Rapid Identification of Carbapenemase Genes Directly from Blood Culture Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting and Study Design
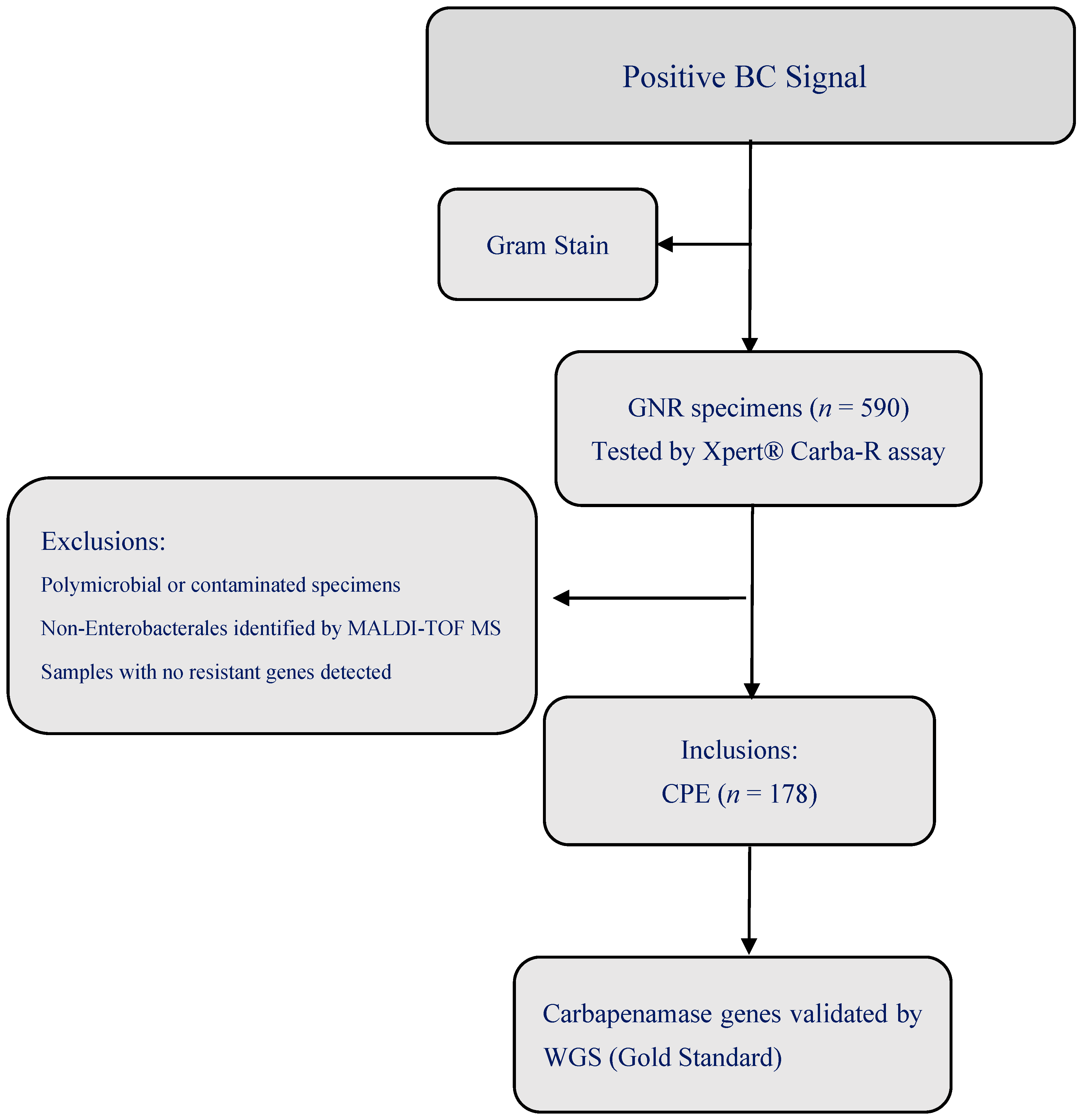
2.2. Study Population
2.3. Microbiological Methods
2.4. Xpert® Carba-R Assay
2.5. Whole-Genome Sequencing (WGS): (Figure 2)
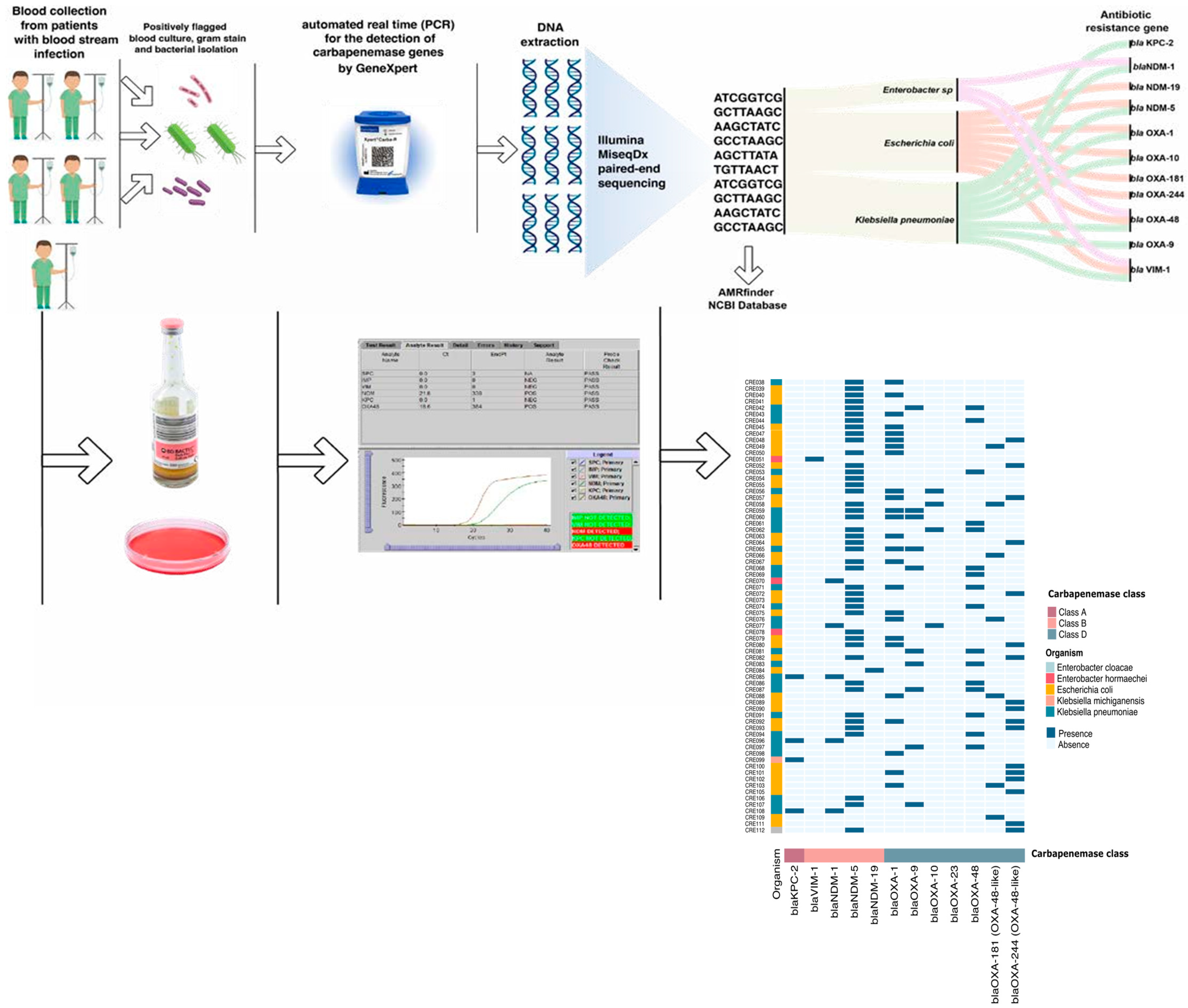
2.6. Statistical Analysis
3. Results
3.1. Overview of Clinical Isolates
3.2. Performance of the Xpert® Carba-R Assay
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ling, W.; Paterson, D.L.; Harris, P.N.A.; Furuya-Kanamori, L.; Edwards, F.; Laupland, K.B. Mortality, hospital length of stay, and recurrent bloodstream infections associated with extended-spectrum beta-lactamase-producing Escherichia coli in a low prevalence region: A 20-year population-based large cohort study. Int. J. Infect. Dis. 2024, 138, 84–90. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Kotb, S.; Lyman, M.; Ismail, G.; Abd El Fattah, M.; Girgis, S.A.; Etman, A.; Hafez, S.; El-Kholy, J.; Zaki, M.E.S.; Rashed, H.G.; et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare-associated Infections Surveillance Data, 2011–2017. Antimicrob. Resist. Infect. Control 2020, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Tawfick, M.M.; Alshareef, W.A.; Bendary, H.A.; Elmahalawy, H.; Abdulall, A.K. The emergence of carbapenemase blaNDM genotype among carbapenem-resistant Enterobacteriaceae isolates from Egyptian cancer patients. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1251–1259. [Google Scholar] [CrossRef]
