Integrating Feature Selection, Machine Learning, and SHAP Explainability to Predict Severe Acute Pancreatitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Study Population and Data Collection
2.3. Outcome Definition
2.4. Feature Selection and Machine Learning Models
2.5. Statistical Analysis
2.6. Proposed Approach
3. Results
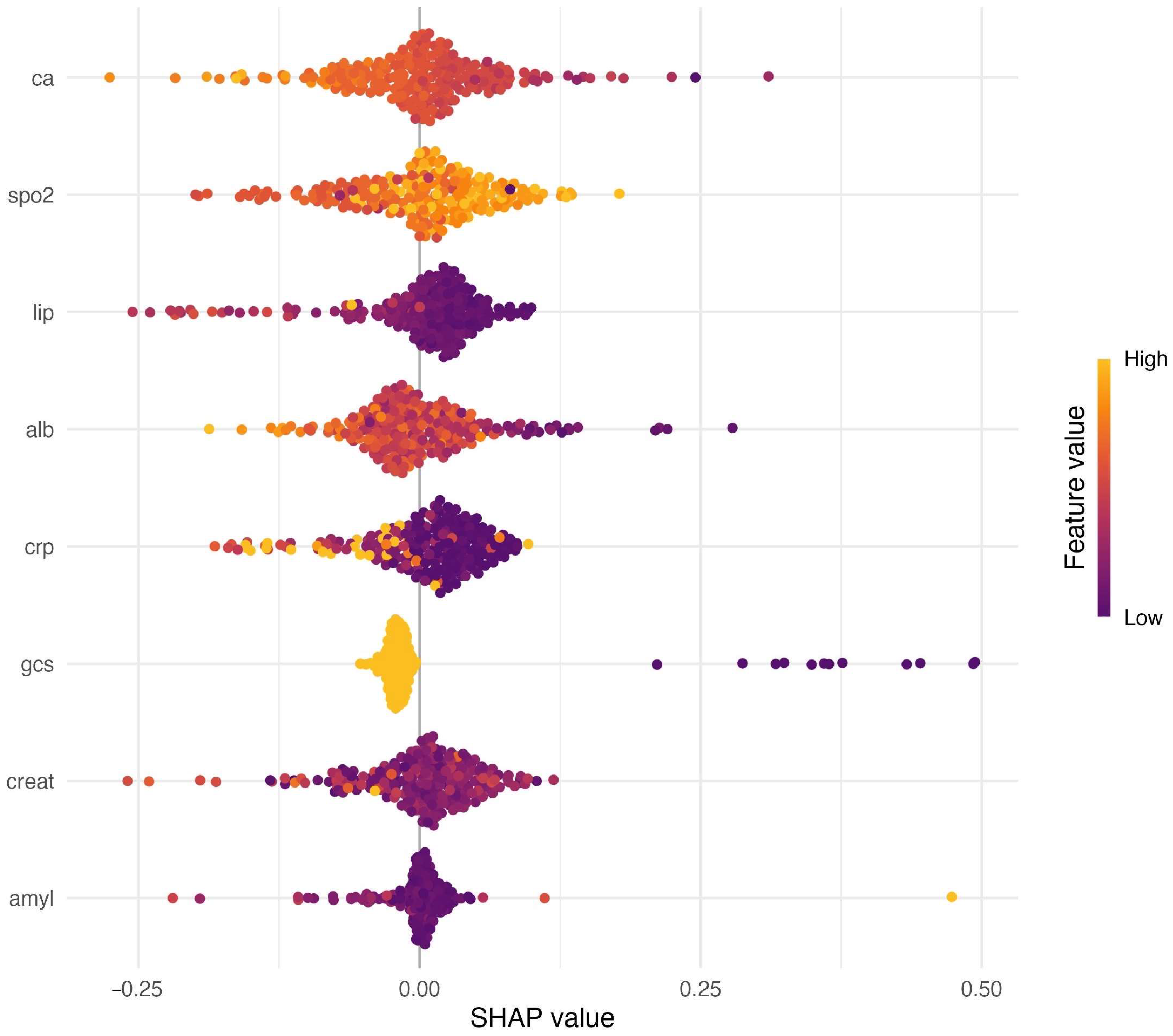
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP | Acute pancreatitis |
| APACHE II | Acute Physiology and Chronic Health Evaluation II |
| AUROC | Area under the receiver operating characteristic curve |
| BISAP | Bedside Index for Severity in Acute Pancreatitis |
| ED | Emergency department |
| GCS | Glasgow Coma Scale |
| ICU | Intensive care unit |
| kNN | k-nearest neighbors |
| LASSO | Least absolute shrinkage and selection operator |
| MARS | Multivariate adaptive regression splines |
| mRMR | Minimum redundancy–maximum relevance |
| RF | Random forest |
| RFE | Recursive feature elimination |
| RFE-RF | Recursive feature elimination using a random-forest estimator |
| ROC | Receiver operating characteristic |
| SAP | Severe acute pancreatitis |
| SHAP | Shapley additive explanations |
| SVM-RBF | Support vector machine with radial basis function kernel |
| XGBoost | Extreme gradient boosting |
References
- Pokras, S.; Ray, M.; Zheng, S.; Ding, Y.; Chen, C.-C. The Short- and Long-Term Burden of Acute Pancreatitis in the United States: A Retrospective Cohort Study. Pancreas 2021, 50, 330–340. [Google Scholar] [CrossRef]
- Boxhoorn, L.; Voermans, R.P.; Bouwense, S.A.; Bruno, M.J.; Verdonk, R.C.; Boermeester, M.A.; van Santvoort, H.C.; Besselink, M.G. Acute Pancreatitis. Lancet 2020, 396, 726–734. [Google Scholar] [CrossRef]
- Li, C.-L.; Jiang, M.; Pan, C.-Q.; Li, J.; Xu, L.-G. The Global, Regional, and National Burden of Acute Pancreatitis in 204 Countries and Territories, 1990–2019. BMC Gastroenterol. 2021, 21, 332. [Google Scholar] [CrossRef]
