. Result Analysis
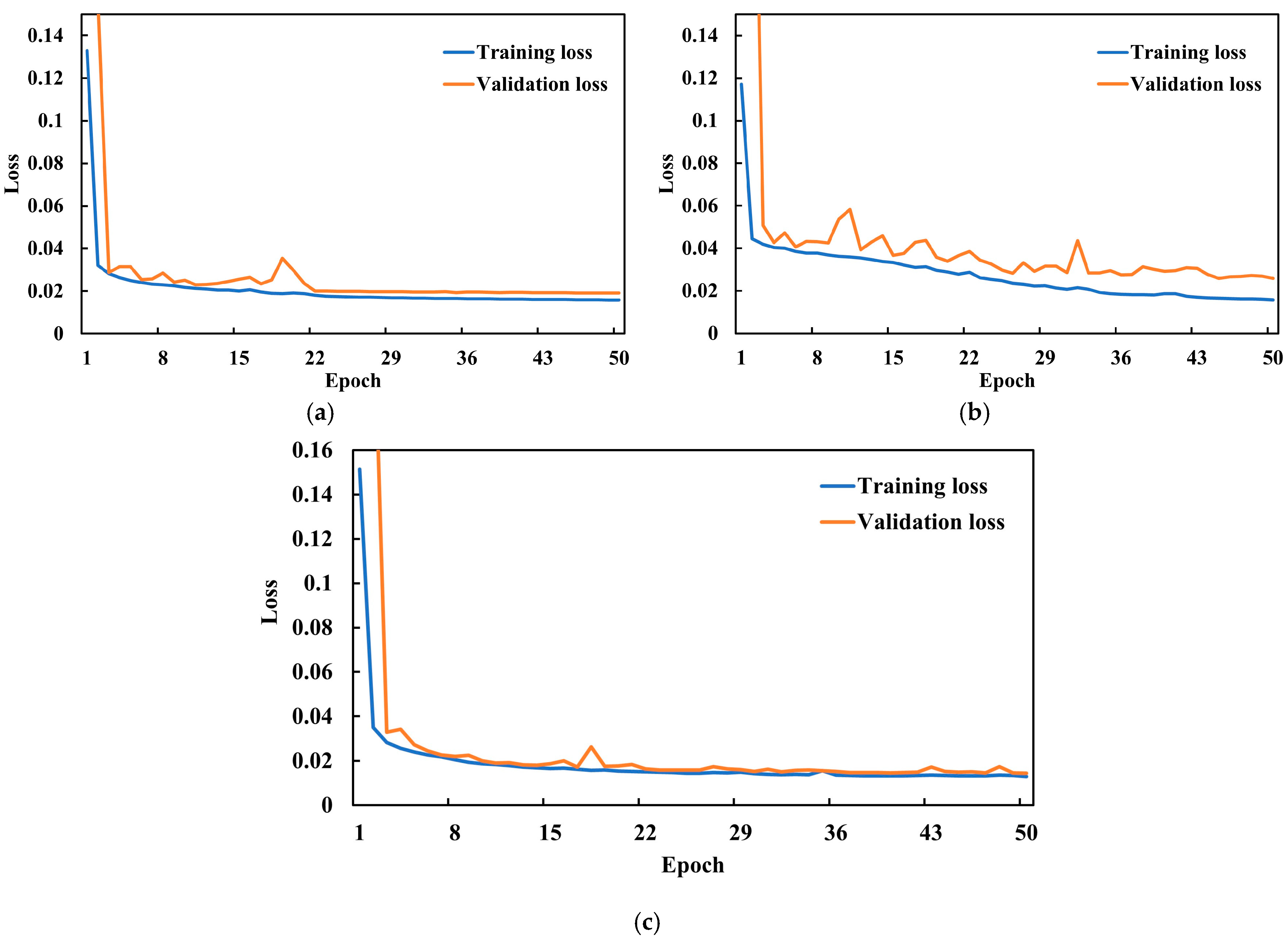
4.1. Hyperparameter Selection and Experimental Configuration
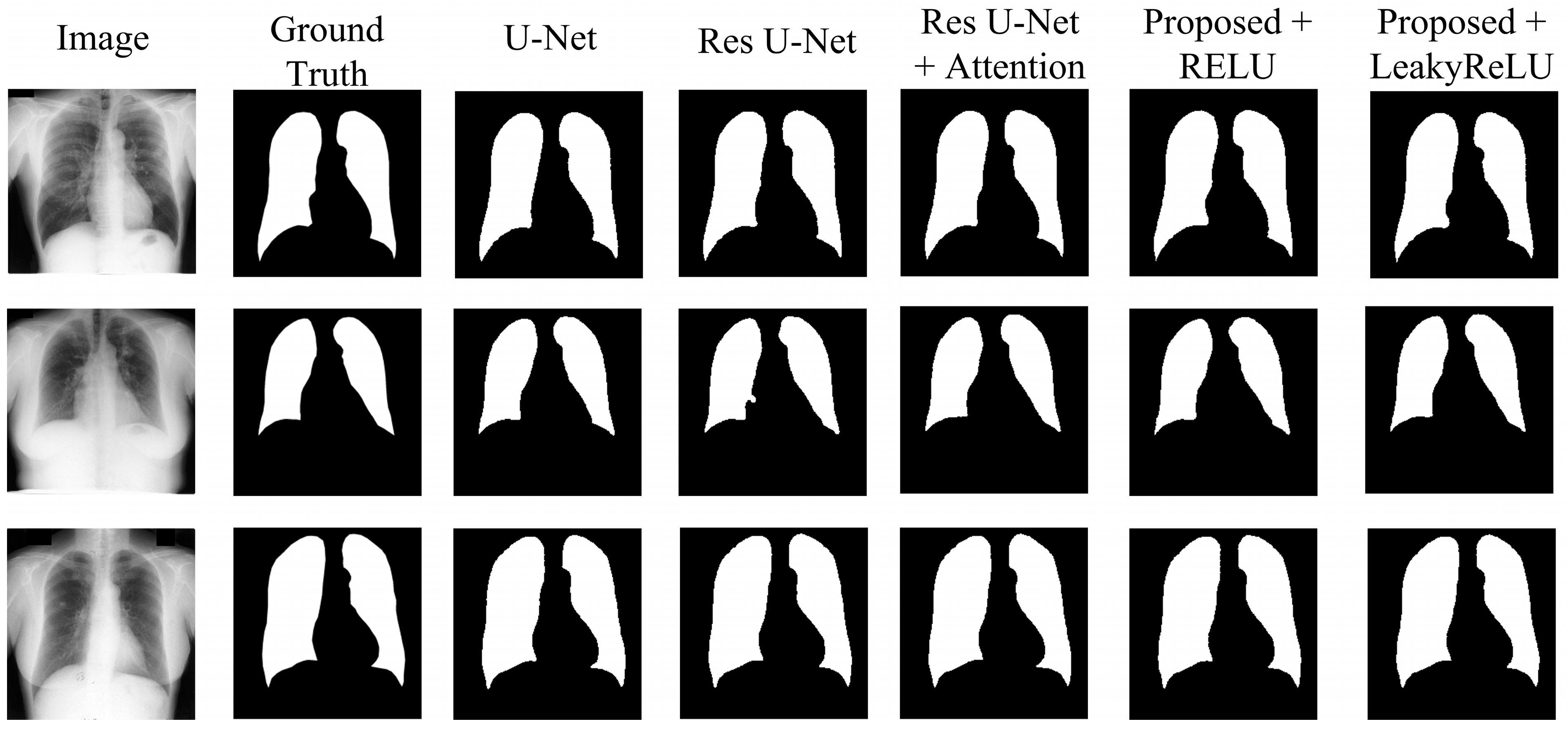
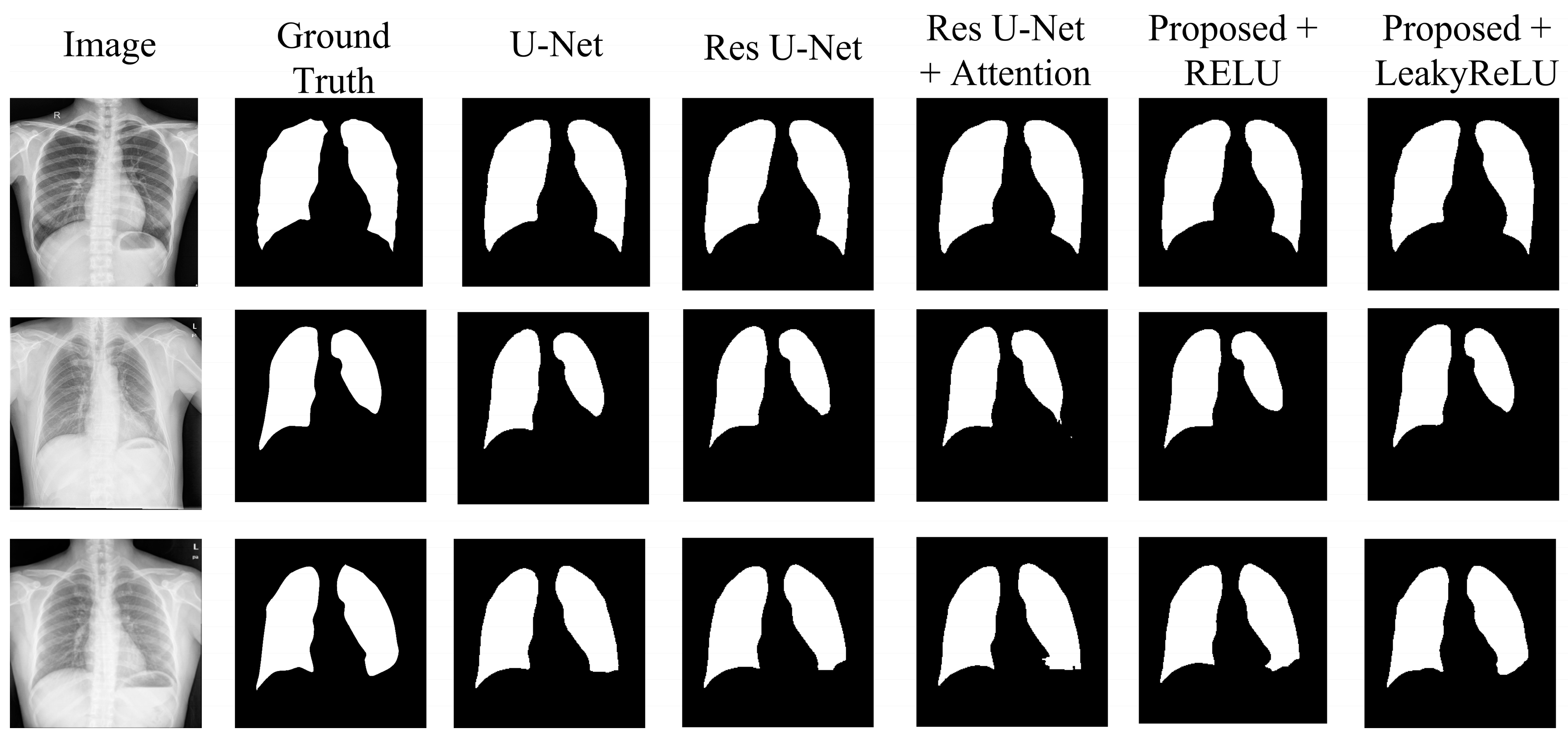
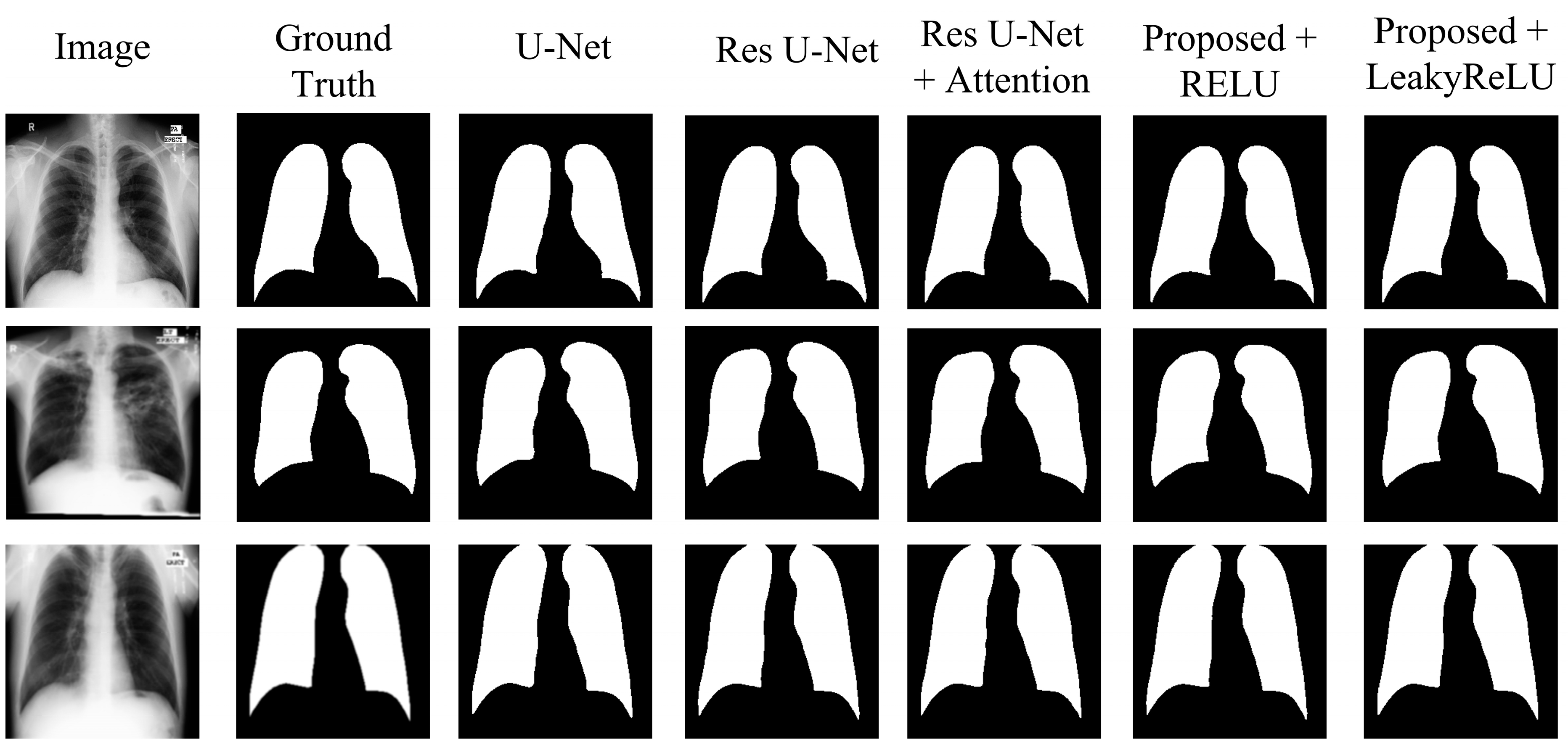
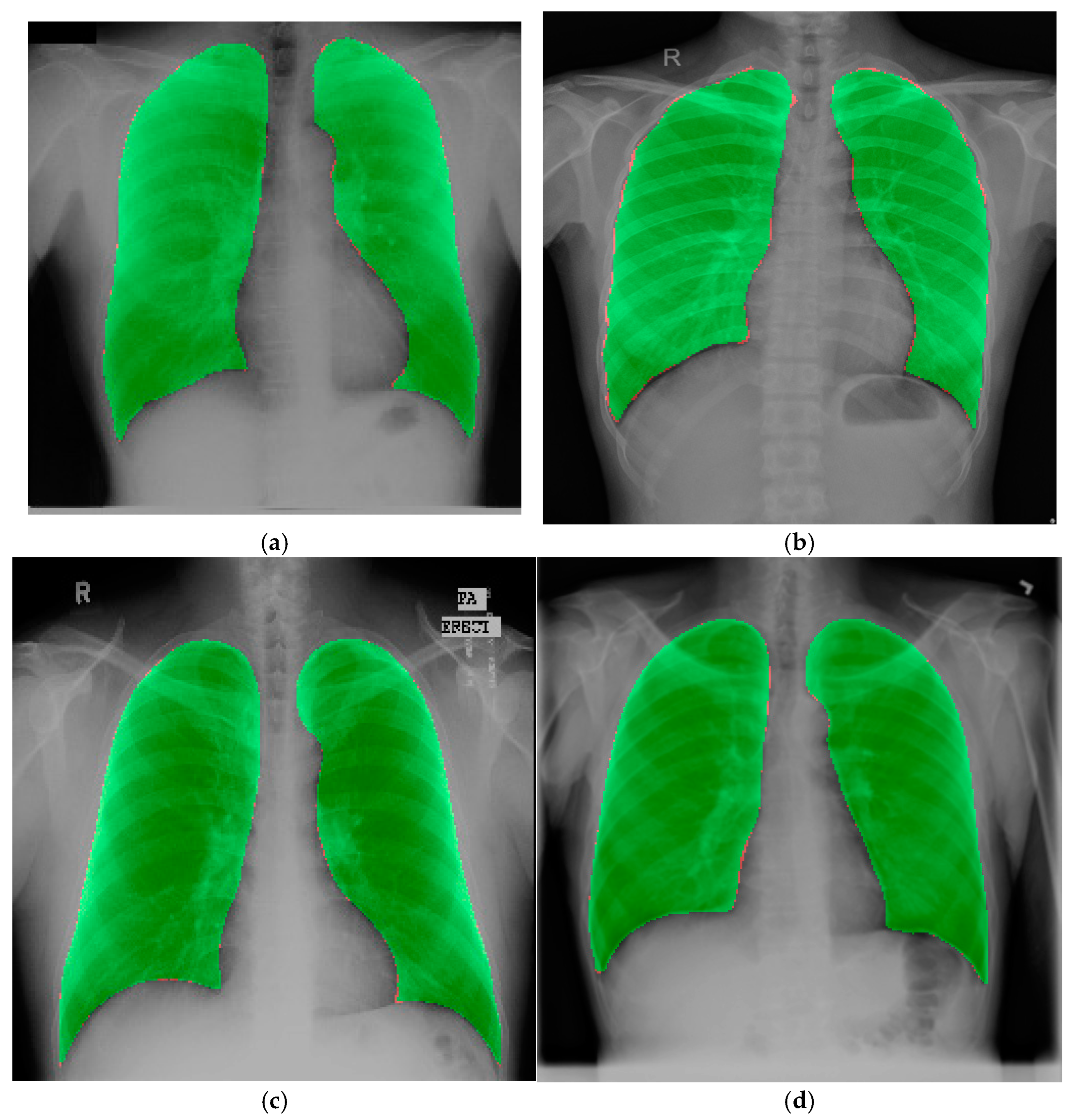
4.2. Experimental Results
- The U-Net model, acting as the original benchmark for semantic segmentation;
- The residual U-Net model, serving as the initial standard for the proposed model;
- Residual U-Net with attention-guided skip connections, without adding CBAM to the network;
- Proposed model with CBAM added after the residual block, with RELU activation;
- Proposed model with CBAM added after the residual block, with LeakyReLU activation.
4.3. Experimental Results on the Chest X-Ray Dataset for External Validation
4.4. Additional Experiments
5. Discussion
5.1. Comparative Analysis
5.2. Strengths, Limitations, and Future Scope
5.3. Clinical Significance