- Li, H.H.; He, Z.J.; Xie, L.M.; Zhang, J.S.; Xie, T.A.; Fan, S.J.; Guo, X.G. Evaluation of Xpert® Carba-R assay for the Detection of Carbapenemase Genes in Gram-Negative Bacteria. Biomed. Res. Int. 2021, 2021, 6614812. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jia, P.; Li, X.; Wang, T.; Zhang, J.; Zhang, G.; Duan, S.; Kang, W.; Xu, Y.; Yang, Q. Carbapenemase detection by NG-Test CARBA 5—A rapid immunochromatographic assay in carbapenem-resistant Enterobacterales diagnosis. Ann. Transl. Med. 2021, 9, 769. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; Romero-Gómez, M.P.; Falces-Romero, I.; Aranda-Diaz, A.; García-Ballesteros, D.; Mingorance, J.; García-Rodríguez, J. Evaluation of a lateral flow immunoassay to detect CTX-M extended-spectrum β-lactamases directly from positive blood cultures for its potential use in Antimicrobial Stewardship programs. Rev. Esp. Quimioter. 2022, 35, 284–287. [Google Scholar] [CrossRef]
- Cointe, A.; Walewski, V.; Hobson, C.A.; Doit, C.; Bidet, P.; Dortet, L.; Bonacorsi, S.; Birgy, A. Rapid Carbapenemase Detection with Xpert® Carba-R assay V2 Directly on Positive Blood Vials. Infect. Drug Resist. 2019, 12, 3311–3316. [Google Scholar] [CrossRef]
- Messacar, K.; Parker, S.; Todd, J.; Dominguez, S. Implementation of Rapid Molecular Infectious Disease Diagnostics: The Role of Diagnostic and Antimicrobial Stewardship. J. Clin. Microbiol. 2017, 55, 5. [Google Scholar] [CrossRef]
