From Lab to Clinic: Artificial Intelligence with Spectroscopic Liquid Biopsies
Abstract
1. Background
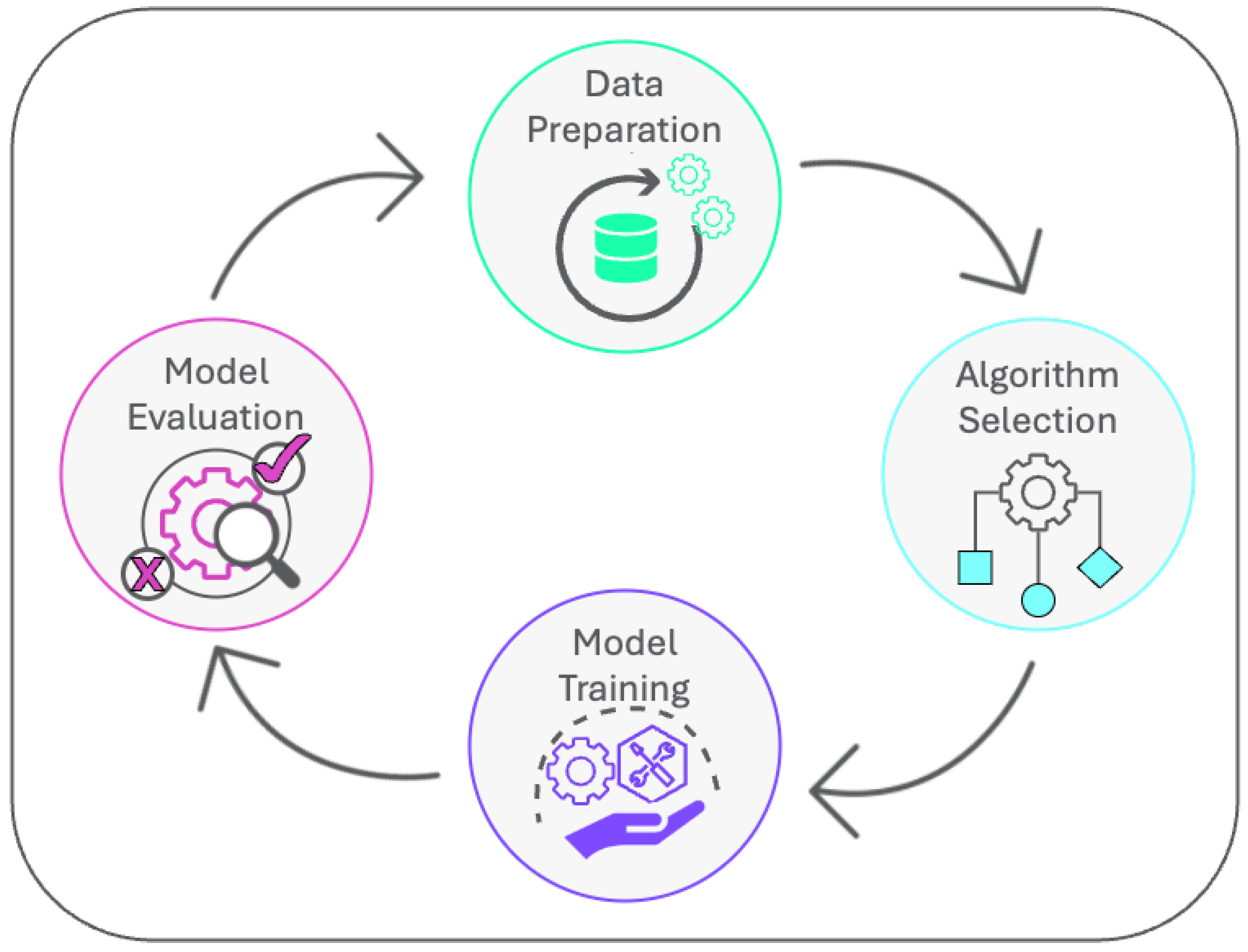
2. Model Development
2.1. Data Preparation
2.2. Algorithm Selection
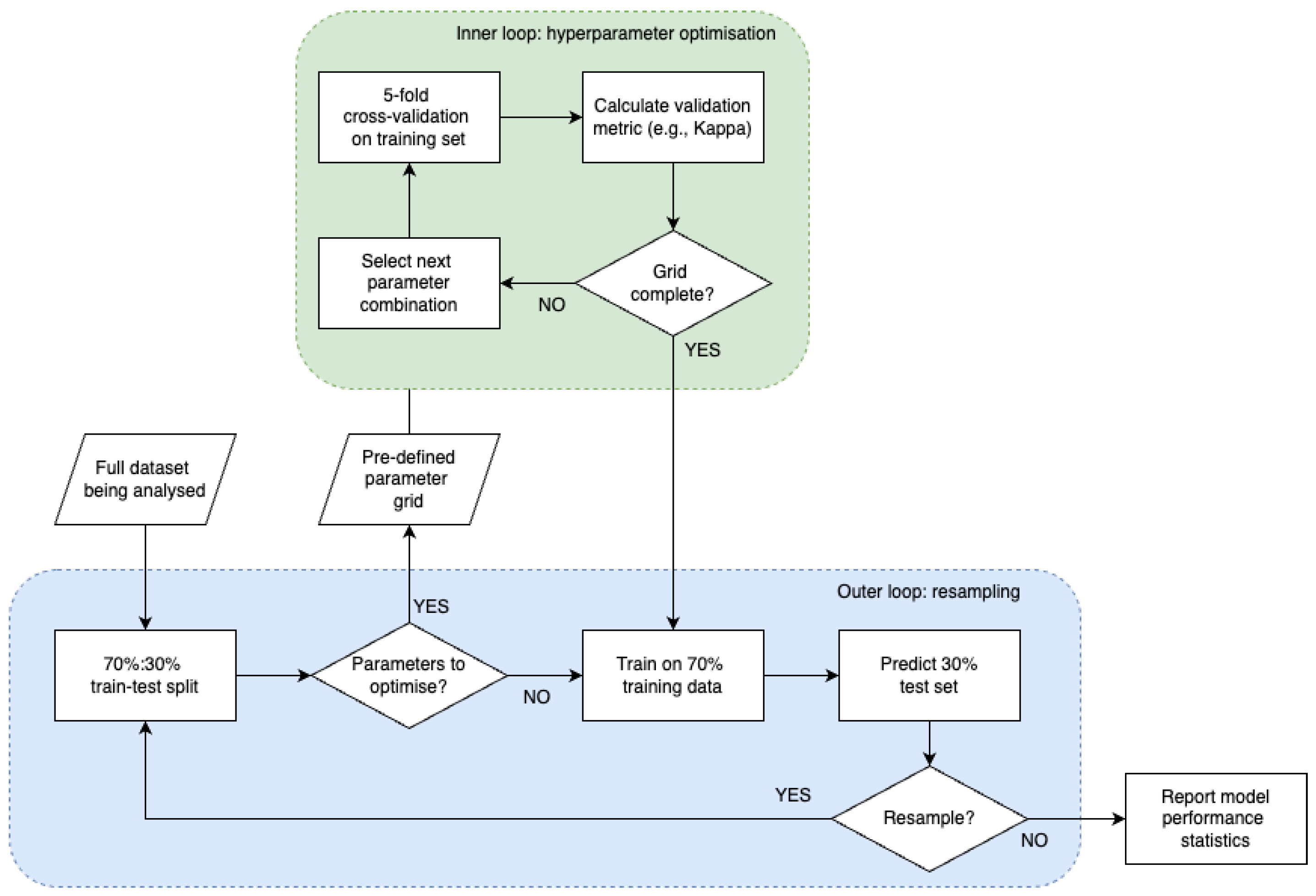
2.3. Model Training
2.4. Model Evaluation
2.4.1. Performance Metrics
2.4.2. Challenges in Model Performance Evaluation
3. Challenges and Opportunities of Applying ML and AI to Spectroscopic Liquid Biopsies
3.1. Challenges
3.2. Opportunities
4. From Lab to Clinic
4.1. Proof-of-Concept Studies
4.2. Clinical Validation
5. Regulation
5.1. Current Regulations for Liquid Biopsies
5.2. Current Regulations for AI/ML Medical Devices
5.3. Currently Approved Liquid Biopsies
6. Future Requirements and Predictions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ATR | Attenuated Total Reflectance |
| AUC | Area Under the Curve |
| cfDNA | Cell Free DNA |
| CLIA | Clinical Laboratory Improvement Amendments |
| CT | Computed Tomography |
| ctDNA | Circulating Tumour DNA |
| CV | Cross-Validation |
| DL | Deep Learning |
| EMA | European Medicines Agency |
| EMSC | Extended Multiplicative Signal Correction |
| FDA | Food and Drug Administration |
| FN | False Negative |
| FP | False Positive |
| FTIR | Fourier Transform Infrared |
| GPU | Graphical Processing Units |
| IMDRF | International Medical Device Regulators Forum |
| IR | Infrared |
| IVD | In Vitro Diagnostic |
| IVDD | In Vitro Diagnostic Medical Devices Directive |
| IVDR | In Vitro Diagnostics Regulation |
| LDT | Laboratory Developed Test |
| MCED | Multicancer Early Detection |
| MDA | Medical Device Amendments |
| MHRA | Medicines and Healthcare products Regulatory Agency |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| NN | Neural Network |
| NPV | Negative Predictive Value |
| PLS | Partial Least Squares |
| PMA | Premarket Approval |
| PPV | Positive Predictive Value |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic |
| SaMD | Software as a Medical Device |
| TN | True Negative |
| TP | True Positive |
| UKCA | UK Conformity Assessed |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, 9040. [Google Scholar] [CrossRef]
- McPhail, S.; Johnson, S.; Greenberg, D.; Peake, M.; Rous, B. Stage at diagnosis and early mortality from cancer in England. Br. J. Cancer 2015, 112, S108–S115. [Google Scholar] [CrossRef]
- Sattlecker, M.; Stone, N.; Bessant, C. Current trends in machine-learning methods applied to spectroscopic cancer diagnosis. Trends Anal. Chem. 2014, 59, 17–25. [Google Scholar] [CrossRef]
- Bleyer, A.; Welch, H.G. Effect of three decades of screening mammography on breast-cancer incidence. N. Engl. J. Med. 2015, 367, 1998–2005. [Google Scholar] [CrossRef]
- Perkins, A.C.; Skinner, E.N. A review of the current cervcal cancer screening guidelines. N. Carol. Med. J. 2016, 77, 420–422. [Google Scholar]
- Huang, K.L.; Wang, S.Y.; Lu, W.C.; Chang, Y.H.; Su, J.; Lu, Y.T. Effects of low-dose computed tomography on lung cancer screening: A systematic review, meta-analysis, and trial sequential analysis. BMC Pulm. Med. 2019, 19, 126. [Google Scholar] [CrossRef] [PubMed]
- Tonini, V.; Zanni, M. Early diagnosis of pancreatic cancer: What strategies to avoid a foretold catastrophe. World J. Gastroenterol. 2022, 28, 4235–4248. [Google Scholar] [CrossRef]
- Nishihara, R.; Wu, K.; Lochhead, P.; Morikawa, T.; Liao, X.; Qian, Z.R.; Inamura, K.; Kim, S.A.; Kuchiba, A.; Yamauchi, M.; et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl. J. Med. 2013, 369, 1095. [Google Scholar] [CrossRef]
- Menna, G.; Guerrato, G.P.; Bilgin, L.; Ceccarelli, G.M.; Olivi, A.; Pepa, G.M.D. Is there a role for machine learning in liquid biopsy for brain tumors? A systematic review. Int. J. Med. Sci. 2023, 24, 9723. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspetcives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef]
- Adashek, J.J.; Janku, F.; Kurzrok, R. Signed in blood: Circulating tumor DNA in cancer diagnosis, treatment and screening. Cancers 2021, 13, 3600. [Google Scholar] [CrossRef]
