Abstract
Background/Objectives: To develop an automated American Joint Committee on Cancer (AJCC) staging system for radical prostatectomy pathology reports using large language model-based information extraction and knowledge graph validation. Methods: Pathology reports from 152 radical prostatectomy patients were used. Five additional parameters (Prostate-specific antigen (PSA) level, metastasis stage (M-stage), extraprostatic extension, seminal vesicle invasion, and perineural invasion) were extracted using GPT-4.1 with zero-shot prompting. A knowledge graph was constructed to model pathological relationships and implement rule-based AJCC staging with consistency validation. Information extraction performance was evaluated using a local open-source large language model (LLM) (Mistral-Small-3.2-24B-Instruct) across 16 parameters. The LLM-extracted information was integrated into the knowledge graph for automated AJCC staging classification and data consistency validation. The developed system was further validated using pathology reports from 88 radical prostatectomy patients in The Cancer Genome Atlas (TCGA) dataset. Results: Information extraction achieved an accuracy of 0.973 and an F1-score of 0.986 on the internal dataset, and 0.938 and 0.968, respectively, on external validation. AJCC staging classification showed macro-averaged F1-scores of 0.930 and 0.833 for the internal and external datasets, respectively. Knowledge graph-based validation detected data inconsistencies in 5 of 150 cases (3.3%). Conclusions: This study demonstrates the feasibility of automated AJCC staging through the integration of large language model information extraction and knowledge graph-based validation. The resulting system enables privacy-protected clinical decision support for cancer staging applications with extensibility to broader oncologic domains.
1. Introduction
The American Joint Committee on Cancer (AJCC) staging manual is the benchmark for determining cancer classification, prognosis, and treatment approaches [1]. The 8th edition of the AJCC manual contains all available information on adult cancer staging [1,2]. The staging of prostate cancer is based on the integration of the Tumor-Node-Metastasis (TNM) staging system, grade group derived from Gleason scores (GS) according to the International Society of Urological Pathology (ISUP) 2014 consensus guidelines, and prostate-specific antigen (PSA) level [2,3].
Radical prostatectomy (RP) has been established as an effective therapy for patients with moderate-risk and high-risk prostate cancer [4,5]. In patients treated with RP as primary therapy, the pathologic features of the prostatectomy specimen are major determinants of prognostic assessment and of the appropriateness of adjuvant therapy [6]. Appropriate pathological examination of surgical specimens by pathologists is critical for accurate prognostic assessment and optimal patient management [7]. For RP specimens, the International Collaboration on Cancer Reporting (ICCR) recommends structured pathology reporting using standardized ICCR datasets with required (core) and recommended (non-core) elements [8]. Core macroscopic elements include specimen weight, seminal vesicles, and lymph nodes, whereas core microscopic elements include histologic tumor type and grade, extraprostatic extension (EPE), and seminal vesicle invasion (SVI) [8,9]. Clinical information and PSA levels are categorized as recommended data elements [8,9].
Most pathology reports are written in free-text format [9,10], which poses significant challenges for information extraction and clinical decision support. Large language models (LLMs) have demonstrated significant advances in medical applications [11,12], including information extraction from electronic health records [13], clinical text summarization [14], and document classification [15]. Prompt engineering utilizing LLMs enables the extraction of structured information from narrative pathology reports without requiring manual annotation or model training [10,16]. Medical knowledge graphs systematically represent information by organizing medical concepts and events as nodes and edges to describe their interrelationships [17]. This study aims to develop an automated AJCC staging system for RP pathology reports by applying prompt engineering with LLMs for information extraction and knowledge graph-based approaches for validation and staging classification. Specifically, key pathological information is extracted from free-text reports, a knowledge graph is constructed from the extracted data, and rule-based AJCC staging is implemented with integrated data consistency verification.
2. Materials and Methods
2.1. Dataset and Expansion of Data Elements
Figure 1 illustrates the overall study workflow. For the purpose of extracting structured data, performing AJCC staging, and validating data consistency, an anonymized, retrospective dataset consisting of 579 English pathology reports from 340 prostate cancer patients was used [10]. The final dataset consisted of 152 cases in which PSA values were documented in the pathology reports. The dataset was publicly released on Zenodo for academic research purposes [18]. For patients with multiple pathology reports, the most recent report was used as the reference. Missing variables were supplemented using earlier reports, and once a value was identified, it was adopted as the final value.
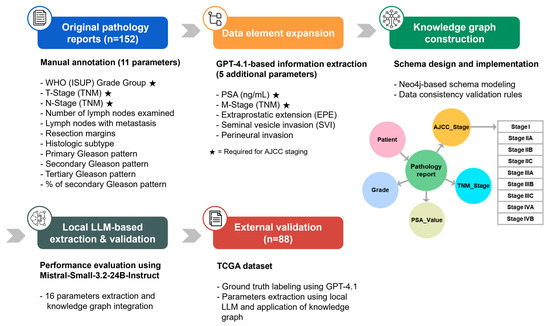
Figure 1.
Overview of the automated AJCC staging system workflow. The integrated system employs a multi-step pipeline involving LLM-based information extraction, missing data imputation, knowledge graph construction, and validation.
The pathology reports were manually annotated with 11 parameters selected based on their clinical and prognostic relevance [10]. In this study, additional extraction was performed using GPT-4.1 to extract five specific data elements not included in the original annotations: PSA level, M-stage, EPE, SVI, and perineural invasion. Information extraction was performed using the GPT-4.1 model accessed through the OpenAI API (OpenAI, L.L.C., San Francisco, CA, USA, 17 July 2025, https://openai.com/) with a zero-shot prompting approach. The model output was provided in a structured JSON format containing the extracted information, the supporting evidence text span, and a confidence score ranging from 0 to 1. All extracted data were manually reviewed to confirm accuracy, and the prompt used for extraction is provided in Appendix A.1.
