Evaluation of the PowerChek™ Respiratory Virus Panel 1/2/3/4 for the Detection of 16 Respiratory Viruses: A Comparative Study with the Allplex™ Respiratory Panel Assay 1/2/3 and BioFire® Respiratory Panel 2.1 plus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Clinical Specimen
2.3. PowerChek™ RVP 1/2/3/4
2.4. Allplex™ RP 1/2/3
2.5. BioFire® Respiratory Panel 2.1 plus (RP 2.1plus)
2.6. Comparison of Three Real-Time PCR Methods for Respiratory Virus Detection
2.7. Further Study for Discordant Results
2.8. Statistical Analysis
3. Results
3.1. Detection of 16 Respiratory Viruses
3.2. Diagnostic Performance of the Powerchek™ RVP 1/2/3/4 Assay
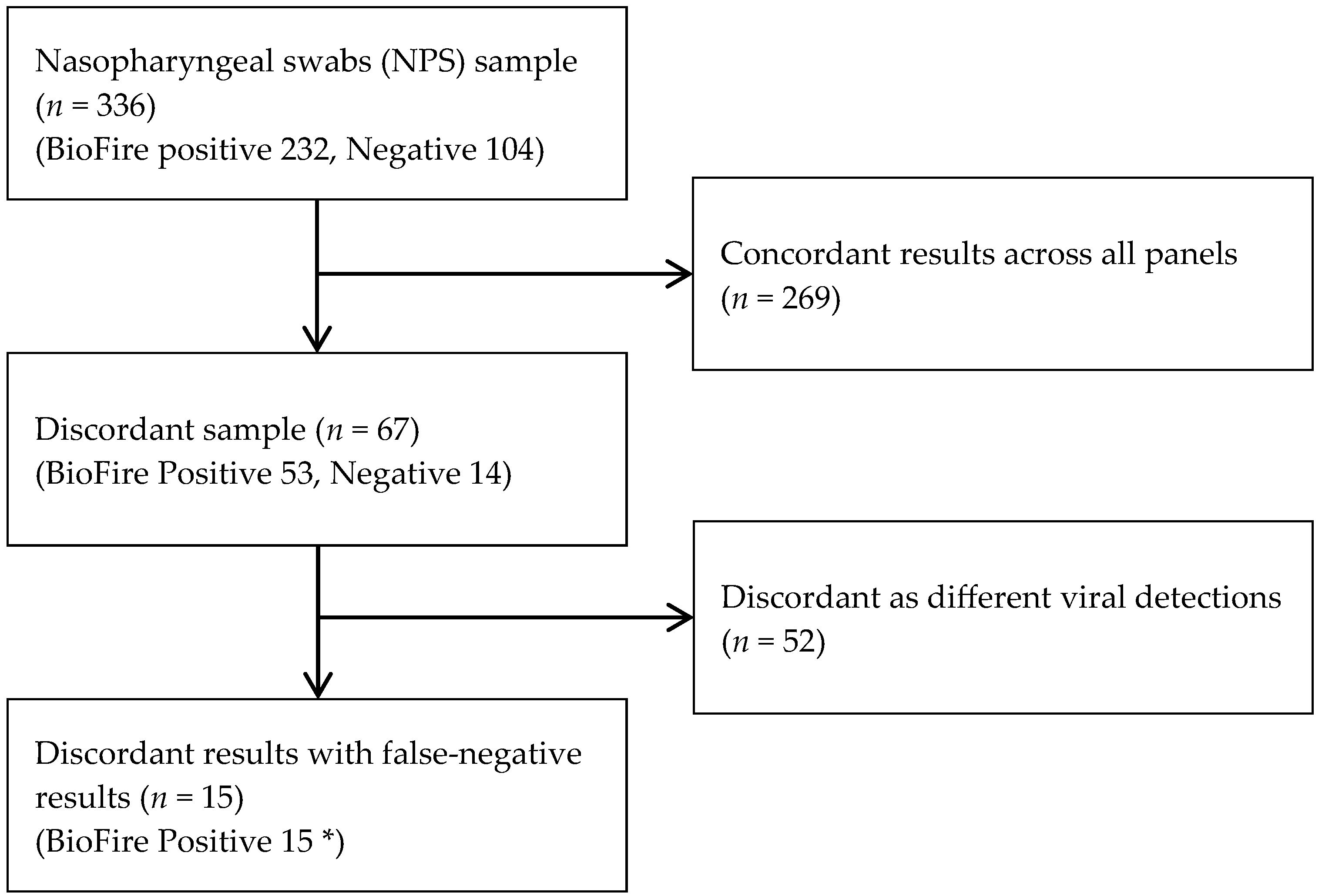
3.3. Further Analysis for Discrepant Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | acute respiratory infections |
| AdV | adenovirus |
| HBoV | human bocavirus |
| CoV | coronavirus |
| RSV | respiratory syncytial virus |
| HEV | human enterovirus |
| HRV | human rhinovirus |
| PIV | parainfluenza virus |
| HMPV | human metapneumovirus |
| TAT | turnaround time |
| PCR | polymerase chain reaction |
| RT-PCR | multiplex reverse transcription PCR |
| RP | respiratory panel |
| IFV | influenza viruses |
| NPS | nasopharyngeal swabs |
| PPV | positive predictive value |
| NPV | negative predictive value |
| Ct | Cycle threshold |
| RVP | respiratory virus panel |
| PoC | point-of-care |
References
- Diseases, G.B.D.; Injuries, C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar]
- Avendaño Carvajal, L.; Perret Pérez, C. Epidemiology of respiratory infections. In Pediatric Respiratory Diseases: A Comprehensive Textbook; Springer: Cham, Switzerland, 2020; pp. 263–272. [Google Scholar]
- Cillóniz, C.; Pericàs, J.M.; Rojas, J.R.; Torres, A. Severe infections due to respiratory viruses. In Seminars in Respiratory and Critical Care Medicine; Thieme Medical Publishers, Inc.: New York, NY, USA, 2022; pp. 60–74. [Google Scholar]
- Ren, L.; Xiang, Z.; Guo, L.; Wang, J. Viral infections of the lower respiratory tract. Curr. Infect. Dis. Rep. 2012, 14, 284–291. [Google Scholar] [CrossRef]
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A. Better tests, better care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013, 57 (Suppl. 3), S139–S170. [Google Scholar] [CrossRef]