- Moser, T.; Kuhberger, S.; Lazzeri, I.; Vlachos, G.; Heitzer, E. Bridging biological cfDNA features and machine learning approaches. Trends Genet. 2023, 39, 285–307. [Google Scholar] [CrossRef]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and prognostic updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a biomarker for pancreatic cancer—A comprehensive review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Wild, N.; Andres, H.; Rollinger, W.; Krause, F.; Dilba, P.; Tacke, M.; Karl, J. A Combination of Serum Markers for the Early Detection of Colorectal Cancer. Clin. Cancer Res. 2010, 16, 6111–6121. [Google Scholar] [CrossRef] [PubMed]
- Salehi, R.; Atapour, N.; Vatandoust, N.; Farahani, N.; Ahangari, F.; Salehi, A.R. Methylation pattern of ALX4 gene promoter as a potential biomaerker for blood-based early detection of colorectal cancer. Adv. Biomed. Res. 2015, 4, 252. [Google Scholar] [CrossRef] [PubMed]
- Traverso, G.; Shuber, A.; Levin, B.; Johnson, C.; Olsson, L.; Schoetz, D.J.; Hamilton, S.R.; Boynton, K.; Kinzler, K.W.; Vogelstein, B. Detection of APC Mutations in Fecal DNA from Patients with Colorectal Tumors. N. Engl. J. Med. 2002, 346, 311–315. [Google Scholar] [CrossRef]
- Yanqing, H.; Cheng, D.; Ling, X. Serum CA72-4 as a biomarker in the diagnosis of colorectal cancer: A meta-analysis. Open Med. 2018, 13, 164–171. [Google Scholar] [CrossRef]
- Cao, H.; Zhu, L.; Li, L.; Wang, W.; Niu, X. Serum CA724 has no diagnostic value for gastorintestinal tumors. Clin. Exp. Med. 2023, 23, 2433–2442. [Google Scholar] [CrossRef] [PubMed]
- Lakemeyer, L.; Sander, S.; Wittau, M.; Henne-Bruns, D.; Kornmann, M.; Lemke, J. Diagnostic and Prognostic Value of CEA and CA19-9 in Colorectal Cancer. Diseases 2021, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhou, Y.; Sheng, S.; Qian, S.Y.; Huo, X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci. Rep. 2018, 8, 2732. [Google Scholar] [CrossRef]
- Thomas, D.S.; Fourkala, E.O.; Apostolidou, S.; Gunu, R.; Ryan, A.; Jacobs, I.; Menon, U.; Alderton, W.; Gentry-Maharaj, A.; Timms, J.F. Evaluation of serum CEA, CYFRA21-1 and CA125 for the early detection of colorectal cancer using longitudinal preclinical samples. Br. J. Cancer 2015, 113, 268–274. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; He, L.; Li, D.; Wang, D.; Wang, M.; Wang, X. Diagnostic value of carcinoembryonic antigen combined with cytokines in seurm of patients with colorectal cancer. Medicine 2022, 101, 37. [Google Scholar] [CrossRef]
- Xie, W.; Xie, L.; Song, X. The diagnostic accuracy of circulating free DNA for the detection of KRAS mutation status in colorectal cancer: A meta-analysis. Cancer Med. 2019, 8, 1218–1231. [Google Scholar] [CrossRef] [PubMed]
- Bu, F.; Cao, S.; Deng, X.; Zhang, Z.; Feng, X. Evaluation of C-reactive protein and fibrinogen in comparison to CEA and CA72-4 as diagnostic biomarkers for colorectal cancer. Heliyon 2023, 9, e16092. [Google Scholar] [CrossRef]
- Koper-Lenkiewicz, O.M.; Dymicka-Piekarska, V.; Milewska, A.J.; Zińczuk, J.; Kamińska, J. The Relationship between Inflammation Markers (CRP, IL-6, sCD40L) and Colorectal Cancer Stage, Grade, Size and Location. Diagnostics 2021, 11, 1382. [Google Scholar] [CrossRef]
- Erlinger, T.P.; Platz, E.A.; Rifai, N.; Helzlsouer, K.J. C-Reactive Protein and the Risk of Incident Colorectal Cancer. JAMA 2004, 291, 585–590. [Google Scholar] [CrossRef]
- Attoye, B.; Baker, M.J.; Thomson, F.; Pou, C.; Corrigan, D.K. Optimisation of an Electrochemical DNA Sensor for Measuring KRAS G12D and G13D Point Mutations in Different Tumour Types. Biosensors 2021, 11, 42. [Google Scholar] [CrossRef]
- Santiago, L.; Castro, M.; Sanz-Pamplona, R.; Garzón, M.; Ramirez-Labrada, A.; Tapia, E.; Moreno, V.; Layunta, E.; Gil-Gómez, G.; Garrido, M.; et al. Extracellular Granzyme A Promotes Colorectal Cancer Development by Enhancing Gut Inflammation. Cell Rep. 2020, 32, 107847. [Google Scholar] [CrossRef]
- Paczek, S.; Łukaszewicz Zajac, M.; Mroczko, B. Granzymes—Their Role in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 5277. [Google Scholar] [CrossRef]