- Banks, P.A.; Bollen, T.L.; Dervenis, C.; Gooszen, H.G.; Johnson, C.D.; Sarr, M.G.; Tsiotos, G.G.; Vege, S.S.; Acute Pancreatitis Classification Working Group. Classification of Acute Pancreatitis—2012: Revision of the Atlanta Classification and Definitions by International Consensus. Gut 2013, 62, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Barreto, S.G.; Kaambwa, B.; Venkatesh, K.; Sasson, S.C.; Andersen, C.; Delaney, A.; Bihari, S.; Pilcher, D.; P-ANZICS Collaborative. Mortality and Costs Related to Severe Acute Pancreatitis in the Intensive Care Units of Australia and New Zealand (ANZ), 2003–2020. Pancreatology 2023, 23, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-X.; Zhao, C.-F.; Wang, S.-L.; Tu, X.-Y.; Huang, W.-B.; Chen, J.-N.; Xie, Y.; Chen, C.-R. Acute Pancreatitis: A Review of Diagnosis, Severity Prediction and Prognosis Assessment from Imaging Technology, Scoring System and Artificial Intelligence. World J. Gastroenterol. 2023, 29, 5268–5291. [Google Scholar] [CrossRef]
- Ong, Y.; Shelat, V.G. Ranson Score to Stratify Severity in Acute Pancreatitis Remains Valid—Old Is Gold. Expert. Rev. Gastroenterol. Hepatol. 2021, 15, 865–877. [Google Scholar] [CrossRef]
- Thapa, R.; Iqbal, Z.; Garikipati, A.; Siefkas, A.; Hoffman, J.; Mao, Q.; Das, R. Early Prediction of Severe Acute Pancreatitis Using Machine Learning. Pancreatology 2022, 22, 43–50. [Google Scholar] [CrossRef]
- Zhou, Y.; Ge, Y.-T.; Shi, X.-L.; Wu, K.-Y.; Chen, W.-W.; Ding, Y.-B.; Xiao, W.-M.; Wang, D.; Lu, G.-T.; Hu, L.-H. Machine Learning Predictive Models for Acute Pancreatitis: A Systematic Review. Int. J. Med. Inform. 2022, 157, 104641. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.A.B.; Alamgir, Z. Improving Mortality Prediction in Acute Pancreatitis by Machine Learning and Data Augmentation. Comput. Biol. Med. 2022, 150, 106077. [Google Scholar] [CrossRef]