- Rattanamas, K.; Taesuji, M.; Kulthonggate, U.; Jantafong, T.; Mamom, T.; Ruenphet, S. Sensitivity of RNA viral nucleic acid-based detection of avian influenza virus, Newcastle disease virus, and African horse sickness virus on flinders technology associates card using conventional reverse-transcription polymerase chain reaction. Vet. World 2022, 15, 2754. [Google Scholar] [CrossRef]
- Liu, Q.; Jin, X.; Cheng, J.; Zhou, H.; Zhang, Y.; Dai, Y. Advances in the application of molecular diagnostic techniques for the detection of infectious disease pathogens. Mol. Med. Rep. 2023, 27, 104. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, D.; Wang, J.; Zhang, R.; Li, J. Design, optimization, and application of multiplex rRT-PCR in the detection of respiratory viruses. Crit. Rev. Clin. Lab. Sci. 2022, 59, 555–572. [Google Scholar] [CrossRef]
- Jiang, X.-W.; Huang, T.-S.; Xie, L.; Chen, S.-Z.; Wang, S.-D.; Huang, Z.-W.; Li, X.-Y.; Ling, W.-P. Development of a diagnostic assay by three-tube multiplex real-time PCR for simultaneous detection of nine microorganisms causing acute respiratory infections. Sci. Rep. 2022, 12, 13306. [Google Scholar] [CrossRef]
- Valones, M.A.A.; Guimarães, R.L.; Brandão, L.A.C.; Souza, P.R.E.d.; Carvalho, A.d.A.T.; Crovela, S. Principles and applications of polymerase chain reaction in medical diagnostic fields: A review. Braz. J. Microbiol. 2009, 40, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Chen, Y.; Han, Y.; Diao, Z.; Zhao, Z.; Zhang, Y.; Huang, T.; Ma, Y.; Li, Z.; Jiang, J. Key performance evaluation of commercialized multiplex rRT-PCR kits for respiratory viruses: Implications for application and optimization. Microbiol. Spectr. 2024, 12, e01641-24. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, B.S.; Tantry, B.A.; Bandy, A.; Ahmad, R.; Khursheed, S.Q.; Ahmad, A.; Hakami, M.A.; Shah, N.N. Simultaneous detection of influenza A/B, respiratory syncytial virus, and SARS-CoV-2 in nasopharyngeal swabs by one-tube multiplex reverse transcription polymerase chain reaction. Trop. Med. Infect. Dis. 2023, 8, 326. [Google Scholar] [CrossRef]
- Kim, J.; Han, E.; Kim, J.; Kim, Y.J.; Bae, M.H. Comparative Evaluation of the PowerChek Respiratory Virus Panel RT-PCR Assay Detecting 16 Respiratory Viruses Including SARS-CoV-2. J. Med. Virol. 2025, 97, e70243. [Google Scholar] [CrossRef]
- Software, G. GraphPad Prism Version 10. Available online: https://www.graphpad.com/quickcalcs/ (accessed on 31 August 2025).
- Kim, T.Y.; Kim, J.-Y.; Shim, H.J.; Yun, S.A.; Jang, J.-H.; Huh, H.J.; Kim, J.-W.; Lee, N.Y. Performance evaluation of the PowerChek SARS-CoV-2, influenza A & B multiplex real-time PCR kit in comparison with the BioFire respiratory panel. Ann. Lab. Med. 2022, 42, 473–477. [Google Scholar] [PubMed]