- Kakourou, A.; Koutsioumpa, C.; Lopez, D.S.; Hoffman-Bolton, J.; Bradwin, G.; Rifai, N.; Helzlsouer, K.J.; Platz, E.A.; Tsilidis, K.K. Interleukin-6 and risk of colorectal cancer: Results from the CLUE II cohort and a meta-analysis of prospective studies. Cancer Causes Control 2015, 26, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Herbst, A.; Rahmig, K.; Stieber, P.; Philipp, A.; Jung, A.; Ofner, A.; Crispin, A.; Neumann, J.; Lamerz, R.; Kolligs, F.T. MEthylation of NEUROG1 in Serum Is a Sensitive Marker for the Detection of Early Colorectal Cancer. Am. J. Gastroenterol. 2011, 106, 1110–1118. [Google Scholar] [CrossRef]
- Cai, Y.; Rattray, N.J.W.; Zhang, Q.; Mironova, V.; Santos-Neto, A.; Muca, E.; Vollmar, A.K.R.; Hsu, K.S.; Rattray, Z.; Cross, J.R.; et al. Tumor Tissue-Specific Biomarkers of Colorectal Cancer by Anatomic Location and Stage. Metabolites 2020, 10, 257. [Google Scholar] [CrossRef]
- Li, X.L.; Zhou, J.; Chen, Z.R.; Chng, W.J. p53 mutations in colorectal cancer—Molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015, 21, 84–93. [Google Scholar] [CrossRef]
- Ma, L.; Qin, G.; Fai, F.; Jiang, Y.; Huang, Z.; Yang, H.; Yao, S.; Du, S.; Cao, Y. A novel method of early detection of colorectal cancer based on detection of methylation of two fragments of syndecan-2 (SDC2) in stool DNA. BMC Gastroenterol. 2022, 22, 191. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef]
- Li, Y.; Li, B.; Jiang, R.; Liao, L.; Zheng, C.; Yuan, J.; Zeng, L.; Hu, K.; Zhang, Y.; Mei, W.; et al. A novel screening method of DNA methylation biomarkers helps to improve the detection of colorectal cancer and precancerous lesions. Cancer Med. 2023, 12, 20626–20638. [Google Scholar] [CrossRef]
- Meng, R.; Wang, Y.; He, L.; He, Y.; Du, Z. Potential diagnostic value of serum p53 antibody for detecting colorectal cancer: A meta-analysis. Oncol. Lett. 2018, 15, 5489–5496. [Google Scholar] [CrossRef] [PubMed]
- Dymicka-Piekarska, V.; Korniluk, A.; Gryko, M.; Siergiejko, E.; Kemona, H. Potential role of soluble CD40 ligand as inflammatory biomarker in colorectal cancer patients. Int. J. Biol. Markers 2014, 29, e261–e267. [Google Scholar] [CrossRef]
- Christensen, I.J.; Brünner, N.; Dowell, B.; Davis, G.; Nielsen, H.J.; Newstead, G.; King, D. Plasma TIMP-1 and CEA as Markers for Detection of Primary Colorectal Cancer: A Prospective Validation Study Including Symptomatic and Non-symptomatic Individuals. Anticancer. Res. 2015, 35, 4935–4942. [Google Scholar] [PubMed]
- Meng, C.; Yin, X.; Liu, J.; Tang, K.; Tang, H.; Liao, J. TIMP-1 is a novel serum biomarker for the diagnosis of colorectal cancer: A meta-analysis. PLoS ONE 2018, 13, e0207039. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Baldassano, S.N.; Loh, P.L.; Kording, K.; Litt, B.; Issadore, D. Machine learning to detect signatures of disease in liquid biopsies—A user’s guide. Lab Chip 2018, 18, 395–405. [Google Scholar] [CrossRef]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef]
- Mokari, A.; Guo, S.; Blocklitz, T. Exploring the steps of infrared (IR) spectral analysis: Pre-processing, (classical) data modelling, and deep learning. Molecules 2023, 28, 6886. [Google Scholar] [CrossRef]
- Ollesch, J.; Drees, S.L.; Heise, H.M.; Behrens, T.; Brüning, T.; Gerwert, K. FTIR spectroscopy of biofluids revisited: An automated approach to spectral biomarker identification. Analyst 2013, 138, 4092–4102. [Google Scholar] [CrossRef]
- Cameron, J.M.; Brennan, P.M.; Antoniou, G.; Butler, H.J.; Christie, L.; Conn, J.J.A.; Curran, T.; Gray, E.; Hegarty, M.G.; Jenkinson, M.D.; et al. Clinical validation of a spectroscopic liquid biopsy for earlier detection of brain cancer. Neuro-Oncol. Adv. 2022, 4, vdac024. [Google Scholar] [CrossRef]
- Gajjar, K.; Trevisan, J.; Owens, G.; Keating, P.J.; Wood, N.J.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Fourier Transform infrared spectroscopy coupled with a classification machine for the analysis of blood plasma or serum: A novel diagnostic approach for ovarian cancer. Analyst 2013, 138, 3917–3926. [Google Scholar] [CrossRef]
- Dong, L.; Sun, X.; Chao, Z.; Zhang, S.; Zheng, J.; Gurung, R.; Du, J.; Shi, J.; Xu, Y.; Zhang, Y.; et al. Evaluation of FTIR spectroscopy as diagnostic tool for colorectal cancer using spectral analysis. Spectroschima Acta Part A Mol. Biomol. Spectrosc. 2014, 122, 288–294. [Google Scholar] [CrossRef]