To evaluate the generalizability of the proposed approach, the publicly available The Cancer Genome Atlas (TCGA)-Reports dataset from Mendeley Data was used as an external validation dataset [19,20]. From 9523 unstructured free-text pathology reports representing 32 tissue or cancer types from a wide variety of institutions, 300 reports containing ‘radical prostatectomy’ were identified. Among these, 88 cases with documented PSA values were selected as the final external validation dataset. All 16 parameters for the external dataset were annotated using GPT-4.1 (accessed 14 September 2025) with a zero-shot prompting approach. The extracted data were manually reviewed, and the prompt used for annotating the external dataset is provided in Appendix A.2.
2.2. AJCC Stage Labeling
M-stage was recorded in only 3 cases (2.0%) in the internal dataset and 27 cases (30.7%) in the external dataset. To determine the stage of metastasis, additional clinical data such as the results of systemic imaging studies (bone scan, CT, or PET/CT) are required [21,22]. Because RP is performed for localized prostate cancer, cases with missing M-stage information were classified as M0 stage [23,24,25]. A rule-based AJCC staging algorithm consistent with the 8th-edition criteria [26] was then applied to assign final stage labels to both the internal and external validation datasets. All assigned labels were manually reviewed.
The overall distribution of AJCC stages between datasets was compared using Fisher’s exact test. For stage-specific comparisons of proportions, either Fisher’s exact test or Pearson’s chi-squared test was applied. For each stage within each dataset, 95% confidence intervals for proportions were calculated using the Wilson method. Statistical analyses were performed using R version 4.2.3 (R Foundation for Statistical Computing, Vienna, Austria).
2.3. Knowledge Graph Construction
Neo4j (Neo4j, Inc., San Mateo, CA, USA, v 5.28.1) was employed to systematically model the relationships between the extracted pathological variables and their AJCC classifications, yielding a knowledge graph that supports rule-based staging classification and automated data consistency validation. The constructed knowledge graph processes pathology report data through a structured pipeline and exports both classification results and consistency errors to a single Excel file.
2.3.1. Schema Modelling and Node Structure
The knowledge graph schema was designed to represent the complex relationships between pathological entities and their staging implications (Figure 2). Five primary node types were implemented: Patient, Pathology Report, TNM_Stage, Grade, PSA_Value, and AJCC_Stage. Patient nodes served as the root entities, connected to Pathology Report nodes through HAS_REPORT relationships. Each Pathology Report node stored comprehensive pathological features, including histologic subtype, resection margins, lymph node examination results, Gleason patterns, and invasion indicators (EPE, SVI, and perineural invasion).
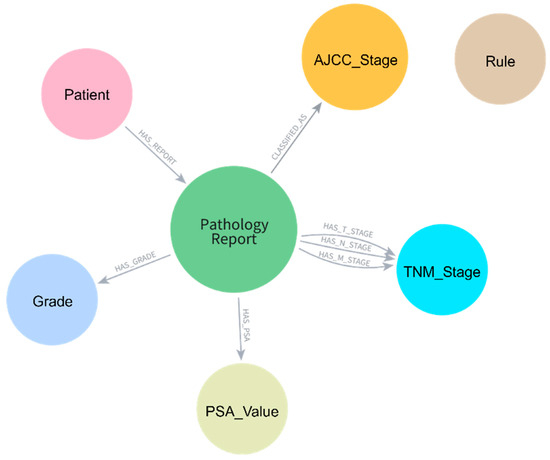
Figure 2.
Knowledge graph schema for pathology report data representation. Central Pathology_Report nodes are connected to Patient, AJCC_Stage, TNM_Stage, Grade, and PSA_Value nodes through specific relationship types, enabling structured data storage and rule-based AJCC staging.
TNM staging information was modeled as separate nodes with unique constraints based on both code and type combinations to ensure proper node uniqueness. The schema employed specific relationship types (HAS_T_STAGE, HAS_N_STAGE, HAS_M_STAGE) to connect Pathology Report nodes to their respective TNM_Stage nodes. Grade Group information was represented through dedicated Grade nodes linked through HAS_GRADE relationships, while PSA values were stored in PSA_Value nodes. The final AJCC stage classification was connected through CLASSIFIED_AS relationships to AJCC_Stage nodes.
2.3.2. AJCC Staging Classification
To construct a knowledge graph capable of classifying AJCC stages, a hierarchical rule-based algorithm was implemented following the AJCC 8th edition criteria [26], with conditions applied in the sequential order specified in Table 1. When M-stage information was missing, M0 was assumed for staging classification. Cases that could not be classified due to missing other essential elements for AJCC staging were labeled as “Unknown” with the missing elements specified in parentheses (e.g., Unknown (Missing N-Stage, Grade Group)).

Table 1.
Prioritized rule-based staging criteria for prostate cancer, according to the 8th edition of the AJCC [23].
2.3.3. Consistency Validation
The knowledge graph included consistency validation rules to identify potential data extraction errors or clinical inconsistencies. The validation rules examined multiple pathological relationships: T-stage consistency with EPE and SVI, N-stage alignment with lymph node metastasis counts, and Gleason score concordance with the World Health Organization (WHO) Grade Groups according to ISUP 2014 guidelines [27].
Specific validation rules detected the following inconsistencies: (1) T2 stages with EPE present; (2) presence of SVI in T2 or T3a stages; (3) N0 stages with positive lymph nodes or N1 stages without positive nodes; (4) mismatches between Gleason sum and Grade Group assignments; (5) T3b stages without SVI or T3a stages without EPE [6]. When inconsistencies were identified, the validation rules generated descriptive error codes (e.g., ‘T2_stage_with_EPE_should_be_T3a’, ‘N0_stage_but_positive_lymph_nodes’) and compiled these into dedicated Excel sheets, facilitating systematic data review and accuracy assessment.