- Kim, T.Y.; Kim, J.-Y.; Shim, H.J.; Yun, S.A.; Jang, J.-H.; Huh, H.J.; Kim, J.-W.; Lee, N.Y. Comparison of the PowerChek SARS-CoV-2, Influenza A&B, RSV Multiplex Real-time PCR Kit and BioFire Respiratory Panel 2.1 for simultaneous detection of SARS-CoV-2, influenza A and B, and respiratory syncytial virus. J. Virol. Methods 2021, 298, 114304. [Google Scholar]
- Suominen, J.; Loginov, R.; Kallio-Kokko, H. Comparative evaluation of STANDARD M10 Flu/RSV/SARS-CoV-2 and Savanna(R) Respiratory Viral Panel-4 assays for the rapid molecular diagnosis of influenza A/B virus, respiratory syncytial virus and SARS-CoV-2. J. Clin. Virol. 2025, 179, 105827. [Google Scholar] [CrossRef]
- Jensen, C.B.; Schneider, U.V.; Madsen, T.V.; Nielsen, X.C.; Ma, C.M.G.; Severinsen, J.K.; Hoegh, A.M.; Botnen, A.B.; Trebbien, R.; Lisby, J.G. Evaluation of the analytical and clinical performance of two RT-PCR based point-of-care tests; Cepheid Xpert(R) Xpress CoV-2/Flu/RSV plus and SD BioSensor STANDARD M10 Flu/RSV/SARS-CoV-2. J. Clin. Virol. 2024, 172, 105674. [Google Scholar] [CrossRef]
- Abdullah, A.; Sam, I.C.; Ong, Y.J.; Theo, C.H.; Pukhari, M.H.; Chan, Y.F. Comparative Evaluation of a Standard M10 Assay with Xpert Xpress for the Rapid Molecular Diagnosis of SARS-CoV-2, Influenza A/B Virus, and Respiratory Syncytial Virus. Diagnostics 2023, 13, 3507. [Google Scholar] [CrossRef]
- Lee, D.H.; Han, E.; Ryu, S.; Choi, H.J.; Kim, J.; Kim, J.; Bae, M.H. Clinical performance of the STANDARD M10 SARS-CoV-2 rapid RT-PCR assay in patients visiting an emergency department. J. Med. Virol. 2023, 95, e29330. [Google Scholar] [CrossRef]
- Kim, Y.K.; Song, S.H.; Ahn, B.; Lee, J.K.; Choi, J.H.; Choi, S.H.; Yun, K.W.; Choi, E.H. Shift in Clinical Epidemiology of Human Parainfluenza Virus Type 3 and Respiratory Syncytial Virus B Infections in Korean Children Before and During the COVID-19 Pandemic: A Multicenter Retrospective Study. J. Korean Med. Sci. 2022, 37, e215. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Rhee, J.E.; Kang, D.; Choi, E.H.; Lee, N.J.; Woo, S.; Lee, J.; Lee, S.W.; Kim, E.J.; Yun, K.W. Epidemiology of Respiratory Viruses in Korean Children Before and After the COVID-19 Pandemic: A Prospective Study From National Surveillance System. J. Korean Med. Sci. 2024, 39, e171. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.J.; Kim, J.