- Lewis, P.D.; Lewis, K.E.; Ghosal, R.; Bayliss, S.; Lloyd, A.J.; Wills, J.; Godfrey, R.; Kloer, P.; Mur, L.A.J. Evaluation of FTIR spectroscopy as a diagnostic tool for lung cancer using sputum. BMC Cancer 2010, 10, 640. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef]
- Cameron, J.M.; Bruno, C.; Parachalil, D.R.; Baker, M.J.; Bonnier, F.; Butler, H.J.; Byrne, H.J. Chapter 10- Vibrational spectroscopic analysis and quanitification of proteins in human blood plasma and serum. Vib. Spectrosc. Protein Res. 2020, 10, 269–314. [Google Scholar]
- Sala, A.; Cameron, J.M.; Jenkins, C.A.; Barr, H.; Christie, L.; Conn, J.J.A.; Evans, T.R.J.; Harris, D.A.; Palmer, D.S.; Rinaldi, C.; et al. Liquid biopsy for pancreatic cancer detection using infrared spectroscopy. Cancers 2022, 14, 3048. [Google Scholar] [CrossRef]
- Sala, A.; Cameron, J.M.; Brennan, P.M.; Crosbie, E.J.; Curran, T.; Gray, E.; Martin-Hirsch, P.; Palmer, D.S.; Rehman, I.U.; Rattray, N.J.W.; et al. Global serum profiling: An opportunity for earlier cancer detection. J. Exp. Clin. Cancer Res. 2023, 42, 207. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Zawaski, J.A.; Francis, K.N.; Qutub, A.A.; Gaber, M.W. Image-based classification of tumor type and growth rate using machine learning: A preclinical study. Sci. Rep. 2019, 9, 12529. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Cheng, M.; Tao, Y.; Wang, Y.; Juengpanich, S.; Jiang, Z.; Jiang, Y.; Yan, Y.; Lu, W.; Lue, J.; et al. Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front. Oncol. 2020, 10, 680. [Google Scholar] [CrossRef]
- Hu, Q.; Whitney, H.M.; Giger, M.L. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci. Rep. 2020, 10, 10536. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zheng, B.; Qian, W. Computer aided lung cancer diagnosis with deep learning algorithms. Proc. SPIE 2016, 9785, 8. [Google Scholar]
- Chamberlin, J.; Kocher, M.R.; Waltz, J.; Snoddy, M.; Stringer, N.F.C.; Stephenson, J.; Sahbaee, P.; Sharma, P.; Rapaka, S.; Schoepf, U.J.; et al. Automated detection of lung nodules and coronary artery calcium using artificial intelligence on low-dose CT scans for lung cancer screening: Accuracy and prognostic value. BMC Med. 2021, 19, 55. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Gige, R.M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Ayer, T.; Alagoz, O.; Chhatwal, J.; Shavlik, J.W.; Kahn, C.E.; Burnside, E.S. Breast cancer risk estimation with artificial neural networks revisited: Discrimination and calibration. Cancer 2010, 116, 3310–3321. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, J.; Niu, J.; Li, D. New convolutional neural network model for screening and diagnosis of mammograms. PLoS ONE 2020, 15, e0237674. [Google Scholar] [CrossRef]
- Cameron, J.M.; Sala, A.; Antoniou, G.; Brennan, P.M.; Butler, H.J.; Conn, J.J.A.; Connal, S.; Curran, T.; Hegarty, M.G.; McHardy, R.G.; et al. A spectroscopic liquid biopsy for the earlier detection of multiple cancer types. Br. J. Cancer 2023, 129, 1658–1666. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Petinrin, O.O.; Zhang, W.; Rahamna, S.; Tang, Z.R.; Wong, K.C. Machine learning protocols in early cancer detection based on liquid biopsy: A survey. Life 2021, 11, 638. [Google Scholar] [CrossRef]
- Fadlemoula, A.; Catarino, S.O.; Minas, G.; Carvalho, V. A review of machine learning methods recently applied to FTIR spectroscopy dtaa for the analysis of human blood cells. Micromachines 2023, 16, 1145. [Google Scholar] [CrossRef] [PubMed]
- Remirez, C.A.M.; Greenop, M.; Ashton, L.; ur Rehman, I. Applications of machine learning in spectroscopy. Appl. Spectrosc. Rev. 2021, 56, 733–763. [Google Scholar] [CrossRef]
- Santos, M.C.D.; Morais, C.L.M.; Lima, K.M.G.; Martin, F.L. Chapter 11—Vibrational Spectroscopy in Protein Research Toward Virus Identification: Challenges, New Research, and Future Perspectives. In Vibrational Spectroscopy in Protein Research; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Butler, H.J.; Smith, B.R.; Fritzsch, R.; Radhakrishnan, P.; Palmer, D.S.; Baker, M.J. Optimised spectral pre-processing for discrimination of biofluids via ATR-FTIR spectroscopy. Analyst 2018, 143, 6121–6134. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.R.; Baker, M.J.; Palmer, D.S. PRFFECT: A versatile tool for spectroscopists. Chemom. Intell. Lab. Syst. 2018, 172, 33–42. [Google Scholar] [CrossRef]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated method for subtraction of fluorescence from biological raman spectra. Appl. Spectrosc. 2003, 57, 1363–1367. [Google Scholar] [CrossRef]