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, C.; Zhou, K.; Zha, M.; Qu, X.; Guo, X.; Chen, H.; Wang, Z.; Xiao, R. An effective deep neural network for lung lesions segmentation from COVID-19 CT images. IEEE Trans. Ind. Inform. 2021, 17, 6528–6538. [Google Scholar]
- Vidal, P.L.; de Moura, J.; Novo, J.; Ortega, M. Multi-stage transfer learning for lung segmentation using portable X-ray devices for patients with COVID-19. Expert Syst. Appl. 2021, 173, 114677. [Google Scholar]
- Candemir, S.; Antani, S. A review on lung boundary detection in chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 563–576. [Google Scholar] [PubMed]
- Chen, L.; Yu, Z.; Huang, J.; Shu, L.; Kuosmanen, P.; Shen, C.; Ma, X.; Li, J.; Sun, C.; Li, Z. Development of lung segmentation method in X-ray images of children based on TransResUNet. Front. Radiol. 2023, 3, 1190745. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4. Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar]
- Xu, X.; Wang, M.; Liu, D.; Lei, M.; Fu, J.; Jia, Y. TransCotANet: A Lung Field Image Segmentation Network with Multidimensional Global Feature Dynamic Aggregation. Symmetry 2023, 15, 1480. [Google Scholar] [CrossRef]
- Akter, S. Generative AI: A Pix2pix-GAN-Based Machine Learning Approach for Robust and Efficient Lung Segmentation. arXiv 2024, arXiv:2412.10826. [Google Scholar]
- Khorasani, M.S.; Babapour Mofrad, F. The effectiveness of FAT-Net in chest X-ray segmentation. J. Eng. 2025, 2025, e70065. [Google Scholar]
- Hao, X.; Zhang, C.; Xu, S. Fast Lung Image Segmentation Using Lightweight VAEL-Unet. EAI Endorsed Trans. Scalable Inf. Syst. 2024, 11. [Google Scholar] [CrossRef]
- Sulaiman, A.; Anand, V.; Gupta, S.; Asiri, Y.; Elmagzoub, M.; Reshan, M.S.A.; Shaikh, A. A convolutional neural network architecture for segmentation of lung diseases using chest X-ray images. Diagnostics 2023, 13, 1651. [Google Scholar] [CrossRef]