2.4. LLM-Based Information Extraction and Knowledge Graph Application
2.4.1. Information Extraction Using Open-Source LLM
In this study, although anonymized data was used, considering the privacy protection aspects of medical data, locally available LLM was used to extract information on 16 parameters from pathology reports included in the internal and external validation datasets and evaluate performance. Considering available resources, the open-source LLM was selected as the Mistral-Small-3.2-24B-Instruct (https://huggingface.co/mistralai/Mistral-Small-3.2-24B-Instruct-2506 (accessed on 17 July 2025)) [28] model, which provides performance comparable to Llama 3.3 70B while utilizing significantly fewer computational resources for extracting cancer-related medical attributes from pathology reports [29]. The model was implemented on dual RTX 3090 GPUs. A zero-shot prompting approach was used to extract 16 parameters from the reports and to return the results in a structured JSON format; the prompt template is provided in Appendix A.3. Extraction performance was assessed against manually curated labels with accuracy, precision, recall, and F1-score: accuracy represents the proportion of correct predictions among all predictions; precision represents the proportion of correctly extracted values among all extracted values; recall represents the proportion of correctly extracted values among all ground truth values; and F1-score is the harmonic mean of precision and recall.
2.4.2. Knowledge Graph–Based AJCC Staging and Consistency Validation
The constructed knowledge graph was utilized to perform automated AJCC staging classification and consistency validation using the data extracted by the Mistral-Small-3.2-24B-Instruct model. LLM-extracted information was applied to the knowledge graph to evaluate the clinical applicability of this automated pipeline. In the internal dataset, two reports failed JSON conversion due to excessive text length and were excluded from knowledge graph processing. The LLM-extracted data were integrated into the knowledge graph system, and both staging classification and data consistency validation were performed. For evaluation purposes, cases labeled as “Unknown” with specific missing parameters in parentheses were reclassified as a unified “Unknown” category. Staging classification performance was evaluated against ground truth annotations using accuracy, precision, recall, and F1-score, while consistency validation rules were applied to detect data extraction errors.
3. Results
3.1. Dataset Characteristics
Table 2 presents the distribution of AJCC staging parameters across the internal (n = 152) and external validation (n = 88) datasets. The WHO Grade Group distribution differed significantly between datasets (p < 0.001), with the internal dataset showing a higher proportion of Grade Group 2 (46.1% vs. 34.1%) and Grade Group 3 (31.6% vs. 25.0%), while the external dataset had higher proportions of Grade Groups 4 (14.8% vs. 2.6%) and 5 (19.3% vs. 13.2%). T-stage distribution also varied significantly (p = 0.014). The internal dataset contained more pT2c cases (47.4% vs. 33.0%), whereas the external dataset showed higher rates of pT3a (38.6% vs. 30.9%) and pT3b (23.9% vs. 14.5%). Notably, three pT4 cases (3.4%) were present only in the external dataset.

Table 2.
Characteristics of the internal and external validation datasets.
N-stage distribution showed no significant difference (p = 0.090), with the majority being pN0 in both datasets (73.7% internal, 81.8% external). Mean serum PSA levels were significantly higher in the internal dataset (11.9 ng/mL, 95% CI: 8.0–17.2) compared to the external dataset (7.11 ng/mL, 95% CI: 5.0–10.1, p < 0.001). M-stage information was rarely recorded in both datasets, with only 3 cases (2.0%) identified in the internal dataset and 27 cases (30.7%) in the external dataset.
Among pathological invasion parameters, EPE was present in 44.1% of internal dataset cases versus 65.9% of external dataset cases (p = 0.002). SVI was documented as present in 15.1% and 26.1% of cases in the internal and external datasets, respectively (p = 0.055). Perineural invasion showed similarly high rates of presence in both datasets (91.4% internal vs. 90.9% external, p = 0.948). Representative examples of GPT-4.1 extraction output with confidence scores and evidence spans for data element expansion are provided in Table A1 (Appendix A). Examples of output for annotating external validation datasets are presented in Table A2 (Appendix A).
Table 3 shows the distribution of AJCC stages between the internal and external validation datasets. The overall stage distribution differed significantly between datasets (p = 0.048). The internal dataset showed no Stage IIA cases, while the external dataset contained 3 cases (4.2%). Stage IIB was more common in the internal dataset (28.7% vs. 20.8%), whereas Stage IIIB showed higher prevalence in the external dataset (43.1% vs. 31.1%). Stage IIIA was present only in the internal dataset (5.7%, p = 0.048). The remaining stages (IIC, IIIC, and IVA) showed similar distributions between datasets.

Table 3.
AJCC stage distribution and PSA value comparison in original versus imputed data.
3.2. Information Extraction Performance
The Mistral-Small-3.2-24B-Instruct model was applied to 150 pathology reports to extract 16 clinical parameters in the internal dataset, resulting in 2400 field-document pairs. Overall micro-averaged performance showed accuracy of 0.973, precision of 0.992, recall of 0.981, and F1-score of 0.986 (Table 4).

Table 4.
LLM information extraction performance by parameters in the internal dataset (n = 150 cases).
M-stage and SVI achieved F1-scores of 1.000, followed by Lymph Nodes with Metastasis (0.997), Tertiary Gleason Pattern (0.997), and N-stage (0.997). Staging-related parameters demonstrated F1-scores of 1.000 (M-stage), 0.997 (N-stage), and 0.980 (T-stage). Percentage of Secondary Gleason Pattern showed the lowest performance, with accuracy of 0.907 and F1-score of 0.951. For example, given “Gleason score 4 + 3 = 7b (60% pattern 4),” the model correctly extracted 60% but failed to compute the secondary proportion of 40%.
The same model was applied to 88 pathology reports to extract 16 clinical parameters from the external validation dataset, resulting in 1408 field-document pairs. Overall micro-averaged performance showed accuracy of 0.938, precision of 0.990, recall of 0.947, and F1-score of 0.968 (Table 5).

Table 5.
LLM information extraction performance by parameters in the external validation dataset (n = 88 cases).