- Deshmukh, F.; Merchant, S.S. Explainable Machine Learning Model for Predicting GI Bleed Mortality in the Intensive Care Unit. Am. J. Gastroenterol. 2020, 115, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Zhang, R.; Zhou, Z.; Liu, L.; Gao, J.; Xu, W.; Yu, C.; Lin, J.; Liu, X.; Xu, C.; et al. Automated Machine Learning for the Early Prediction of the Severity of Acute Pancreatitis in Hospitals. Front. Cell. Infect. Microbiol. 2022, 12, 886935. [Google Scholar] [CrossRef]
- Hong, W.; Lu, Y.; Zhou, X.; Jin, S.; Pan, J.; Lin, Q.; Yang, S.; Basharat, Z.; Zippi, M.; Goyal, H. Usefulness of Random Forest Algorithm in Predicting Severe Acute Pancreatitis. Front. Cell. Infect. Microbiol. 2022, 12, 893294. [Google Scholar] [CrossRef] [PubMed]
- Zerem, E.; Kurtcehajic, A.; Kunosić, S.; Zerem Malkočević, D.; Zerem, O. Current Trends in Acute Pancreatitis: Diagnostic and Therapeutic Challenges. World J. Gastroenterol. 2023, 29, 2747–2763. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Wang, M.; Wen, Y.; Du, F.; Jiang, L.; Geng, X.; Tang, L.; Yan, H. Predicting Acute Pancreatitis Severity with Enhanced Computed Tomography Scans Using Convolutional Neural Networks. Sci. Rep. 2023, 13, 17514. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Y.; Zhang, H.; Yin, H.; Hu, C.; Huang, Z.; Tan, Q.; Song, B.; Deng, L.; Xia, Q. Deep Learning Models for Severity Prediction of Acute Pancreatitis in the Early Phase From Abdominal Nonenhanced Computed Tomography Images. Pancreas 2023, 52, e45–e53. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification Using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature Selection Based on Mutual Information Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Society. Ser. B Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Friedman, J.H. Multivariate Adaptive Regression Splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- López Gordo, S.; Ramirez-Maldonado, E.; Fernandez-Planas, M.T.; Bombuy, E.; Memba, R.; Jorba, R. AI and Machine Learning for Precision Medicine in Acute Pancreatitis: A Narrative Review. Medicina 2025, 61, 629. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Schepers, N.J.; Bakker, O.J.; Besselink, M.G.; Ahmed Ali, U.; Bollen, T.L.; Gooszen, H.G.; van Santvoort, H.C.; Bruno, M.J.; Dutch Pancreatitis Study Group. Impact of Characteristics of Organ Failure and Infected Necrosis on Mortality in Necrotising Pancreatitis. Gut 2019, 68, 1044–1051. [Google Scholar] [CrossRef]
- Zhu, J.; Wu, L.; Wang, Y.; Fang, M.; Liu, Q.; Zhang, X. Predictive Value of the Ranson and BISAP Scoring Systems for the Severity and Prognosis of Acute Pancreatitis: A Systematic Review and Meta-Analysis. PLoS ONE 2024, 19, e0302046. [Google Scholar] [CrossRef]
- Tan, Z.; Li, G.; Zheng, Y.; Li, Q.; Cai, W.; Tu, J.; Jin, S. Advances in the Clinical Application of Machine Learning in Acute Pancreatitis: A Review. Front. Med. 2024, 11, 1487271. [Google Scholar] [CrossRef]
- Zhou, H.; Mei, X.; He, X.; Lan, T.; Guo, S. Severity Stratification and Prognostic Prediction of Patients with Acute Pancreatitis at Early Phase: A Retrospective Study. Medicine 2019, 98, e15275. [Google Scholar] [CrossRef]
- Ding, N.; Guo, C.; Li, C.; Zhou, Y.; Chai, X. An Artificial Neural Networks Model for Early Predicting In-Hospital Mortality in Acute Pancreatitis in MIMIC-III. BioMed Res. Int. 2021, 2021, 6638919. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tian, Y.; Li, S.; Wu, H.; Wang, T. Interpretable Prediction of 30-Day Mortality in Patients with Acute Pancreatitis Based on Machine Learning and SHAP. BMC Med. Inform. Decis. Mak. 2024, 24, 328. [Google Scholar] [CrossRef]
- Ning, C.; Ouyang, H.; Xiao, J.; Wu, D.; Sun, Z.; Liu, B.; Shen, D.; Hong, X.; Lin, C.; Li, J.; et al. Development and Validation of an Explainable Machine Learning Model for Mortality Prediction among Patients with Infected Pancreatic Necrosis. eClinicalMedicine 2025, 80, 103074. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ji, M.; Wang, S.; Wen, X.; Huang, P.; Shen, L.; Xu, J. Machine Learning Model Identifies Aggressive Acute Pancreatitis within 48 h of Admission: A Large Retrospective Study. BMC Med. Inform. Decis. Mak. 2022, 22, 312. [Google Scholar] [CrossRef] [PubMed]