- Alam, M.S.; Wang, D.; Arzhaeva, Y.; Ende, J.A.; Kao, J.; Silverstone, L.; Yates, D.; Salvado, O.; Sowmya, A. Attention-based multi-residual network for lung segmentation in diseased lungs with custom data augmentation. Sci. Rep. 2024, 14, 28983. [Google Scholar]
- Gite, S.; Mishra, A.; Kotecha, K. Enhanced lung image segmentation using deep learning. Neural Comput. Appl. 2023, 35, 22839–22853. [Google Scholar]
- Zhang, Q.; Min, B.; Hang, Y.; Chen, H.; Qiu, J. A full-scale lung image segmentation algorithm based on hybrid skip connection and attention mechanism. Sci. Rep. 2024, 14, 23233. [Google Scholar]
- Yuan, H.; Hong, C.; Tran, N.T.A.; Xu, X.; Liu, N. Leveraging anatomical constraints with uncertainty for pneumothorax segmentation. Health Care Sci. 2024, 3, 456–474. [Google Scholar] [PubMed]
- Hasan, D.; Abdulazeez, A.M. Lung segmentation from chest X-ray images using deeplabv3plus-based cnn model. Indones. J. Comput. Sci. 2024, 13. [Google Scholar] [CrossRef]
- Turk, F.; Kılıçaslan, M. Lung image segmentation with improved U-Net, V-Net and Seg-Net techniques. PeerJ Comput. Sci. 2025, 11, e2700. [Google Scholar]
- Xu, X.; Lei, M.; Liu, D.; Wang, M.; Lu, L. Lung segmentation in chest X-ray image using multi-interaction feature fusion network. IET Image Process. 2023, 17, 4129–4141. [Google Scholar]
- Ji, Z.; Zhao, Z.; Zeng, X.; Wang, J.; Zhao, L.; Zhang, X.; Ganchev, I. ResDSda_U-Net: A novel U-net-based residual network for segmentation of pulmonary nodules in lung CT images. IEEE Access 2023, 11, 87775–87789. [Google Scholar]
- Khehrah, N.; Farid, M.S.; Bilal, S.; Khan, M.H. Lung nodule detection in CT images using statistical and shape-based features. J. Imaging 2020, 6, 6. [Google Scholar] [CrossRef]
- Din, S.; Shoaib, M.; Serpedin, E. CXR-Seg: A Novel Deep Learning Network for Lung Segmentation from Chest X-ray Images. Bioengineering 2025, 12, 167. [Google Scholar] [CrossRef]
- Rajaraman, S.; Zamzmi, G.; Folio, L.; Alderson, P.; Antani, S. Chest X-ray bone suppression for improving classification of tuberculosis-consistent findings. Diagnostics 2021, 11, 840. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, J.; Zhang, Y.; Tian, X. A Segmentation Method Based on PDNet for Chest X-rays with Targets in Different Positions and Directions. Appl. Sci. 2023, 13, 5000. [Google Scholar] [CrossRef]
- Fu, H.; Song, G.; Wang, Y. Improved YOLOv4 marine target detection combined with CBAM. Symmetry 2021, 13, 623. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Liu, W.; Luo, J.; Yang, Y.; Wang, W.; Deng, J.; Yu, L. Automatic lung segmentation in chest X-ray images using improved U-Net. Sci. Rep. 2022, 12, 8649. [Google Scholar]
- Alirezaie, J.; Tam, W.; Babyn, P. Robust Lung Segmentation in Chest X-Ray Images Using Modified U-Net with Deeper Network and Residual Blocks. SSRN. Available online: http://dx.doi.org/10.2139/ssrn.5007476 (accessed on 15 November 2024).