- Daubechies, I. Orthonormal bases of compactly supported wavelets. Commun. Pure Appl. Math. 1988, 41, 909–996. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Raulf, A.P.; Butke, J.; Menzen, L.; Küpper, C.; Groberüschkamp, F.; Gerwert, K.; Mosig, A. A representation learning approach for recovering scatter-corrected spectra from Fourier-transform infrared spectra of tissue samples. J. Biophotonics 2021, 14, e202000385. [Google Scholar] [CrossRef]
- Guo, S.; Mayerhöfer, T.; Pahlow, S.; Hübner, U.; Popp, J.; Bocklitz, T. Deep learning for artefact removal in infrared spectroscopy. Analyst 2020, 145, 5213–5220. [Google Scholar] [CrossRef]
- Donalek, C. Supervised and Unsupervised Learning; Caltech Astronomy: Pasadena, CA, USA, 2011. [Google Scholar]
- Baştanlar, Y.; Özuysal, M. Introduction to Machine Learning. miRNomics MicroRNA Biol. Comput. Anal. 2013, 1107, 105–128. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Lavecchia, A. Deep learning in drug discovery: Opportunities, challenges and future prospects. Drug Discov. Today 2019, 24, 2017–2032. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Nielsen, M.A. Neural Networks and Deep Learning; Determination Press: Los Angeles, CA, USA, 2015. [Google Scholar]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Durg Discov. Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Khan, E.B.M.; Bashier, M.B. Machine Learning Algorithms and Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Akobeng, A.K. Understanding diagnostic tests 2: Likelihood ratios, pre- and post-test probabilities and their use in clinical practice. Acta Paediatr. 2007, 96, 487–491. [Google Scholar] [CrossRef]
- Platt, J. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods. Adv. Large Margin Classif. 1999, 10, 61–74. [Google Scholar]
- Haijian-Tilaki, K. Receiver Operating Characteristic (ROC) curve analysis for medical diagnostic test evalutation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Park, S.H.; Goo, J.M.; Jo, C.H. Receiver Operating Characteristic (ROC) Curve: A Practical Review for Radiologists. Korean J. Radiol. 2004, 5, 11–18. [Google Scholar] [CrossRef]
- Varoquaux, G.; Colliot, O. Evaluating machine learning models and their diagnostic value. Mach. Learn. Brain Disord. 2023, 197, 601–630. [Google Scholar]
- Lammers, D.; Eckert, C.M.; Ahmad, M.A.; Bingham, J.R.; Eckert, M.J. A surgeon’s guide to machine learning. Ann. Surg. Open 2021, 2, e091. [Google Scholar] [CrossRef]
- Chua, M.; Kim, D.; Choi, J.; Lee, N.G.; Deshpande, V.; Schwab, J.; Lev, M.H.; Gonzalez, R.G.; Gee, M.S.; Do, S. Tackling prediction uncertainty in machine learning for healthcare. Nat. Biomed. Eng. 2023, 7, 711–718. [Google Scholar] [CrossRef]
- Bender, A.; Schneider, N.; Segler, M.; Walters, W.P.; Engkvist, O.; Rodrigues, T. Evaluation guidelines for machine learning tools in the chemical sciences. Nat. Rev. Chem. 2022, 6, 428–442. [Google Scholar] [CrossRef]
- McDermott, M.B.A.; Wang, S.; Marinesk, N.; Ranganath, R.; Foschini, L.; Ghassemi, M. Reproducibility in machine learning for health research: Still a ways to go. Sci. Transl. Med. 2021, 13, eabb1655. [Google Scholar] [CrossRef]
- Rajula, H.S.R.; Varlato, G.; Manchia, M.; Antonucci, N.; Fanos, V. Comparison of conventional statistical methods with machine learning in medicine: Diagnosis, drug development, and treatment. Medicina 2020, 56, 455. [Google Scholar] [CrossRef]
- Dinsdale, N.K.; Bluemke, E.; Sundaresan, V.; Jenkinson, M.; Smith, S.M.; Namburete, A.I.L. Challenges for machine learning in lcinical translation of big data imaging studies. Neuron 2022, 110, 3866–3881. [Google Scholar] [CrossRef]
- Singh, S.; Maurya, M.K.; Singh, N.P.; Kumar, R. Survey of AI-driven techniques for ovarian cancer detection: State-of-the-art methods and open challenges. Netw. Model. Anal. Health Inform. Bioinform. 2024, 13, 56. [Google Scholar] [CrossRef]