Multiple parameters achieved F1-scores of 1.000: Lymph Nodes with Metastasis, N-stage, Number of Lymph Nodes examined, Perineural Invasion, SVI, and T-stage. M-stage showed an F1-score of 0.819, while Percentage of Secondary Gleason Pattern had the lowest performance at 0.803.
3.3. AJCC Staging Classification Performance
AJCC staging classification performance in the internal dataset showed macro-averaged precision of 0.922, recall of 0.944, and F1-score of 0.930 (Table 6). Stage IIIC achieved the highest F1-score of 1.000, followed by IVA (0.960), IIB (0.957), and IIIB (0.950). Stage IIC showed lower performance with an F1-score of 0.897, while “Unknown” cases demonstrated the lowest performance with an F1-score of 0.815.

Table 6.
AJCC staging classification performance by stage using knowledge graph-based rule application in the internal dataset.
In the external validation dataset, macro-averaged performance metrics for AJCC staging classification were precision 0.830, recall 0.883, and F1-score 0.833 (Table 7). Stage IIIB demonstrated the highest F1-score of 0.939, followed by IIC (0.923) and IVA (0.900). Stage IIA achieved an F1-score of 0.857, while IIB and IIIC showed F1-scores of 0.867 and 0.889, respectively. “Unknown” cases exhibited notably reduced performance with an F1-score of 0.455, representing the lowest performance across all categories. This was mainly attributable to discrepancies in M-stage labeling, where the ground truth was recorded as “pMx” but the open-source LLM extracted the value as “not mentioned,” resulting in misclassification as “Unknown.”

Table 7.
AJCC staging classification performance by stage using knowledge graph-based rule application in the external validation dataset.
3.4. Consistency Validation of Extracted Data
Knowledge graph-based validation applied to LLM-extracted data from 150 internal dataset cases identified 5 cases (3.3%) with staging and grade inconsistencies (Table 8). The most frequent inconsistency was the T3a stage documented without evidence of EPE (2 cases, 40.0%). Other inconsistencies included Gleason 7 inconsistent with Grade Group 3 (1 case, 20.0%), presence of SVI that should correspond to T3b or higher stage (1 case, 20.0%), and Gleason 7 inconsistent with Grade Group 2 (1 case, 20.0%). No staging or grade inconsistencies were detected in the external validation dataset.

Table 8.
Types and frequency of staging and grade inconsistencies detected by knowledge graph validation in the internal dataset.
The T3a_stage_but_no_EPE cases involved complex pathological descriptions where frozen section findings documented “small capsular defect” with “tumor infiltrates forming margins,” indicating EPE that is classified as pT3a according to the AJCC system [30]. However, the LLM did not reflect these microscopic findings and extracted EPE as “Absent.” The Gleason_7_inconsistent_with_GG3 case occurred when the LLM correctly extracted Primary and Secondary Gleason patterns (3 and 4) but incorrectly identified Grade Group 3 instead of Grade Group 2 from a tabulated format where the correct grade was marked with an “X” symbol, demonstrating challenges in interpreting structured table formats within narrative reports.
4. Discussion
This study presents an integrated automated AJCC staging system that combines LLM-based information extraction, missing data imputation, and knowledge graph construction for radical prostatectomy pathology reports. The approach utilized prompt engineering with an open-source LLM to extract structured data, achieving a micro-averaged F1-score of 0.986 in the internal dataset and 0.968 in the external dataset. Grothey et al. [10] demonstrated that open-source LLMs achieve comparable accuracy to third-party managed models such as GPT-4 in extracting structured information from pathology reports. These results suggest that open-source LLMs can effectively extract information from unstructured pathology reports while mitigating privacy concerns. Knowledge graph-based validation identified data errors in 5 of 150 internal datasets (3.3%). These errors were found in cases with complex microscopic descriptions or tabulated formats. For the AJCC staging classification, the system achieved a macro-averaged F1-score of 0.930 in the internal dataset and 0.833 in the external dataset. This pipeline provides a framework for automated clinical decision support systems in cancer staging workflows, processing unstructured free-text medical data into structured formats by validating data accuracy within pathology reports and performing AJCC classification, thereby contributing to systematic clinical data management.
Richter et al. [9] analyzed pathology reports for RP specimens, comparing narrative reports with standardized synoptic reports, and found that PSA values were documented in fewer than half of cases in both formats. In this study, PSA values were available in only 152 of 340 internal pathology reports (44.7%) and 88 of 300 external reports (29.3%), and only these subsets were included in the study. The presence of PSA within pathology reports is not determined by the reporting format [9]. Clinical parameters such as PSA depend primarily on whether the ordering clinician supplies them on the histopathology requisition [9,31]. Therefore, AJCC staging based on pathology reports could be further improved by enhancing documentation practices and information transfer within clinician–pathologist communication workflows, or by integrating pre-diagnostic PSA data from patients into the system.
This study has several limitations. First, although both internal and external validation were performed, the system has not yet been integrated into real clinical workflows. In particular, only pathology reports were available, and relevant clinical data such as imaging findings could not be incorporated. Cases without documented M-stage in pathology reports were classified as M0, given that radical prostatectomy is performed for localized prostate cancer [23,24,25]. Future research should integrate clinical data, such as systemic imaging results or PSA levels, into the pipeline for application in clinical workflows. Additionally, only one open-source LLM was utilized, and performance comparisons with other models were not conducted. In the future, benchmarking across multiple LLMs or optimizing prompt design tailored to the specific characteristics of pathology reports may further improve extraction accuracy. Furthermore, integrating probabilistic reasoning into the knowledge graph, such as converting microscopic findings into probability values, could enhance the processing of detailed findings from unstructured pathology reports within the knowledge graph framework. Despite these limitations, our approach demonstrates significant potential for supporting standardized AJCC staging and reducing medical errors through automated consistency validation. This methodology provides a foundation for developing comprehensive clinical decision support systems that can process unstructured medical text while maintaining patient privacy through local deployment strategies.