| Variable | Non-SAP (n = 676) | SAP (n = 67) | p | Mean Difference (95% CI) |
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | 49 ± 17 | 49 ± 19 | 0.980 | |
| Male sex (n [%]) | 342 (50.6%) | 40 (59.7%) | 0.155 | |
| Comorbidities | ||||
| Coronary artery disease | 55 (8.1%) | 10 (14.9%) | 0.061 | |
| Chronic obstructive pulmonary disease | 34 (5.0%) | 1 (1.5%) | 0.357 | |
| Diabetes mellitus | 113 (16.7%) | 14 (20.9%) | 0.386 | |
| Hypertension | 183 (27.1%) | 26 (38.8%) | 0.042 * | |
| Malignancy | 49 (7.2%) | 13 (19.4%) | <0.001 * | |
| Etiology | ||||
| Biliary etiology | 279 (41.3%) | 21 (31.3%) | 0.114 | |
| Neurological/Score | ||||
| Glasgow Coma Scale (median [IQR]) | 15.0 [15.0–15.0] | 15.0 [14.0–15.0] | <0.001 * | |
| Vital Signs | ||||
| Heart rate (beats/min) | 85 ± 15 | 93 ± 20 | 0.004 * | Δ −7.45 (95% CI −12.48 to −2.41) |
| Respiratory rate (breaths/min) | 18 ± 4 | 21 ± 6 | <0.001 * | Δ −2.68 (95% CI −4.13 to −1.22) |
| Systolic blood pressure (mmHg) | 120 ± 15 | 115 ± 17 | 0.022 * | Δ 5.10 (95% CI 0.75 to 9.45) |
| Diastolic blood pressure (mmHg) | 75 ± 10 | 72 ± 14 | 0.148 | |
| Oxygen saturation (%) (median [IQR]) | 97 [95–98] | 96 [94–98] | 0.018 * | |
| Temperature (°C) | 36.8 ± 0.5 | 37.0 ± 0.6 | 0.029 * | Δ −0.17 (95% CI −0.31 to −0.02) |
| Shock Index (median [IQR]) | 0.7 [0.6–0.8] | 0.8 [0.7–1.0] | <0.001 * | |
| Clinical/Imaging Findings | ||||
| Peripancreatic fluid (n [%]) | 114 (16.9%) | 21 (31.3%) | 0.003 * | |
| Pleural effusion (n [%]) | 118 (17.5%) | 29 (43.3%) | <0.001 * |
| Variable | Non-SAP (n = 676) | SAP (n = 67) | p | Mean Difference (95% CI) |
|---|---|---|---|---|
| Proteins/Enzymes & Hepatobiliary | ||||
| Albumin (g/L) | 37.8 ± 4.9 | 33.6 ± 6.2 | <0.001 * | Δ 4.18 (95% CI 2.63 to 5.72) |
| Alkaline phosphatase (U/L) | 76.0 [52.0–104.0] | 91.0 [55.5–131.0] | 0.033 * | |
| Alanine aminotransferase (U/L) | 27.0 [9.0–82.0] | 36.0 [14.5–111.0] | 0.171 | |