- Sarkiss, C.A.; Germano, I.M. Machine learning in neuro-oncology: Can data analysis from 5346 patients change decision-making paradigms? World Neurosurg. 2019, 124, 286–294. [Google Scholar] [CrossRef]
- Hunter, B.; Hindocha, S.; Lee, R.W. The role of artificial intelligence in early cancer diagnosis. Cancers 2022, 14, 1524. [Google Scholar] [CrossRef] [PubMed]
- Painuli, D.; Bhardwaj, S.; Köse, U. Recent advancement in cancer diagnosis using machine learning and deep learning techniques: A comprehensive review. Comput. Biol. Med. 2022, 146, 105580. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Deep learning approaches fro data augmentation and classification of breast masses using ultrasound images. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 618–627. [Google Scholar]
- Gao, X.; Wang, X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: A preliminary study. Diagn. Interv. Imaging 2020, 101, 91–100. [Google Scholar] [CrossRef]
- Wickramaratne, S.d.; Mahmud, S. Conditional-GAN based data augmentation for deep learning task classifier improvment using fNIRs data. Front. Big Data 2021, 4, 659146. [Google Scholar] [CrossRef]
- Nagasawa, T.; Sato, T.; Nambu, I.; Wada, Y. fNIRs-GANs: Data augmentation using generative adversarial networks for classifying motor tasks from functinal near-infrared spectroscopy. J. Neural Eng. 2020, 17, 016068–016078. [Google Scholar] [CrossRef]
- McHardy, R.G.; Antoniou, G.; Conn, J.J.A.; Baker, M.J.; Palmer, D.S. Augmentation of FTIR spectral datasets using Wasserstein generative adversarial networks for cancer liquid biopsies. Analyst 2023, 148, 3860–3869. [Google Scholar] [CrossRef]
- Sala, A.; Anderson, D.J.; Brennan, P.M.; Butler, H.J.; Cameron, J.M.; Jenkinson, M.D.; Rinaldi, C.; Theakstone, A.G.; Baker, M.J. Biofluid diagnostics by FTIR spectroscopy: A platform technology for cancer detection. Cancer Lett. 2020, 477, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Noor, J.; Chaudhry, A.; Noor, R.; Batool, S. Advancements and applications of liquid biopsies in oncology: A narrative review. Cureus 2023, 15, e42731. [Google Scholar] [CrossRef]
- Bratulic, S.; Gatto, F.; Nielsen, J. The translational status of cancer liquid biopsies. Regen. Eng. Transl. Med. 2021, 7, 312–352. [Google Scholar] [CrossRef]
- Crosby, D. Delivering on the promise of early detection with liquid biopsies. Br. J. Cancer 2022, 126, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Richard-Kortum, R.; Lorenzoni, C.; Bagnato, V.S.; Schmeler, K. Optical imaging for screening and early cancer diagnosis in low-resource settings. Nat. Rev. Bioeng. 2023, 2, 25–43. [Google Scholar] [CrossRef]
- Castillo, J.M.; Arif, M.; Niessen, W.J.; Schoots, I.G.; Veenland, J.F. Automated classification of significant prostate cancer on MRI: A systematic review on the performance of machine learning applications. Cancers 2020, 12, 1606. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Boukovala, M.; Westphalen, C.B.; Probst, V. Liquid biopsy into the clinics: Current evidence and future perspectives. J. Liq. Biopsy 2024, 4, 100146. [Google Scholar] [CrossRef]
- Genzen, J.R.; Mohlman, J.S.; Lynch, J.L.; Squires, M.W.; Weiss, R.L. Laboratory-developed test: A legislative and regulatory review. Clin. Chem. 2017, 63, 1575–1584. [Google Scholar] [CrossRef]
- Zhao, Z.; Fan, J.; Hsu, Y.M.S.; Lyon, C.J.; Ning, B.; Hu, T.Y. Extracellular vesicles as cancer liquid biopsies: From discovery, validation, to clinical application. Lab Chip 2019, 19, 1114. [Google Scholar] [CrossRef] [PubMed]
- Strotman, L.N.; Millner, L.M.; Valdes, R., Jr.; Linder, M.W. Liquid biopsies in oncology and the current regulatory landscape. Mol. Diagn. Ther. 2016, 20, 429–436. [Google Scholar] [CrossRef] [PubMed]
- The Public Health Evidence for FDA Oversight of Laboratory Developed Tests: 20 Case Studies. Office of Public Health and Strategy and Analysis, Office of Commissioner, Food and Drug Administration. 16 November 2015. Available online: http://wayback.archive-it.org/7993/20171114205911/https:/www.fda.gov/AboutFDA/ReportsManualsForms/Reports/ucm472773.htm (accessed on 21 March 2024).