- Murugappan, M.; Bourisly, A.K.; Prakash, N.; Sumithra, M.; Acharya, U.R. Automated semantic lung segmentation in chest CT images using deep neural network. Neural Comput. Appl. 2023, 35, 15343–15364. [Google Scholar]
| Dataset | Original Image Samples | Training Images | Augmented Training Images | Validation Images | Testing Images |
|---|---|---|---|---|---|
| JSRT | 247 | 198 | 8000 | 25 | 25 |
| SZ | 662 | 530 | 8000 | 66 | 66 |
| MC | 138 | 110 | 8000 | 14 | 14 |
| Hyperparameters | Value |
|---|---|
| Batch size | 32 |
| LeakyReLU negative slope | 0.01 |
| Optimizer | Adam |
| Initial learning rate | 0.001 |
| Epochs | 50 |
| LR reduce factor | 0.1 |
| Loss function | Dice Loss |
| Tools | Configuration |
|---|---|
| Programming Language | Python |
| Backend | Keras with TensorFlow |
| Disk Space | 78.2 GB |
| GPU RAM | 15 GB |
| GPU | Nvidia Tesla T4 |
| System RAM | 12.72 GB |
| Operating system | windows 10 |
| Input | Lung images |
| Input Size | 256 × 256 |
| Model | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| I. | 99.03 | 98.34 | 96.74 | 98.38 | 98.32 | 99.22 |
| II. | 99.04 | 98.39 | 96.83 | 98.39 | 98.39 | 99.30 |
| III. | 99.06 | 98.44 | 96.94 | 98.42 | 98.47 | 99.34 |
| IV. | 99.11 | 98.51 | 97.06 | 98.50 | 98.52 | 99.36 |
| V. | 99.24 | 98.72 | 97.48 | 98.68 | 98.76 | 99.46 |
| Model | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| I. | 98.33 | 96.32 | 93.00 | 95.69 | 97.13 | 99.17 |
| II. | 98.47 | 96.65 | 93.56 | 96.83 | 96.56 | 98.99 |
| III. | 98.37 | 96.41 | 93.15 | 95.79 | 97.17 | 99.18 |
| IV. | 98.83 | 97.41 | 94.98 | 97.12 | 97.74 | 99.33 |
| V. | 98.88 | 97.49 | 95.13 | 97.29 | 98.04 | 99.42 |
| Model | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| I. | 98.15 | 96.27 | 92.93 | 96.17 | 96.55 | 98.86 |
| II. | 98.29 | 96.57 | 93.47 | 96.11 | 97.17 | 99.07 |
| III. | 98.22 | 96.42 | 93.20 | 95.87 | 97.13 | 99.07 |
| IV. | 98.26 | 96.51 | 93.36 | 96.05 | 97.12 | 99.06 |
| V. | 99.56 | 99.08 | 98.18 | 99.34 | 98.83 | 99.62 |
| Models | Total Parameters (Millions) | Trainable Parameters (Millions) | Size (MB) |
|---|---|---|---|
| U-Net | 3.27 | 3.27 | 12.50 |
| Res U-Net | 5.06 | 5.05 | 19.32 |
| Res U-Net+ Attention | 5.86 | 5.85 | 22.35 |
| Proposed | 3.24 | 3.23 | 12.37 |
| 5-Fold CV | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| Fold-1 | 99.490 | 98.902 | 97.893 | 98.824 | 99.027 | 99.703 |
| Fold-2 | 99.509 | 98.951 | 97.975 | 98.876 | 99.067 | 99.714 |
| Fold-3 | 99.510 | 98.931 | 97.950 | 98.835 | 99.086 | 99.727 |
| Fold-4 | 99.467 | 98.842 | 97.806 | 98.721 | 99.044 | 99.712 |
| Fold-5 | 99.490 | 98.899 | 97.892 | 98.834 | 99.025 | 99.710 |
| Mean | 99.493 | 98.905 | 97.903 | 98.818 | 99.050 | 99.713 |
| Std. Dev | 0.018 | 0.041 | 0.065 | 0.058 | 0.026 | 0.008 |
| Loss | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| BCE loss | 99.45 | 98.81 | 97.73 | 98.659 | 99.030 | 99.723 |
| Dice loss | 99.56 | 99.08 | 98.18 | 99.34 | 98.83 | 99.62 |
| Focal loss | 98.612 | 98.641 | 96.381 | 98.554 | 99.177 | 98.535 |