5. Conclusions
This study presents an integrated automated AJCC staging system for radical prostatectomy pathology reports by combining large language model-based information extraction with knowledge graph validation. The developed framework provides automated, privacy-preserving clinical decision support in cancer staging workflows and has potential for application to other oncologic domains.
Author Contributions
Conceptualization, E.J. and H.J.J.; methodology, E.J., T.I.N. and H.J.J.; software, E.J.; validation, E.J., T.I.N. and H.J.J.; formal analysis, E.J.; investigation, E.J.; resources, E.J. and H.J.J.; data curation, E.J.; writing—original draft preparation, E.J. and H.J.J.; writing—review and editing, E.J., T.I.N. and H.J.J.; visualization, E.J.; supervision, H.J.J.; project administration, H.J.J.; funding acquisition, H.J.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the IITP (Institute of Information & Communications Technology Planning & Evaluation)-ICAN (ICT Challenge and Advanced Network of HRD) grant funded by the Korean government (Ministry of Science and ICT) (IITP-2025-RS-2022-00156439) and the National Institute of Health (NIH) Research Project (Project no. 2024-ER0807-00).
Institutional Review Board Statement
Not applicable. This study analyzed publicly available, fully de-identified pathology report data and did not involve human participants or the use of identifiable private information.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Zenodo at https://doi.org/10.5281/zenodo.14175293, reference number [18], and Mendeley Data at https://data.mendeley.com/datasets/hyg5xkznpx/1, reference number [20]. These data were derived from the following resources available in the public domain: The Cancer Genome Atlas (TCGA, https://www.cancer.gov/ccg/research/genome-sequencing/tcga) for reference number [20].
Acknowledgments
During the preparation of this study, the authors used the OpenAI API (GPT-4.1; accessed on 17 July 2025 and 14 September 2025) to assist in data labeling. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction to the Data Availability Statement. This change does not affect the scientific content of the article.
Abbreviations
| AJCC | American Joint Committee on Cancer |
| EPE | Extraprostatic extension |
| FN | False Negative |
| FP | False Positive |
| GS | Gleason scores |
| ICCR | International Collaboration on Cancer Reporting |
| ISUP | International Society of Urological Pathology |
| LLM | Large language model |
| PSA | Prostate-specific antigen |
| RP | Radical prostatectomy |
| SVI | Seminal vesicle invasion |
| TCGA | The Cancer Genome Atlas |
| TP | True Positive |
| TNM | Tumor-Node-Metastasis |
| WHO | World Health Organization |
Appendix A
Appendix A.1
Prompt for automated pathological parameter extraction using GPT-4.1
You are a senior genitourinary pathologist. Extract ONLY the five parameters listed below from the user supplied pathology report and return a **STRICTLY valid single line JSON object** (no markdown, no commentary).
──────── OUTPUT FORMAT ────────
{
“extracted” : {
“Serum_PSA_ng_per_mL” : <float> | “Not mentioned”,
“M_Stage” : “cM0” | “cM1” | “pM0” | “pM1” | “Not mentioned”,
“Extraprostatic_Extension_EPE” : “Present” | “Absent” | “Not mentioned”,
“Seminal_Vesicle_Invasion_SVI” : “Present” | “Absent” | “Not mentioned”,
“Perineural_Invasion_Pn” : “Present” | “Absent” | “Not mentioned”
},
“evidence” : {
“Serum_PSA_ng_per_mL” : “<quote or null>“,
“M_Stage” : “<quote or null>“,
“Extraprostatic_Extension_EPE” : “<quote or null>“,
“Seminal_Vesicle_Invasion_SVI” : “<quote or null>“,
“Perineural_Invasion_Pn” : “<quote or null>“
},
“confidence” : {
“Serum_PSA_ng_per_mL” : 0.00 1.00,
“M_Stage” : 0.00 1.00,
“Extraprostatic_Extension_EPE” : 0.00 1.00,
“Seminal_Vesicle_Invasion_SVI” : 0.00 1.00,
“Perineural_Invasion_Pn” : 0.00 1.00
}
}
──────── EXTRACTION RULES ────────
- 1
- Use **only** the allowed categories/number formats above.
- 2
- PSA : choose the **pretreatment serum PSA** value; strip “ng/mL”; output as float.
- 3
- If the report lacks clear info → set value = “Not mentioned”, evidence = null, confidence = 0.0.
- 4
- Quote evidence verbatim; maximum 25 words.
- 5
- Return JSON only—no other text.
Appendix A.2
Prompt for Parameter Extraction for External Validation Dataset Annotation Using GPT-4.1
You are a senior genitourinary pathologist.
Extract ONLY the parameters listed below from the user-supplied pathology report
and return a **STRICTLY valid single-line JSON object** (no markdown, no commentary).