-Y.; Kwon, H.J.; Yun, S.A.; Lee, M.-K.; Lee, N.Y.; Kim, J.-W.; Ki, C.-S. Performance evaluation of Allplex respiratory panels 1, 2, and 3 for detection of respiratory viruses and influenza A virus subtypes. J. Clin. Microbiol. 2017, 55, 479–484. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Phung, L.T.; Thanh Tran, H.T.; Ly, H.T.T.; Vo, A.H.M.; Dinh, N.P.; Doan, P.M.; Nguyen, A.T.; Dang, L.D.; Doan, T.T.; et al. Molecular subtypes of Adenovirus-associated acute respiratory infection outbreak in children in Northern Vietnam and risk factors of more severe cases. PLoS Negl. Trop. Dis. 2023, 17, e0011311. [Google Scholar] [CrossRef]
- Komiazyk, M.; Walory, J.; Kozinska, A.; Wasko, I.; Baraniak, A. Impact of the Nucleic Acid Extraction Method and the RT-qPCR Assay on SARS-CoV-2 Detection in Low-Viral Samples. Diagnostics 2021, 11, 2247. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Shin, S.; Kim, M.; Lee, Y.K.; Kang, H.J.; Kim, H.S.; Kim, J.-S.; Song, W.; Kim, H.-S. Evaluation of the Real-Q RV Detection Kit for the Identification of Viruses That Result in Respiratory Infections. Lab. Med. Online 2019, 9, 17–21. [Google Scholar] [CrossRef]
- BioMérieux. BIOFIRE® Respiratory Panel 2.1 Plus (RP2.1plus): Instructions for Use; BioMérieux: Salt Lake City, UT, USA, 2023; Volume BFR0000-8307-04. [Google Scholar]
- BioMérieux. 2023–2024 Influenza A Sequence Surveillance Assessment for BIOFIRE FILMARRAY and BIOFIRE SPOTFIRE Respiratory Solutions; BioMérieux: Salt Lake City, UT, USA, 2024. [Google Scholar]
- Lade, H.; Kim, J.-M.; Chung, Y.; Han, M.; Mo, E.-K.; Kim, J.-S. Comparative evaluation of allplex respiratory panels 1, 2, 3, and BioFire FilmArray respiratory panel for the detection of respiratory infections. Diagnostics 2021, 12, 9. [Google Scholar] [CrossRef] [PubMed]
| Specification | PowerChek™ RVP (Kogene, Seoul, Republic of Korea) | Allplex™ RP (Seegene, Seoul, Republic of Korea) | Biofire® RP 2.1plus (BioMérieux, Salt Lake City, UT, USA) |
|---|---|---|---|
| Main detection principle | Multiplex Real-time RT-PCR (TaqMan probe) | Multiplex Real-time RT-PCR (MuDT™) | Nested multiplex PCR with syndromic panel |
| Number of virus targets | 16 | 19 | 19 |
| Test number in one run | 22 specimens can be processed per run | 30 specimens can be processed per run | Single cartridge per test; 23 targets per test |
| Turnaround time (TAT) | 2 h 20 min | 3 h 10 min | 50 min |