- Christians, F.C.; Akhund-Zade, J.; Jarman, K.; Venkatasubrahmanyam, S.; Noll, N.; Blauwkamp, T.A.; Bercovici, S.; Zielinska, A.; Carr, A.L.; Craney, A.; et al. Analytical and clinical validation of direct detection of antimicrobial resistance markers by plasma microbial cell-free DNA sequencing. J. Clin. Microbiol. 2024, 16, 62. [Google Scholar] [CrossRef]
- Xu, Y.; Song, W.; Huang, P.; Mei, Y.; Zhang, Y.; Xu, T. A Rapid Carbapenemase Genes Detection Method with Xpert® Carba-R assay from Positive Blood Cultures Compared with NG-Test Carba 5 and Sequencing. Infect. Drug Resist. 2022, 15, 7719–7725. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Principles and Procedures for Blood Cultures, 2nd ed.; CLSI Guideline M47; CLSI: Wayne, PA, USA, 2022. [Google Scholar]
- Rose, R.; Nolan, D.J.; Ashcraft, D.; Feehan, A.K.; Velez-Climent, L.; Huston, C.; Lain, B.; Rosenthal, S.; Miele, L.; Fogel, G.B.; et al. Comparing antimicrobial resistant genes and phenotypes across multiple sequencing platforms and assays for Enterobacterales clinical isolates. BMC Microbiol. 2023, 23, 225. [Google Scholar] [CrossRef]
- Cepheid. Xpert® Carba-R Kit Insert; Cepheid: Sunnyvale, CA, USA, 2024. [Google Scholar]
- Toumi, M.; Pulikottil-Jacob, R.; Watts, K.; Odeyemi, I.; Butler, K. An Economic Model to Assess the Cost Impact of Using Xpert® Carba-R assay to Screen Carbapenemase-Producing Enterobacterales in Comparison with Standard of Care, in a National Health Service Setting. Infect. Dis. Ther. 2025, 14, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Carbapenem-Resistant Enterobacterales, Third Update; ECDC: Stockholm, Sweden, 2025.