| Aspartate aminotransferase (U/L) | 26.0 [15.0–51.0] | 33.0 [16.5–73.0] | 0.048 * | |
| Gamma-glutamyl transferase (U/L) | 38.0 [25.0–52.0] | 52.0 [32.0–74.0] | <0.001 * | |
| Total bilirubin (µmol/L) | 17.4 [11.7–27.9] | 16.5 [11.4–23.5] | 0.249 | |
| Direct bilirubin (µmol/L) | 6.0 [4.3–8.6] | 7.0 [5.4–9.9] | 0.002 * | |
| Pancreatic Enzymes | ||||
| Amylase (U/L]) | 576.0 [347.2–1008.2] | 648.0 [365.5–1248.5] | 0.172 | |
| Lipase (U/L) | 635.0 [352.5–1103.0] | 655.0 [362.0–1387.0] | 0.438 | |
| Renal/Electrolytes & Acid–Base | ||||
| Blood urea nitrogen (mmol/L) | 4.8 [3.7–6.4] | 5.6 [4.2–8.3] | 0.001 * | |
| Creatinine (µmol/L) | 60.5 [50.1–75.6] | 74.7 [56.0–84.8] | <0.001 * | |
| Sodium (mmol/L) | 138.0 ± 3.2 | 137.1 ± 4.4 | 0.080 | |
| Potassium (mmol/L) | 4.1 ± 0.4 | 4.3 ± 0.5 | 0.009 * | Δ −0.15 (95% CI −0.27 to −0.04) |
| Chloride (mmol/L) | 100.0 ± 5.0 | 100.5 ± 5.9 | 0.483 | |
| Calcium (mmol/L) | 2.2 [2.1–2.3] | 2.0 [1.8–2.2] | <0.001 * | |
| Bicarbonate (mmol/L) | 23.8 ± 3.2 | 22.7 ± 3.4 | 0.014 * | Δ 1.10 (95% CI 0.23 to 1.96) |
| Hematology/Inflammation & Coagulation | ||||
| White blood cells (109/L) | 10.8 [7.8–15.4] | 13.2 [9.4–17.4] | 0.023 * | |
| Neutrophils (109/L) | 6.5 [4.6–9.0] | 8.6 [5.6–11.5] | 0.005 * | |
| Lymphocytes (109/L) | 1.1 [0.8–1.7] | 1.0 [0.7–1.5] | 0.164 | |
| Hematocrit (%) | 41.1 ± 6.1 | 42.2 ± 6.3 | 0.151 | |
| Platelets (109/L) | 214.6 ± 70.9 | 202.5 ± 77.8 | 0.224 | |
| C-reactive protein (mg/L) | 31.6 [6.4–149.1] | 23.3 [6.9–338.3] | 0.886 | |
| Procalcitonin (ng/mL]) | 0.07 [0.01–0.55] | 0.07 [0.01–0.55] | 0.598 | |
| Lactate dehydrogenase (U/L) | 245.9 ± 102.9 | 270.7 ± 134.7 | 0.148 | |
| Prothrombin time (s) | 11.8 ± 2.1 | 12.4 ± 2.2 | 0.040 * | Δ −0.59 (95% CI −1.15 to −0.03) |
| Activated partial thromboplastin time (s) | 30.1 ± 5.1 | 31.4 ± 5.6 | 0.060 | |
| International normalized ratio | 1.00 ± 0.18 | 1.04 ± 0.18 | 0.087 | |
| D-dimer (mg/L FEU) | 0.88 [0.24–2.85] | 1.77 [0.42–5.62] | 0.011 * |