| Primer information | Target-specific primers for key respiratory viruses | Multiplex primer sets for multiple viral targets | Preloaded primers in a closed system |
| Estimated cost per test | USD 30~40 | USD 40~50 | >USD 200 (Higher owing to single-use cartridge) |
| Hands-on time | 40 min: Sample prep and RNA extraction | 40 min: Sample prep and RNA extraction | 5 min |
| PCR run time | 100 min | 150 min | 45 min |
| Batch throughput | 22 samples | 30 samples | N/A |
| Internal control (IC) | GAPDH | Bacteriophage MS2 | RNA process control (Schizosaccharomyces pombe) Array PCR control |
| Others | Detection of 16 targets, including SARS-CoV-2 Pre-Mix type | Automated workflow available | Fully automated, minimal hands-on time, individually testable |
| BioFire® RP 2.1plus (BioMérieux) | Allplex™ RP (Seegene) | PowerChek™ RVP (Kogene) | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Target | Positive | Negative | Target | Positive | Negative | Target | Positive | Negative | |
| AdV | 20 | 0 | AdV | 20 | 0 | AdV | 19 | 1 | 20 |
| CoV-229E | 3 | 0 | CoV-229E | 3 | 0 | CoV-229E | 3 | 0 | 3 |
| CoV-NL63 | 10 | 0 | CoV-NL63 | 8 | 2 | CoV-NL63 | 9 | 1 | 10 |
| CoV-OC43 | 10 | 0 | CoV-OC43 | 10 | 0 | CoV-OC43 | 10 | 0 | 10 |
| Flu A-H3 | 10 | 0 | Flu A-H3 | 10 | 0 | Flu A | 20 | 2 | 10 |
| FluA-H1-2009 | 10 | 0 | Flu A-H1-2009 | 10 | 0 | 10 | |||
| Flu A, NS | 2 | 0 | Flu A, NS | 0 | 2 | 2 | |||
| Flu B | 20 | 0 | Flu B | 18 | 2 | Flu B | 19 | 1 | 20 |
| hMPV | 10 | 0 | hMPV | 8 | 2 | hMPV | 10 | 0 | 10 |
| PIV1 | 9 | 0 | PIV1 | 8 | 1 | PIV1 | 9 | 0 | 9 |
| PIV2 | 10 | 0 | PIV2 | 9 | 1 | PIV2 | 10 | 0 | 10 |
| PIV3 | 10 | 0 | PIV3 | 9 | 1 | PIV3 | 9 | 1 | 10 |
| PIV4 | 10 | 0 | PIV4 | 8 | 2 | PIV4 | 10 | 0 | 10 |
| HBoV | - | - | HBoV | 8 | 0 | HBoV | 8 | 0 | 8 |
| RSV | 30 | 0 | RSV A | 15 | 0 | RSV | 30 | 0 | 30 |
| RSV B | 14 | 1 | |||||||
| HRV/HEV | 30 | 0 | HRV | 15 | 0 | HRV/HEV | 30 | 0 | 30 |
| HEV | 15 | 0 | |||||||
| SARS-CoV-2 | 30 | 0 | SARS-CoV-2 | - | - | SARS-CoV-2 | 30 | 0 | 30 |
| Total No. | 224 | 0 | 188 | 14 | 226 | 6 | 232 | ||
| Pathogen | Relative Sensitivity | Relative Specificity | Relative Accuracy (*) | |||
|---|---|---|---|---|---|---|
| Target | PowerChek™ RVP | Allplex™ RP | PowerChek™ RVP | Allplex™ RP | PowerChek™ RVP | Allplex™ RP |
| AdV | 95.0% | 100% | 98.1% | 100% | 97.6% | 100% |
| CoV-229E | 100% | 100% | 100% | 100% | 100% | 100% |
| CoV-NL63 | 90% | 80.0% | 100% | 100% | 99.1% | 98.2% |
| CoV-OC43 | 100% | 100% | 100% | 100% | 100% | 100% |