- Tompkins, K.; van Duin, D. Treatment for carbapenem-resistant Enterobacterales infections: Recent advances and future directions. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2053–2068. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jeon, K.; Kym, D.; Jung, J.; Jang, Y.J.; Han, S.B.; Antimicrobial Resistance & Infection Control. Carbapenem-resistant Enterobacterales infection and colonization in patients with severe burns: A retrospective cohort study in a single burn center. Antimicrob. Resist. Infect. Control 2025, 14, 3. [Google Scholar] [CrossRef]
- Mahmoud, F.M.; Khedr, R.A.; Ebeid, E.; El-Mahallawy, H.A.; Hassan, S.S. Impact of Rapid Molecular Diagnostic Technique on Time to Optimal Antimicrobial Therapy and Hospital Outcomes in Pediatric Cancer Patients with Sepsis. Asian Pac. J. Cancer Prev. 2023, 24, 2465–2471. [Google Scholar] [CrossRef]
- Seo, H.; Lee, J.Y.; Ryu, S.H.; Kwak, S.H.; Kim, E.O.; Bae, S.; Kim, M.J.; Chong, Y.P.; Kim, S.-H.; Lee, S.-O.; et al. Comparison of the Clinical Outcomes of Patients with Positive Xpert® Carba-R assay Tests for Carbapenemase-Producing Enterobacterales According to Culture Positivity. Open Forum Infect. Dis. 2022, 9, ofab594. [Google Scholar] [CrossRef]
- El-Kholy, A.A.; Girgis, S.A.; Shetta, M.A.F.; Abdel-Hamid, D.H.; Elmanakhly, A.R. Molecular characterization of multi-drug-resistant Gram-negative pathogens in three tertiary hospitals in Cairo, Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 987–992. [Google Scholar] [CrossRef]
- Duze, S.T.; Thomas, T.; Pelego, T.; Jallow, S.; Perovic, O.; Duse, A. Evaluation of Xpert® Carba-R assay for detecting carbapenemase-producing organisms in South Africa. Afr. J. Lab. Med. 2023, 12, 1898. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Naas, T.; Bonnin, R.A.; Patino, R.; Glaser, P.; Fortineau, N.; Dortet, L. OXA-244-Producing Escherichia coli Isolates, a Challenge for Clinical Microbiology Laboratories. Antimicrob. Agents Chemother. 2017, 61, e00818-17. [Google Scholar] [CrossRef] [PubMed]
- MacVane, S.H.; Dwivedi, H.P. Evaluating the impact of rapid antimicrobial susceptibility testing for bloodstream infections: A review of actionability, antibiotic use and patient outcome metrics. J. Antimicrob. Chemother. 2024, 79 (Suppl. 1), i13–i25. [Google Scholar] [CrossRef]
- Jin, S.; Lee, J.Y.; Park, J.Y.; Jeon, M.J. Xpert® Carba-R assay for detection of carbapenemase-producing organisms in patients admitted to emergency rooms. Medicine 2020, 99, e21117. [Google Scholar] [CrossRef]
- Lee, C.-H.; Cao, H.; Jiang, S.; Wong, T.T.-Y.; Tse, C.W.-S.; Ho, P.-L. Inoculum Size and False-Positive Detection of NDM- and OXA-48OXA-48-Type Carbapenemases Using Two Multiplex Lateral Flow Assays. Diagnostics 2024, 14, 1274. [Google Scholar] [CrossRef]