──────── OUTPUT FORMAT ────────
{
“extracted”: {
“WHO_ISUP_Grade_Group”: “GG1” | “GG2” | “GG3” | “GG4” | “GG5” | “Not mentioned”,
“T_Stage_TNM”: “pT0” | “pT2a” | “pT2b” | “pT2c” | “pT3a” | “pT3b” | “pT4” | “Not mentioned”,
“N_Stage_TNM”: “pNx” | “pN0” | “pN1” | “Not mentioned”,
“Number_of_Lymph_Nodes_examined”: <integer> | “Not mentioned”,
“Lymph_Nodes_with_Metastasis”: <integer> | “Not mentioned”,
“Resection_Margins”: “R0” | “R1” | “Rx” | “Not mentioned”,
“Histologic_Subtype”: “acinar” | “ductal” | “mixed” | “Not mentioned”,
“Primary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Secondary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Tertiary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Percentage_of_Secondary_Gleason_Pattern”: <integer> | “Not mentioned”,
“Serum_PSA_ng_per_mL”: <float> | “Not mentioned”,
“M_Stage_TNM”: “cM0” | “cM1” | “pM0” | “pM1” | “Not mentioned”,
“Extraprostatic_Extension_EPE”: “Present” | “Absent” | “Not mentioned”,
“Seminal_Vesicle_Invasion_SVI”: “Present” | “Absent” | “Not mentioned”,
“Perineural_Invasion_Pn”: “Present” | “Absent” | “Not mentioned”
},
“evidence”: {
“WHO_ISUP_Grade_Group”: “<quote or null>“,
“T_Stage_TNM”: “<quote or null>“,
“N_Stage_TNM”: “<quote or null>“,
“Number_of_Lymph_Nodes_examined”: “<quote or null>“,
“Lymph_Nodes_with_Metastasis”: “<quote or null>“,
“Resection_Margins”: “<quote or null>“,
“Histologic_Subtype”: “<quote or null>“,
“Primary_Gleason_Pattern”: “<quote or null>“,
“Secondary_Gleason_Pattern”: “<quote or null>“,
“Tertiary_Gleason_Pattern”: “<quote or null>“,
“Percentage_of_Secondary_Gleason_Pattern”: “<quote or null>“,
“Serum_PSA_ng_per_mL”: “<quote or null>“,
“M_Stage_TNM”: “<quote or null>“,
“Extraprostatic_Extension_EPE”: “<quote or null>“,
“Seminal_Vesicle_Invasion_SVI”: “<quote or null>“,
“Perineural_Invasion_Pn”: “<quote or null>“
},
“confidence”: {
“WHO_ISUP_Grade_Group”: 0.00–1.00,
“T_Stage_TNM”: 0.00–1.00,
“N_Stage_TNM”: 0.00–1.00,
“Number_of_Lymph_Nodes_examined”: 0.00–1.00,
“Lymph_Nodes_with_Metastasis”: 0.00–1.00,
“Resection_Margins”: 0.00–1.00,
“Histologic_Subtype”: 0.00–1.00,
“Primary_Gleason_Pattern”: 0.00–1.00,
“Secondary_Gleason_Pattern”: 0.00–1.00,
“Tertiary_Gleason_Pattern”: 0.00–1.00,
“Percentage_of_Secondary_Gleason_Pattern”: 0.00–1.00,
“Serum_PSA_ng_per_mL”: 0.00–1.00,
“M_Stage_TNM”: 0.00–1.00,
“Extraprostatic_Extension_EPE”: 0.00–1.00,
“Seminal_Vesicle_Invasion_SVI”: 0.00–1.00,
“Perineural_Invasion_Pn”: 0.00–1.00
}
}
──────── EXTRACTION RULES ────────
- Use **only** the allowed categories/number formats above.
- **WHO/ISUP Grade Group**: Look for “Grade Group”, “ISUP grade”, “GG1-5”, or equivalent grading terminology.
- **T-Stage**: Focus on pathologic T-stage (pT). Look for tumor extent and capsular involvement.
- **N-Stage**: Extract nodal involvement status (pNx, pN0, pN1).
- **Lymph Nodes**:
- -
- Count total examined nodes separately from positive nodes
- -
- Extract exact numbers when available
- **Resection Margins**:
- -
- R0 = negative/clear margins
- -
- R1 = positive margins
- -
- Rx = cannot be assessed
- **Histologic Subtype**: Look for adenocarcinoma subtypes (acinar, ductal, mixed).
- **Gleason Patterns**:
- -
- Extract primary (predominant), secondary, and tertiary patterns
- -
- Look for percentage of secondary pattern when mentioned
- **PSA**: Use **pretreatment serum PSA** value; strip “ng/mL” units; output as float.
- **M-Stage**: Look for metastasis status (clinical cM or pathologic pM).
- **Extensions/Invasions**: Determine presence or absence of:
- -
- Extraprostatic extension (EPE)
- -
- Seminal vesicle invasion (SVI)
- -
- Perineural invasion (Pn)
- **General Rules**:
- -
- If report lacks clear information → set value = “Not mentioned”, evidence = null, confidence = 0.0
- -
- Quote evidence verbatim; maximum 25 words per quote
- -
- Return JSON only—no other text or formatting
- -
- Confidence should reflect certainty of extraction (1.0 = completely certain, 0.0 = not found)
Appendix A.3
Mistral-Small-3.2-24B-Instruct prompt for extracting 16 parameters from pathology reports
You are a senior genitourinary pathologist with expertise in prostate cancer. Extract ONLY the 16 parameters listed below from the pathology report and return a STRICTLY valid JSON object.
CRITICAL INSTRUCTIONS:
- 1.
- Use EXACTLY the specified answer options for each parameter
- 2.
- If information is not clearly stated, use “Not mentioned”
- 3.
- For integer fields, provide only the numeric value
- 4.
- For float fields (PSA), provide only the numeric value
- 5.
- Return ONLY the JSON object - no explanations, no markdown, no commentary
OUTPUT FORMAT:
{
“WHO_ISUP_Grade_Group”: “GG1” | “GG2” | “GG3” | “GG4” | “GG5” | “Not mentioned”,
“T_Stage_TNM”: “pT0” | “pT2a” | “pT2b” | “pT2c” | “pT3a” | “pT3b” | “pT4” | “Not mentioned”,
“N_Stage_TNM”: “pNx” | “pN0” | “pN1” | “Not mentioned”,
“Number_of_Lymph_Nodes_examined”: <integer> | “Not mentioned”,
“Lymph_Nodes_with_Metastasis”: <integer> | “Not mentioned”,
“Resection_Margins”: “R0” | “R1” | “Rx” | “Not mentioned”,
“Histologic_Subtype”: “acinar” | “ductal” | “mixed” | “Not mentioned”,
“Primary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Secondary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Tertiary_Gleason_Pattern”: 3 | 4 | 5 | “Not mentioned”,
“Percentage_of_Secondary_Gleason_Pattern”: <integer> | “Not mentioned”,
“Serum_PSA_ng_per_mL”: <float> | “Not mentioned”,
“M_Stage_TNM”: “cM0” | “cM1” | “pM0” | “pM1” | “Not mentioned”,
“Extraprostatic_Extension_EPE”: “Present” | “Absent” | “Not mentioned”,
“Seminal_Vesicle_Invasion_SVI”: “Present” | “Absent” | “Not mentioned”,
“Perineural_Invasion_Pn”: “Present” | “Absent” | “Not mentioned”
}
EXTRACTION GUIDELINES:
- -
- WHO/ISUP Grade Group: Look for Grade Group, ISUP grade, or equivalent grading
- -
- T-Stage: Focus on pathologic T-stage (pT), primary tumor extent
- -
- N-Stage: Look for nodal involvement status
- -
- Lymph nodes: Count examined nodes and positive nodes separately
- -
- Margins: R0 = negative, R1 = positive, Rx = cannot be assessed
- -
- Gleason: Extract primary, secondary, and tertiary patterns with percentages
- -
- PSA: Use pretreatment serum PSA value, remove units
Extensions/Invasions: Determine presence or absence of specific invasions
Analyze the following pathology report:

Table A1.