| Flu A-H3 | 90.9% | 100% | 100% | 100% | 98.4% | 100% |
| FluA-H1-2009 | 100% | 100% | 100% | |||
| Flu A, NS | 0% | 100% | 98.1% | |||
| Flu B | 95.0% | 90.0% | 100% | 100% | 99.2% | 98.4% |
| hMPV | 100% | 80.0% | 100% | 100% | 100% | 98.2% |
| PIV1 | 100% | 88.9% | 99.0% | 100% | 99.1% | 99.1% |
| PIV2 | 100% | 90.0% | 100% | 100% | 100% | 99.1% |
| PIV3 | 90.0% | 90.0% | 100% | 100% | 99.1% | 99.1% |
| PIV4 | 100% | 80.0% | 99.0% | 100% | 99.1% | 98.2% |
| HBoV | - | - | - | - | - | - |
| RSV A | 100% | 100% | 100% | 100% | 100% | 100% |
| RSV B | 93.3% | 100% | 99.2% | |||
| HRV | 100% | 100% | 92.3% | 99.0% | 94.0% | 99.2% |
| HEV | 100% | 99.0% | 99.2% | |||
| SARS-CoV-2 | 100% | - | 100% | - | 100% | - |
| Overall | 97.4% | 93.1% | 88.5% | 97.1% | 94.6% | 94.4% |
| Pathogen | BioFire® RP 2.1plus vs. PowerChek™ RVP | BioFire® RP 2.1plus vs. Allplex™ RP | ||
|---|---|---|---|---|
| Kappa | 95% C.I. | Kappa | 95% C.I. | |
| AdV | 0.912 | 0.815–1.000 | 1.000 | 1.000–1.000 |
| CoV-229E | 1.000 | 1.000–1.000 | 1.000 | 1.000–1.000 |
| CoV-NL63 | 0.943 | 0.831–1.000 | 0.879 | 0.715–1.000 |
| CoV-OC43 | 1.000 | 1.000–1.000 | 1.000 | 1.000–1.000 |
| Flu A-H3 | 0.943 | 0.864–1.000 | 1.000 | 1.000–1.000 |
| FluA-H1-2009 | 1.000 | 1.000–1.000 | ||
| Flu A, NS | 0 | 0–0 | ||
| Flu B | 0.970 | 0.910–1.000 | 0.938 | 0.853–1.000 |
| hMPV | 1.000 | 1.000–1.000 | 0.879 | 0.715–1.000 |
| PIV1 | 0.943 | 0.831–1.000 | 0.936 | 0.813–1.000 |
| PIV2 | 1.000 | 1.000–1.000 | 0.943 | 0.831–1.000 |
| PIV3 | 0.943 | 0.831–1.000 | 0.943 | 0.831–1.000 |
| PIV4 | 0.948 | 0.845–1.000 | 0.879 | 0.715–1.000 |
| HBoV | - | - | - | - |
| RSV A | 1.000 | 1.000–1.000 | 1.000 | 1.000–1.000 |
| RSV B | 0.961 | 0.884–1.000 | ||
| HRV | 0.843 | 0.739–0.947 | 0.963 | 0.891–1.000 |
| HEV | 0.963 | 0.891–1.000 | ||
| SARS-CoV-2 | 1.000 | 1.000–1.000 | - | - |
| Overall | 0.873 | 0.816–0.930 | 0.879 | 0.824–0.935 |
| Specimen No. | Target Virus | BioFire®RP 2.1plus | PowerChek™ RVP | Allplex™ RP | Virus Sequencing | Interpretation | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Results | Results | Ct | IC (Ct) | Results | Ct | IC (Ct) | Target | Result | |||
| 8 | AdV | AdV | Neg | - | 21.31 | AdV | 37.4 | 28.74 | AdV | N/A | Unidentified |
| 28 | CoV-NL63 | CoV-NL63 | CoV-NL63 | 31.19 | 19.74 | Neg | - | 29.85 | CoV-NL63 | N/A | Unidentified |
| 29 | CoV-NL63 | CoV-NL63 | Neg | - | 20.49 | Neg | - | 29.48 | CoV-NL63 | N/A | Unidentified |