| BCE + Dice | 99.480 | 98.864 | 97.845 | 98.824 | 98.978 | 99.696 |
| BCE + Focal | 99.409 | 98.721 | 97.538 | 98.891 | 98.604 | 99.575 |
| Dice + Focal | 99.464 | 98.848 | 97.804 | 98.865 | 98.899 | 99.659 |
| BCE + Dice + Focal | 99.414 | 98.721 | 97.577 | 98.721 | 98.812 | 99.645 |
| Loss | Accuracy | Dice | IoU | Recall | Precision | Specificity |
|---|---|---|---|---|---|---|
| Adam | 99.45 | 98.81 | 97.73 | 98.659 | 99.030 | 99.723 |
| RMSprop | 97.32 | 96.85 | 94.56 | 96.21 | 97.45 | 98.65 |
| SGD | 94.27 | 93.89 | 91.12 | 93.45 | 94.12 | 96.32 |
| Model | p-Value (Accuracy) | p-Value (Dice) | p-Value (IoU) |
|---|---|---|---|
| I. | 0.032 | 0.041 | 0.028 |
| II. | 0.015 | 0.048 | 0.039 |
| III. | 0.035 | 0.036 | 0.027 |
| IV. | 0.022 | 0.033 | 0.018 |
| Reference | Model | Dataset Used | Accuracy | Dice | IoU | No. of Total Paramters (M) |
|---|---|---|---|---|---|---|
| Xu et al. [8] | TransCotANet | JSRT SZ MC | 99.14 98.46 98.91 | 99.03 97.66 98.02 | 98.76 94.41 97.89 | - |
| Sharmin et al. [9] | Pix2pix-GAN | SZ MC | 95.87 98.25 | - 98.05 | - | 11.5 |
| Khorasani et al. [10] | FAT-Net | SZ, MC (Merged) | 98.12 | 96.10 | - | - |
| Hao et al. [11] | VAEL-Unet | Chest X-ray | 97.69 | - - | 93.65 | 1.1 |
| Alam et al. [13] | AMRU++ | MC + JSRT SZ | - - | 96.28 93.38 | 93.46 87.97 | 10.65 |
| Gite et al. [14] | U-Net++ | SZ MC | 98.74 97.71 | 97.96 96.30 | 95.96 92.93 | 36.64 |
| Hasan et al. [17] | DeeplabV3+ | SZ | 97.42 | 96.63 | 93.49 | 11.85 |
| Liu et al. [29] | Efficientnet-b4 encoder + Residual blocks + LeakyReLU | JSRT MC | 98.55 98.94 | 97.92 97.82 | 95.73 95.55 | - |
| Tam et al. [30] | DDRU-Net | SZ MC | 98.23 99.35 | 94.84 98.87 | 90.30 97.77 | 64.65 |
| Din et al. [22] | CXR-Seg | MC SZ | 98.77 96.69 | 97.76 96.32 | 95.63 92.97 | 5.98 |
| Proposed | Residual U-Net with CBAM+ LeakyReLU | JSRT SZ MC Chest X-ray | 99.24 98.88 99.56 99.51 | 98.72 97.49 99.08 98.93 | 97.48 95.13 98.18 97.95 | 3.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jannat, M.; Birahim, S.A.; Hasan, M.A.; Roy, T.; Sultana, L.; Sarker, H.; Fairuz, S.; Abdallah, H.A. Lung Segmentation with Lightweight Convolutional Attention Residual U-Net. Diagnostics 2025, 15, 854. https://doi.org/10.3390/diagnostics15070854
Jannat M, Birahim SA, Hasan MA, Roy T, Sultana L, Sarker H, Fairuz S, Abdallah HA. Lung Segmentation with Lightweight Convolutional Attention Residual U-Net. Diagnostics. 2025; 15(7):854. https://doi.org/10.3390/diagnostics15070854
Chicago/Turabian StyleJannat, Meftahul, Shaikh Afnan Birahim, Mohammad Asif Hasan, Tonmoy Roy, Lubna Sultana, Hasan Sarker, Samia Fairuz, and Hanaa A. Abdallah. 2025. "Lung Segmentation with Lightweight Convolutional Attention Residual U-Net" Diagnostics 15, no. 7: 854. https://doi.org/10.3390/diagnostics15070854
APA StyleJannat, M., Birahim, S. A., Hasan, M. A., Roy, T., Sultana, L., Sarker, H., Fairuz, S., & Abdallah, H. A. (2025). Lung Segmentation with Lightweight Convolutional Attention Residual U-Net. Diagnostics, 15(7), 854. https://doi.org/10.3390/diagnostics15070854