Representative examples of GPT-4.1 extraction output with confidence scores and supporting evidence for data element expansion.
Table A1.
Representative examples of GPT-4.1 extraction output with confidence scores and supporting evidence for data element expansion.
| Parameter | Extracted Value | Confidence | Evidence Span |
|---|---|---|---|
| PSA level | 14 | 1.00 | PSA 14 ng/mL. |
| M-stage | cM0 | 0.95 | Staging without metastases. |
| EPE | Present | 1.00 | Right-sided capsular perforation with infiltration of the adjacent fatty/connective tissue. |
| SVI | Absent | 1.00 | No infiltration of the seminal vesicles and the ductus deferens segments on both sides. |
| Perineural invasion | Present | 1.00 | Extensive perineural sheath invasion. |

Table A2.
Representative examples of GPT-4.1 output for annotating external validation datasets.
Table A2.
Representative examples of GPT-4.1 output for annotating external validation datasets.
| Parameter | Extracted Value | Confidence | Evidence Span |
|---|---|---|---|
| EPE | Present | 1.00 | EXTRAPROSTATIC EXTENSION: Yes-Established (>0.8 mm). |
| Histologic Subtype | acinar | 0.95 | TUMOR HISTOLOGY: Adenocarcinoma NOS. |
| Lymph Nodes with Metastasis | 0 | 1.00 | LYMPH NODES POSITIVE: o. |
| M-Stage | pMx | 0.90 | M STAGE, PATHOLOGIC: pMX. |
| N-Stage | pN0 | 1.00 | N STAGE, PATHOLOGIC: pNO |
| Number of Lymph Nodes examined | 11 | 1.00 | LYMPH NODES EXAMINED: 11 |
| Percentage of Secondary Gleason Pattern | 10 | 1.00 | Gleason pattern 5 constitutes approximately 10% of the tissues evaluated. |
| Perineural Invasion | Present | 1.00 | MULTIFOCAL PERINEURAL INVASION BY THE CARCINOMA IS SEEN. |
| Primary Gleason Pattern | 4 | 1.00 | PRIMARY GLEASON GRADE: 4. |
| Resection Margins | R0 | 1.00 | All surgical margins free of tumor. RESIDUAL TUMOR: R0. |
| Secondary Gleason Pattern | 5 | 1.00 | SECONDARY GLEASON GRADE: 5. |
| SVI | Absent | 1.00 | BILATERAL SEMINAL VESICLES, NO EVIDENCE OF CARCINOMA SEEN. |
| PSA | 35 | 1.00 | PSA value: 35. |
| T-Stage | pT3a | 1.00 | T STAGE, PATHOLOGIC: pT3a |
| Tertiary Gleason Pattern | 3 | 1.00 | Gleason pattern 3 constitutes approximately 35% |
| WHO Grade Group | GG5 | 1.00 | Gleason score 4 + 5 a 9 |
References
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population—Based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the eighth edition of the tumor-node-metastasis staging classification for urologic cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Offermann, A.; Hohensteiner, S.; Kuempers, C.; Ribbat-Idel, J.; Schneider, F.; Becker, F.; Hupe, M.C.; Duensing, S.; Merseburger, A.S.; Kirfel, J. Prognostic value of the new prostate cancer international society of urological pathology grade groups. Front. Med. 2017, 4, 157. [Google Scholar] [CrossRef] [PubMed]
- Costello, A.J. Considering the role of radical prostatectomy in 21st century prostate cancer care. Nat. Rev. Urol. 2020, 17, 177–188. [Google Scholar] [CrossRef]
- Chung, B.H. The role of radical prostatectomy in high-risk prostate cancer. Prostate Int. 2013, 1, 95–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grignon, D.J. Prostate cancer reporting and staging: Needle biopsy and radical prostatectomy specimens. Mod. Pathol. 2018, 31, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Montironi, R.; van der Kwast, T.; Boccon-Gibod, L.; Bono, A.V.; Boccon-Gibod, L. Handling and pathology reporting of radical prostatectomy specimens. Eur. Urol. 2003, 44, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Kench, J.G.; Judge, M.; Delahunt, B.; Humphrey, P.A.; Kristiansen, G.; Oxley, J.; Rasiah, K.; Takahashi, H.; Trpkov, K.; Varma, M. Dataset for the reporting of prostate carcinoma in radical prostatectomy specimens: Updated recommendations from the International Collaboration on Cancer Reporting. Virchows Arch. 2019, 475, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Mezger, E.; Schüffler, P.J.; Sommer, W.; Fusco, F.; Hauner, K.; Schmid, S.C.; Gschwend, J.E.; Weichert, W.; Schwamborn, K. Pathological reporting of radical prostatectomy specimens following ICCR recommendation: Impact of electronic reporting tool implementation on quality and interdisciplinary communication in a large university hospital. Curr. Oncol. 2022, 29, 7245–7256. [Google Scholar] [CrossRef]
- Grothey, B.; Odenkirchen, J.; Brkic, A.; Schömig-Markiefka, B.; Quaas, A.; Büttner, R.; Tolkach, Y. Comprehensive testing of large language models for extraction of structured data in pathology. Commun. Med. 2025, 5, 96. [Google Scholar] [CrossRef]
- Yu, E.; Chu, X.; Zhang, W.; Meng, X.; Yang, Y.; Ji, X.; Wu, C. Large Language Models in Medicine: Applications, Challenges, and Future Directions. Int. J. Med. Sci. 2025, 22, 2792. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Yan, X.; Zhang, K.; Liu, D.; Cui, X.; Yang, Y.; Zhang, M.; Cao, C.; Wang, J.; Wang, X. The application of large language models in medicine: A scoping review. Iscience 2024, 27, 109713. [Google Scholar] [CrossRef] [PubMed]