- Breakthrough Devices Program. Available online: https://www.fda.gov/medical-devices/how-study-and-market-your-device/breakthrough-devices-program (accessed on 12 April 2024).
- Holmes, D.R.; Farb, A.; Dib, N.; Jacques, L.; Rowe, S.; DeMaria, A.; King, S.; Zuckerman, B. The medical device development ecosystem: Current regulatory state and challenges for future development: A review. Cardiovasc. Revascularization Med. 2024, 60, 95–101. [Google Scholar] [CrossRef]
- Verbaanderd, C.; Jimeno, A.T.; Engelbergs, J.; Zander, H.; Reischl, I.; Oliver, A.M.; Vamvakas, S.; Vleminckx, C.; Bouygues, C.; Girard, T.; et al. Biomarker-driven developments in the context of the new regulatory framework for companion diagnostics in the European Union. Clin. Pharmacol. Ther. 2023, 114, 316–324. [Google Scholar] [CrossRef]
- Ritzhaupt, A.; Hayes, I.; Ehmann, F. Implementing the EU in vitro diagnostic regulation—A European regulatory perspective on companion diagnostics. Expert Rev. Mol. Diagn. 2020, 20, 565–567. [Google Scholar] [CrossRef]
- Valla, V.; Alzabin, S.; Koukoura, A.; Lewis, A.; Nielsen, A.A.; Vassiliadis, E. Companion diagnostics: State of the art and new regulations. Biomark. Insights 2021, 16, 11772719211047763. [Google Scholar] [CrossRef]
- Fraga-García, M.; Taléns-Visconti, R.; Nácher-Alonso, A.; Díez-Sales, O. New scenario in the field of medical devices in the European Union: Switzerland and the United Kingdom become third countries. Farm. Hosp. 2022, 46, 244–250. [Google Scholar] [PubMed]
- Marshall, J.; Morrill, K.; Gobbe, M.; Blanchard, L. The difference between approval processes for medicinal products and medical devices in Europe. Opthalmologica 2021, 244, 368–378. [Google Scholar] [CrossRef]
- Drabiak, K.; Kyzer, S.; Nemov, V.; Naqa, I.E. AI in machine learning ethics, law, diversity, and global impact. Br. J. Radiol. 2023, 96, 20220934. [Google Scholar] [CrossRef]
- Gerke, S.; Babic, B.; Evgeniou, T.; Cohen, I.G. The need for a system view to regulate artificial intelligence/machine learning-based software as a medical device. npj Digit. Med. 2020, 3, 53. [Google Scholar] [CrossRef]
- Wu, E.; Wu, K.; Daneshjou, R.; Ouyang, D.; Ho, D.E.; Zou, J. How AI medical devices are evaluated: Limitations and recommendations from an analysis of FDA approvals. Nat. Med. 2021, 27, 576–584. [Google Scholar] [CrossRef] [PubMed]
- FDA; Health Canada; MHRA. Good Machine Learning Practice for Medical Device Development: Guiding Principles. 2021. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles (accessed on 8 October 2025).
- Muehlematter, U.J.; Daniore, P.; Vokinger, K.N. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): A comparative analysis. Lancet Digit. Health 2021, 3, 195–203. [Google Scholar] [CrossRef]
- Hwang, T.J.; Sokolov, E.; Franklin, J.M.; Kesselheim, A.S. Comparison of rates of safety issues and reporting of trial outcomes for medical devices approved in the European Union and United States: Cohort study. Br. Med. J. 2016, 353, 3323. [Google Scholar] [CrossRef] [PubMed]
- EU AI Act. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32024R1689 (accessed on 26 September 2024).
- Prainsack, B.; Forgó, N. New AI regulation in the AU seeks to reduce risk without assessing public benefit. Nat. Med. 2024, 30, 1235–1237. [Google Scholar] [CrossRef] [PubMed]
- MHRA. Software and AI as a Medical Device Change Programme. 2021. Available online: https://www.gov.uk/government/publications/software-and-artificial-intelligence-ai-as-a-medical-device/software-and-artificial-intelligence-ai-as-a-medical-device (accessed on 8 October 2025).
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progession, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Nedin, B.P.; Cohen, S.J. Ciculating tumour cells in colorectal cancer: Past, present, and future challenges. Curr. Treat. Options Oncol. 2020, 11, 1–13. [Google Scholar]
- Folkersma, L.R.; Gómez, C.O.; Manso, S.J.; de Castro, S.V.; Romo, I.G.; Lázaro, M.V.; de la Orden, G.V.; Fernández, M.A.; Rubio, E.D.; Moyano, A.S.; et al. Immunomagnetic quantification of circulating tumoral cells in patients with prostate cancer: Clinical and pathological correlation. Arch. Esp. Urol. 2010, 63, 23–31. [Google Scholar]
- Milner, L.M.; Linder, M.W.; Valdes, R. Circulating tumor cells: A review of present methods and the need to identify heterogeneous phenotypes. Ann. Clin. Lab. Sci. 2013, 43, 295–304. [Google Scholar]
- Lamb, Y.N.; Dhillon, S. Epi procolon 2.0 CE: A blood-based screening test for colorectal cancer. Mol. Diagn. Ther. 2017, 21, 225–232. [Google Scholar] [CrossRef]
- Shirley, M. Epi procolon for colorectal cancer screening: A profile of its use in the USA. Mol. Diagn. Ther. 2020, 24, 497–503. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Lu, S.; et al. Erlotnib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (optimal, ctong-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Kwapisz, D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small-cell lung cancer? Ann. Transl. Med. 2017, 5, 46. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.B.; Kopetz, E.S.; et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and analytical validation of Foundation One liwuid CDx, a novel 324-gene cdDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Thompson, J.C.; Carpenter, E.L.; Elkhouly, E.; Aggarwal, C. Plasma cell-free DNA genotyping: From an emerging concept to a standard-of-care tool in metastatic non-small-cell lung cancer. Oncologist 2021, 26, 1812–1821. [Google Scholar] [CrossRef]
- Freenome Announces Topline Results for PREEMPT CRC to Validate the First Version of Its Blood-Based Test for the Early Detection of Colorectal Cancer. Available online: https://www.freenome.com/newsroom/freenome-announces-topline-results-for-preempt-crc-to-validate-the-first-version-of-its-blood-based-test-for-the-early-detection-of-colorectal-cancer/ (accessed on 9 April 2024).