- Dortet, L.; Fusaro, M.; Naas, T. Improvement of the Xpert® Carba-R assay kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.; Kim, Y.A.; Kim, M.; Kim, B.; Choi, J.Y.; Park, Y.S.; Yong, D. Evaluation of Xpert® Carba-R assay v.2 to Detect Carbapenemase Genes in Two Hospitals in Korea. Ann. Lab. Med. 2020, 40, 209–215. [Google Scholar] [CrossRef]
- Cañada-García, J.E.; Grippo, N.; de Arellano, E.R.; Bautista, V.; Lara, N.; Navarro, A.M.; Cabezas, T.; Martínez-Ramírez, N.M.; García-Cobos, S.; Calvo, J.; et al. Phenotypic and molecular characterization of IMP-producing Enterobacterales in Spain: Predominance of IMP-8 in Klebsiella pneumoniae and IMP-22 in Enterobacter roggenkampii. Front. Microbiol. 2022, 13, 1000787, Erratum in: Front. Microbiol. 2023, 14, 1331683. [Google Scholar] [CrossRef] [PubMed]
| Pathogen | Number of Isolates | Percentage (%) |
|---|---|---|
| Klebsiella pneumoniae | 72 | 40.4 |
| Escherichia coli | 97 | 54.5 |
| Enterobacter spp. | 7 | 3.9 |
| Citrobacter spp. | 1 | 0.6 |
| Serratia marcescens | 1 | 0.6 |
| Total | 178 | 100.0 |
| Resistance Gene(s) | Number of Isolates | Percentage (%) |
|---|---|---|
| blaNDM & blaOXA-48-like | 59 | 33.1 |
| blaNDM | 49 | 27.5 |
| blaOXA-48-like | 53 | 29.8 |
| blaKPC-2 & blaNDM-1 | 8 | 4.5 |
| blaVIM | 7 | 3.9 |
| blaKPC-2 | 1 | 0.6 |
| blaVIM & blaOXA-48 | 1 | 0.6 |
| blaIMP | – | – |
| Total | 178 | 100.0 |
| Bacterial Species | Genotype by WGS | No. of Isolates | No. (% Concordant on Xpert® Carba-R Assay) | |
|---|---|---|---|---|
| Klebsiella pneumoniae | blaKPC | blaKPC-2 | 1 | 1 (100) |
| blaNDM | blaNDM-1 | 3 | 3 (100) | |
| blaNDM-5 | 10 | 10 (100) | ||
| blaNDM-9 | 2 | 2 (100) | ||
| blaOXA-48-like | blaOXA-48 | 10 | 10 (100) | |
| blaOXA-181 | 5 | 5 (100) | ||
| blaIMP | blaIMP | 0 | 0 (100) | |
| blaVIM | blaVIM-1 | 1 | 1 (100) | |
| blaKPC/blaNDM | blaKPC-2/blaNDM-1 | 8 | 7 (87.5) | |
| blaNDM/blaOXA-48 | blaNDM-1/blaOXA-48 | 2 | 2 (100) | |
| blaNDM-5/blaOXA-48 | 29 | 29 (100) | ||
| Escherichia coli | blaKPC | blaKPC | 0 | 0 (100) |
| blaNDM | blaNDM-5 | 29 | 27 (93.1) | |
| blaNDM-19 | 2 | 2 (100) | ||
| blaOXA-48-like | blaOXA-244 | 23 | 22 (95.6) | |
| blaOXA-181/blaOXA-1 | 2 | 2 (100) | ||
| blaOXA-181 | 6 | 6 (100) | ||
| blaOXA-244/blaOXA-1 | 4 | 4 (100) | ||
| blaOXA-484 | 3 | 2 (66.6) | ||
| blaNDM/blaOXA-48-like | blaNDM-5/blaOXA-48 | 2 | 2 (100) | |
| blaNDM-5/blaOXA-244 | 21 | 20 (95.2) | ||
| blaNDM-5/blaOXA-181 | 5 | 5 (100) | ||
| Enterobacter spp. | blaVIM | blaVIM-1 | 4 | 4 (100) |
| blaNDM | blaNDM-1 | 1 | 1 (100) | |
| blaNDM-5 | 1 | 1 (100) | ||