- Kweon, S.; Kim, J.; Kwak, H.; Cha, D.; Yoon, H.; Kim, K.; Yang, J.; Won, S.; Choi, E. Ehrnoteqa: An llm benchmark for real-world clinical practice using discharge summaries. In Proceedings of the 38th Conference on Neural Information Processing Systems (NeurIPS 2024), Vancouver, BC, Canada, 10–15 December 2024; Volume 37, pp. 124575–124611. [Google Scholar]
- Van Veen, D.; Van Uden, C.; Blankemeier, L.; Delbrouck, J.-B.; Aali, A.; Bluethgen, C.; Pareek, A.; Polacin, M.; Reis, E.P.; Seehofnerova, A. Clinical text summarization: Adapting large language models can outperform human experts. Res. Sq. 2023, rs.3.rs-3483777. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, Q.; Chen, A.; Peng, C.; Hu, Y.; Lin, F.; Peng, X.; Huang, J.; Zhang, J.; Keloth, V. Medical foundation large language models for comprehensive text analysis and beyond. npj Digit. Med. 2025, 8, 141. [Google Scholar] [CrossRef]
- Huang, J.; Yang, D.M.; Rong, R.; Nezafati, K.; Treager, C.; Chi, Z.; Wang, S.; Cheng, X.; Guo, Y.; Klesse, L.J. A critical assessment of using ChatGPT for extracting structured data from clinical notes. npj Digit. Med. 2024, 7, 106. [Google Scholar] [CrossRef]
- Murali, L.; Gopakumar, G.; Viswanathan, D.M.; Nedungadi, P. Towards electronic health record-based medical knowledge graph construction, completion, and applications: A literature study. J. Biomed. Inform. 2023, 143, 104403. [Google Scholar] [CrossRef]
- Grothey, B.; Tolkach, Y. Large Language Models for Extraction of Structured Data in Pathology. Zenodo 2024. [Google Scholar] [CrossRef]
- Kefeli, J.; Tatonetti, N. TCGA-Reports: A machine-readable pathology report resource for benchmarking text-based AI models. Patterns 2024, 5, 100933. [Google Scholar] [CrossRef] [PubMed]
- Kefeli, J.; Tatonetti, N. TCGA-Reports: A Machine-Readable Pathology Report Resource for Benchmarking Text-Based AI Models Kefeli et al. In Mendeley Data, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2024; Available online: https://data.mendeley.com/datasets/hyg5xkznpx/1 (accessed on 10 September 2025).
- Sable, N.P.; Bakshi, G.K.; Raghavan, N.; Bakshi, H.; Sharma, R.; Menon, S.; Kumar, P.; Katdare, A.; Popat, P. Imaging Recommendations for Diagnosis, Staging, and Management of Prostate Cancer. Indian J. Med. Paediatr. Oncol. 2023, 44, 130–137. [Google Scholar] [CrossRef]
- Combes, A.D.; Palma, C.A.; Calopedos, R.; Wen, L.; Woo, H.; Fulham, M.; Leslie, S. PSMA PET-CT in the diagnosis and staging of prostate cancer. Diagnostics 2022, 12, 2594. [Google Scholar] [CrossRef]
- Rosario, E.; Rosario, D.J. Localized Prostate Cancer; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Eastham, J.; Auffenberg, G.; Barocas, D. Clinically Localized Prostate Cancer: AUA/ASTRO Guideline 2022. 2023. Available online: https://www.auanet.org/guidelines-and-quality/guidelines/clinically-localized-prostate-cancer-aua/astro-guideline-2022 (accessed on 19 September 2025).
- Burt, L.M.; Shrieve, D.C.; Tward, J.D. Factors influencing prostate cancer patterns of care: An analysis of treatment variation using the SEER database. Adv. Radiat. Oncol. 2018, 3, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, O. Assessment of the prognostic value of the 8th AJCC staging system for patients with clinically staged prostate cancer; A time to sub-classify stage IV? PLoS ONE 2017, 12, e0188450. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A.; Moch, H.; Cubilla, A.L.; Ulbright, T.M.; Reuter, V.E. The 2016 WHO classification of tumours of the urinary system and male genital organs—Part B: Prostate and bladder tumours. Eur. Urol. 2016, 70, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Team, M.A. Mistral Small 3. Available online: https://mistral.ai/news/mistral-small-3 (accessed on 17 July 2025).
- Bartels, S.; Carus, J. From text to data: Open-source large language models in extracting cancer related medical attributes from German pathology reports. Int. J. Med. Inform. 2025, 203, 106022. [Google Scholar] [CrossRef]
- Teramoto, Y.; Numbere, N.; Wang, Y.; Miyamoto, H. The clinical significance of either extraprostatic extension or microscopic bladder neck invasion alone versus both in men with pT3a prostate cancer undergoing radical prostatectomy: A proposal for a new pT3a subclassification. Am. J. Surg. Pathol. 2022, 46, 1682–1687. [Google Scholar] [CrossRef]
- Abbasi, F.; Asghari, Y.; Niazkhani, Z. Information adequacy in histopathology request forms: A milestone in making a communication bridge between confusion and clarity in medical diagnosis. Turk. J. Pathol. 2023, 39, 185. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).