| 79 | Flu B | Flu B | Neg | - | 22.91 | Neg | - | 28.71 | Flu B | N/A | Unidentified |
| 83 | Flu B | Flu B | Flu B | 31.29 | 25.22 | Neg | - | 28.55 | Flu B | N/A | Unidentified |
| 90 | hMPV | hMPV | hMPV | 29.4 | 20.28 | Neg | - | 27.96 | hMPV | N/A | Unidentified |
| 92 | hMPV | hMPV | hMPV | 29.2 | 21.6 | Neg | - | 30.9 | hMPV | hMPV | hMPV |
| 100 | PIV1 | PIV1 | PIV1 | 18.96 | 25.75 | Neg | - | 29.03 | PIV1 | PIV1 | PIV1 |
| 103 | PIV2 | PIV2 | PIV2 | 31.18 | 21.44 | Neg | - | 30.32 | PIV1 | N/A | Unidentified |
| 122 | PIV3 | PIV3 | Neg | - | 20.35 | Neg | - | 29.55 | PIV3 | N/A | Unidentified |
| 125 | PIV4 | PIV4 | PIV4 | 26.33 | 21.34 | Neg | - | 30.36 | PIV4 | PIV4 | PIV4 |
| 132 | PIV4 | PIV4 | PIV4 | 19.57 | 23.73 | Neg | - | 34.46 | PIV4 | PIV4 | PIV4 |
| 141 | Flu A,NS | Flu A | Neg | - | 26.46 | Neg | - | 32.57 | Flu A | N/A | Unidentified |
| 142 | Flu A, NS | Flu A | Neg | - | 21.06 | Neg | - | 31 | Flu A | N/A | Unidentified |
| 179 | RSV | RSV | RSV | 32.17 | 18.58 | Neg | - | 29.99 | RSV | N/A | Unidentified |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Akter, R.; Lee, J.-H.; Ryu, S.W. Evaluation of the PowerChek™ Respiratory Virus Panel 1/2/3/4 for the Detection of 16 Respiratory Viruses: A Comparative Study with the Allplex™ Respiratory Panel Assay 1/2/3 and BioFire® Respiratory Panel 2.1 plus. Diagnostics 2025, 15, 2713. https://doi.org/10.3390/diagnostics15212713
Lee H, Akter R, Lee J-H, Ryu SW. Evaluation of the PowerChek™ Respiratory Virus Panel 1/2/3/4 for the Detection of 16 Respiratory Viruses: A Comparative Study with the Allplex™ Respiratory Panel Assay 1/2/3 and BioFire® Respiratory Panel 2.1 plus. Diagnostics. 2025; 15(21):2713. https://doi.org/10.3390/diagnostics15212713
Chicago/Turabian StyleLee, Hyeongyu, Rokeya Akter, Jong-Han Lee, and Sook Won Ryu. 2025. "Evaluation of the PowerChek™ Respiratory Virus Panel 1/2/3/4 for the Detection of 16 Respiratory Viruses: A Comparative Study with the Allplex™ Respiratory Panel Assay 1/2/3 and BioFire® Respiratory Panel 2.1 plus" Diagnostics 15, no. 21: 2713. https://doi.org/10.3390/diagnostics15212713
APA StyleLee, H., Akter, R., Lee, J.-H., & Ryu, S. W. (2025). Evaluation of the PowerChek™ Respiratory Virus Panel 1/2/3/4 for the Detection of 16 Respiratory Viruses: A Comparative Study with the Allplex™ Respiratory Panel Assay 1/2/3 and BioFire® Respiratory Panel 2.1 plus. Diagnostics, 15(21), 2713. https://doi.org/10.3390/diagnostics15212713