| blaVIM/blaOXA-48-like | blaVIM-1/blaOXA-48 | 1 | 1 (100) | |
| blaVIM/blaOXA-48/blaNDM | blaVIM-1/blaOXA-48/blaNDM | 0 | 1 (0) | |
| Citrobacter spp. | blaVIM | blaVIM-1 | 1 | 1 (100) |
| Serratia marcescens | blaVIM | blaVIM-4 | 1 | 1 (100) |
| Gene Target | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) | Cohen’s Kappa (κ) | p-Value |
|---|---|---|---|---|---|---|
| blaNDM | 100.0 (97.2–100.0) | 96.8 (92.4–100) | 98.3 (96.0–100) | 100.0 (95.0–100.0) | 0.98 | <0.0001 |
| blaKPC | 88.9 (68.4–100.0) | 100 (98.2–100) | 100 (62.6–100) | 99.4 (98.3–100.0) | 0.94 | <0.0001 |
| blaOXA-48 | 99.1 (97.4–100.0) | 95.4 (90.3–100) | 97.4 (94.5–100) | 98.4 (95.4–100.0) | 0.95 | <0.0001 |
| blaVIM | 100.0 (62.6–100.0) | 98.8 (97.2–100) | 80.0 (55.2–100) | 100.0 (98.2–100.0) | 0.99 | <0.0001 |
| blaIMP | N/A * | 100.0 (98.3–100.0) | N/A * | 100.0 (98.3–100.0) | N/A * | |
| Overall | 99.2 (98.1–100) | 98.9 (98.1–99.7) | 97.2 (95.2–99.2) | 99.7 (99.3–100) | 0.98 | <0.0001 |
| Bacterial Species | Xpert® Carba-R Assay | WGS |
|---|---|---|
| Klebsiella pneumoniae | blaNDM | blaNDM-1/blaKPC-2 |
| blaNDM/blaOXA-48-like/blaVIM | blaNDM-5/blaOXA-48 | |
| blaNDM/blaOXA-48-like | blaNDM-9 | |
| Escherichia coli | BlaNDM/OXA-48-like | blaNDM-5 |
| blaNDM/blaOXA-48-like | blaNDM-5 | |
| blaNDM | blaNDM-5/blaOXA-244 | |
| blaNDM/blaOXA-48-like | blaOXA-244 | |
| Enterobacter sp. | blaOXA-48-like/blaVIM | blaNDM-5/blaOXA-48/blaVIM-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziad, G.A.; Jalal, D.; Hashem, M.; Sayed, A.A.; Mahfouz, S.; Bayoumi, A.; Lotfi, M.; Hassanain, O.; Tolba, M.; Madney, Y.; et al. Rapid Identification of Carbapenemase Genes Directly from Blood Culture Samples. Diagnostics 2025, 15, 2480. https://doi.org/10.3390/diagnostics15192480
Ziad GA, Jalal D, Hashem M, Sayed AA, Mahfouz S, Bayoumi A, Lotfi M, Hassanain O, Tolba M, Madney Y, et al. Rapid Identification of Carbapenemase Genes Directly from Blood Culture Samples. Diagnostics. 2025; 15(19):2480. https://doi.org/10.3390/diagnostics15192480
Chicago/Turabian StyleZiad, Ghada A., Deena Jalal, Mohamed Hashem, Ahmed A. Sayed, Sally Mahfouz, Ahmed Bayoumi, Maryam Lotfi, Omneya Hassanain, May Tolba, Youssef Madney, and et al. 2025. "Rapid Identification of Carbapenemase Genes Directly from Blood Culture Samples" Diagnostics 15, no. 19: 2480. https://doi.org/10.3390/diagnostics15192480
APA StyleZiad, G. A., Jalal, D., Hashem, M., Sayed, A. A., Mahfouz, S., Bayoumi, A., Lotfi, M., Hassanain, O., Tolba, M., Madney, Y., Shalaby, L., & Elanany, M. (2025). Rapid Identification of Carbapenemase Genes Directly from Blood Culture Samples. Diagnostics, 15(19), 2480. https://doi.org/10.3390/diagnostics15192480