- Duffy, M.J.; Crown, J. Circulating tumor DNA (ctDNA): Can it be used as a pan-cancer early detection test? Crit. Rev. Clin. Lab. Sci. 2023, 7, 1–13. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 23, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020, 369, eabb9601. [Google Scholar] [CrossRef]
- Gainullin, V.; Hwang, H.J.; Hogstrom, L.; Arvai, K.; Best, A.; Gray, M.; Kumar, M.; Manesse, M.; Chen, X.; Uren, P.; et al. Performance of a multi-analyte, multi-cancer early detection (MCED) blood test in a prospectively-collected cohort. In Proceedings of the AACR Annual Meeting 2024, San Diego, CA, USA, 5–10 April 2024. [Google Scholar]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 2022, 40, 1537–1549. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Tohka, J.; van Gils, M. Evaluation of machine learning algorithms for health and wellness applications: A tutorial. Comput. Biol. Med. 2021, 132, 104324. [Google Scholar] [CrossRef]
- Gray, E.; Butler, H.J.; Board, R.; Brennan, P.M.; Chalmers, A.J.; Dawson, T.; Goodden, J.; Hamilton, W.; Hegarty, M.G.; James, A.; et al. HEalth economic evaluation of a serum-based blood test for brain tumour diagnosis: Exploration of two clinical scenarios. BMJ Open 2018, 8, e017593. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Niraula, D.; Gates, E.D.H.; Fu, J.; Luo, Y.; Nyflot, M.J.; Bowen, S.R.; El Naqa, I.M.; Cui, S. Artificial Intelligence (AI) and machine learning (ML) in precision oncology: A review on enhancing discoverability through multiomics integration. Br. J. Radiol. 2023, 96, 20230211. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Sousa, J.; Fonseca, J.; Negrão, E.; Flor de Lima, B.; et al. Towards machine learning-aided lung cancer clinical routines: Approaches and open challenges. J. Pers. Med. 2022, 12, 480. [Google Scholar] [CrossRef]
- Adlung, L.; Cohen, Y.; Mor, U.; Elinav, E. Machine learning in clinical decision making. Med 2021, 2, 642–665. [Google Scholar] [CrossRef]
- Kleppe, A.; Skrede, O.J.; De Raedt, S.; Liestol, K.; Kerr, D.J.; Danielsen, H.E. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 2021, 21, 199–211. [Google Scholar] [CrossRef]
- Markowetz, F. All models are wrong and yours are useless: Making clinical prediction models impactful for patients. Precis. Oncol. 2024, 8, 54. [Google Scholar] [CrossRef] [PubMed]


| Preprocessing | Brief Description | Benefit | Example Algorithms and References |
|---|---|---|---|
| Spectral truncation | Also known as spectral cutting, removes regions of the spectrum to leave only purpose-relevant bands. | Removes noise from the spectrum and reduces dimensionality. | [71,72]. |
| Baseline correction | Baseline of spectrum is fitted with a function and the new fitted baseline subtracted from the other spectral baselines. | Remove slopes in the baseline caused by interference and background effects to effectively compare peak intensities. | First and second derivative, rubberband, and polynomial [73,74]. |
| Smoothing | Reduces the resolution of spectra by fitting a function to a pre-defined window of spectral points. | Reduces/removes spectral noise to enhance the biologically-relevant peaks. | Wavelet denoising, Savitzky-Golay filtering, and local polynomial fitting with Gaussian weighting [53,73,75,76]. |
| Normalisation | Normalise spectra to allow the spectral intensities to be on the same, or similar, scales. | Allows multiple spectra to be effectively compared to each other. | Scaling of each spectrum between 0 and 1, vector normalisation, normalisation to Amide I band [73]. |
| EMSC | Uses a reference spectrum and polynomial functions to correct baseline slopes and spectral noise. | Corrects additive baseline effects, multiplicative scaling effects, and interference effects. | [77]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McHardy, R.G.; Cameron, J.M.; Eustace, D.A.; Baker, M.J.; Palmer, D.S. From Lab to Clinic: Artificial Intelligence with Spectroscopic Liquid Biopsies. Diagnostics 2025, 15, 2589. https://doi.org/10.3390/diagnostics15202589
McHardy RG, Cameron JM, Eustace DA, Baker MJ, Palmer DS. From Lab to Clinic: Artificial Intelligence with Spectroscopic Liquid Biopsies. Diagnostics. 2025; 15(20):2589. https://doi.org/10.3390/diagnostics15202589
Chicago/Turabian StyleMcHardy, Rose G., James M. Cameron, David Andrew Eustace, Matthew J. Baker, and David S. Palmer. 2025. "From Lab to Clinic: Artificial Intelligence with Spectroscopic Liquid Biopsies" Diagnostics 15, no. 20: 2589. https://doi.org/10.3390/diagnostics15202589
APA StyleMcHardy, R. G., Cameron, J. M., Eustace, D. A., Baker, M. J., & Palmer, D. S. (2025). From Lab to Clinic: Artificial Intelligence with Spectroscopic Liquid Biopsies. Diagnostics, 15(20), 2589. https://doi.org/10.3390/diagnostics15202589