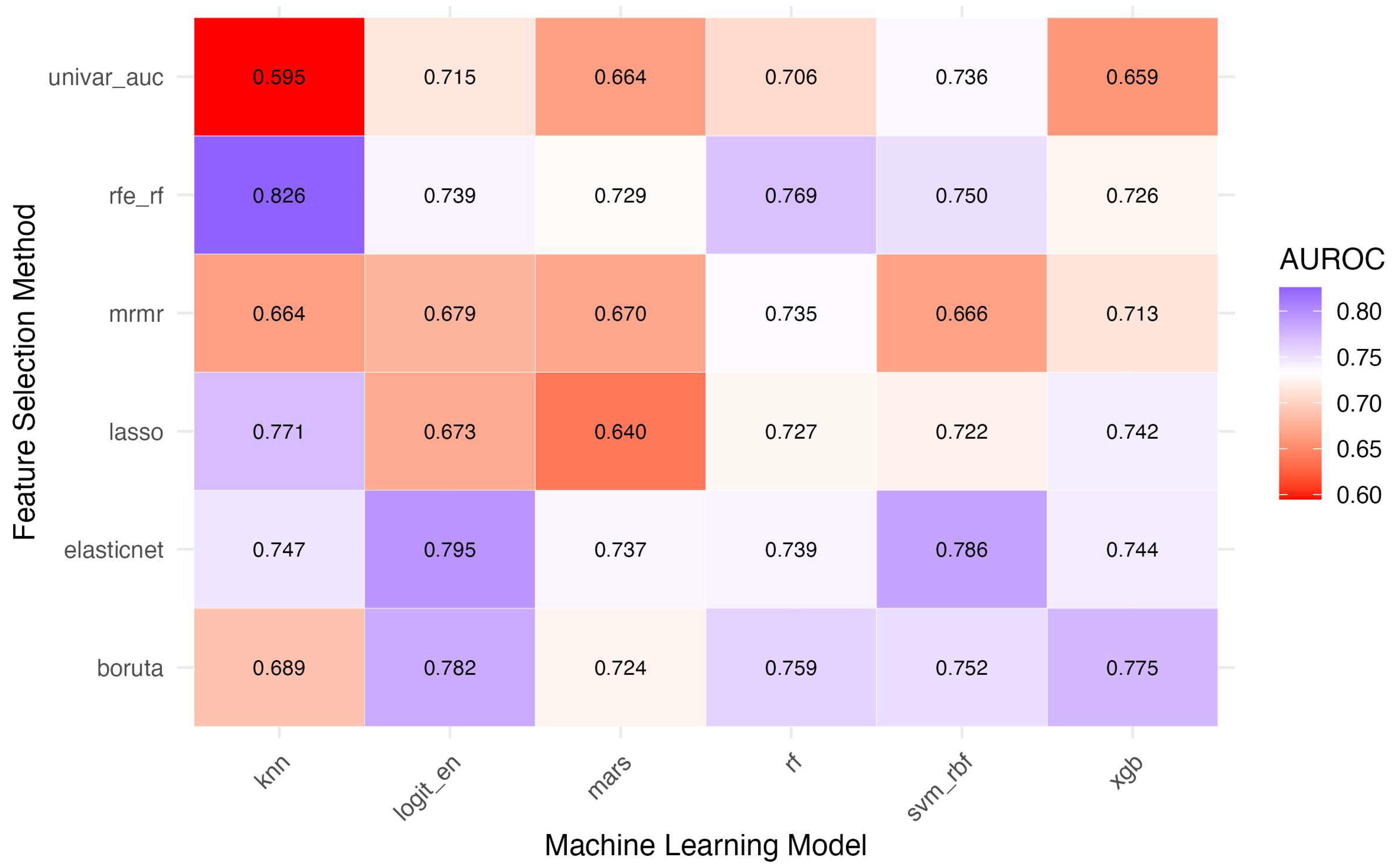
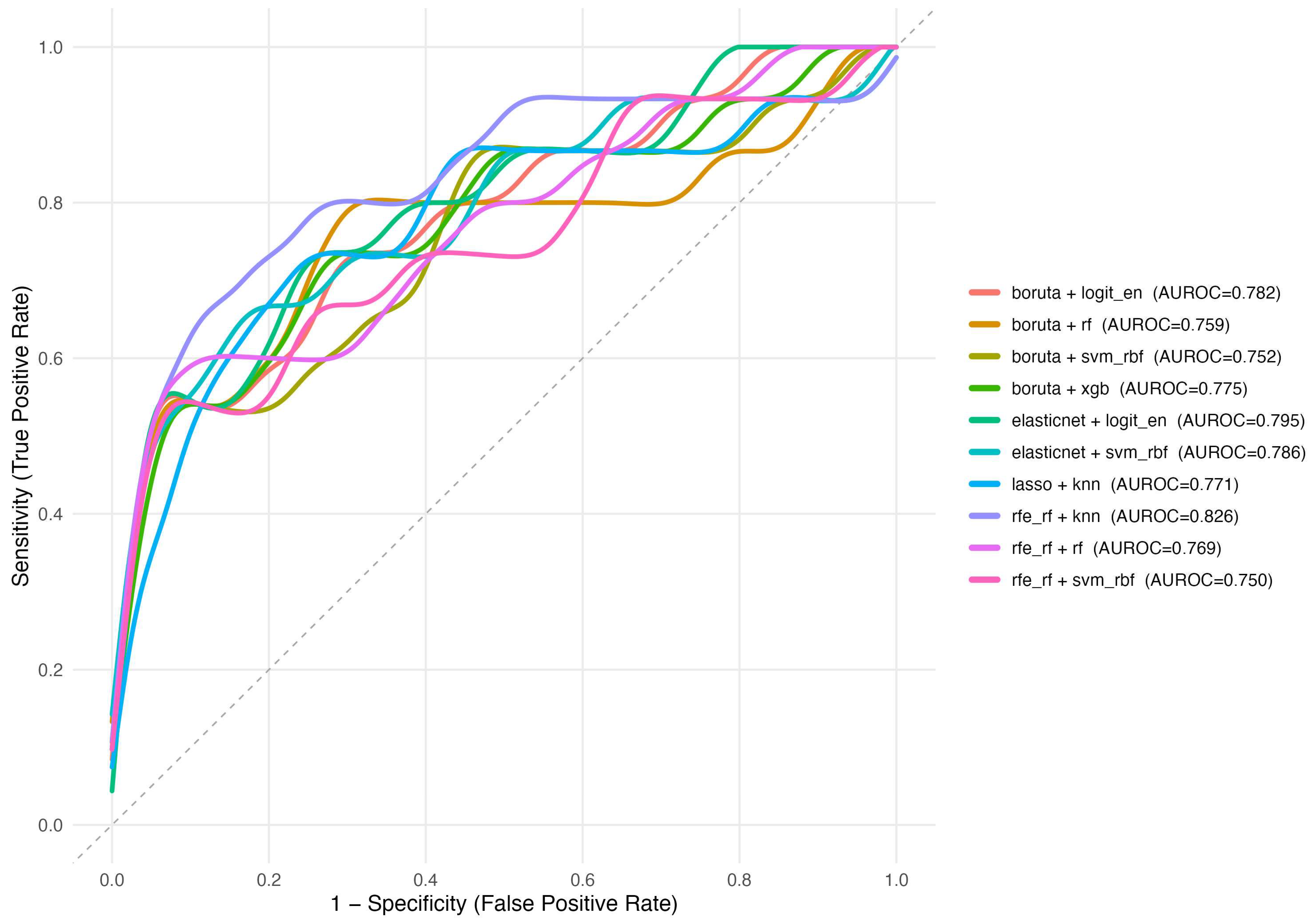
| Feature Selection | Model | AUROC (95% CI) | F1 | Precision | Recall | Log Loss | Brier |
|---|---|---|---|---|---|---|---|
| Recursive Feature Elimination (RF) | k-Nearest Neighbors | 0.826 (0.686–0.965) | 0.319 | 0.204 | 0.733 | 0.504 | 0.159 |
| Elastic Net Selection | Logistic Regression (Elastic Net) | 0.795 (0.661–0.929) | 0.302 | 0.211 | 0.533 | 0.503 | 0.154 |
| Elastic Net Selection | Support Vector Machine (RBF) | 0.786 (0.637–0.936) | 0.421 | 0.348 | 0.533 | 0.399 | 0.111 |
| Boruta | Logistic Regression (Elastic Net) | 0.782 (0.642–0.922) | 0.320 | 0.229 | 0.533 | 0.521 | 0.162 |
| Boruta | Extreme Gradient Boosting (XGBoost) | 0.775 (0.628–0.921) | 0.348 | 0.258 | 0.533 | 0.663 | 0.235 |
| LASSO (L1) Selection | k-Nearest Neighbors | 0.771 (0.616–0.927) | 0.317 | 0.208 | 0.667 | 0.505 | 0.158 |
| Recursive Feature Elimination (RF) | Random Forest (ranger) | 0.769 (0.622–0.916) | 0.444 | 0.381 | 0.533 | 0.284 | 0.077 |
| Boruta | Random Forest (ranger) | 0.759 (0.589–0.928) | 0.485 | 0.444 | 0.533 | 0.316 | 0.085 |
| Boruta | Support Vector Machine (RBF) | 0.752 (0.596–0.909) | 0.421 | 0.348 | 0.533 | 0.360 | 0.098 |
| Recursive Feature Elimination (RF) | Support Vector Machine (RBF) | 0.750 (0.593–0.907) | 0.308 | 0.216 | 0.533 | 0.470 | 0.139 |
| Elastic Net Selection | k-Nearest Neighbors | 0.747 (0.583–0.911) | 0.286 | 0.182 | 0.667 | 1.037 | 0.157 |
| Elastic Net Selection | Extreme Gradient Boosting (XGBoost) | 0.744 (0.582–0.905) | 0.367 | 0.265 | 0.600 | 0.664 | 0.235 |
| LASSO (L1) Selection | Extreme Gradient Boosting (XGBoost) | 0.742 (0.581–0.904) | 0.389 | 0.333 | 0.467 | 0.288 | 0.080 |
| Elastic Net Selection | Random Forest (ranger) | 0.739 (0.570–0.908) | 0.471 | 0.421 | 0.533 | 0.330 | 0.090 |
| Recursive Feature Elimination (RF) | Logistic Regression (Elastic Net) | 0.739 (0.580–0.898) | 0.356 | 0.267 | 0.533 | 0.584 | 0.196 |
| Elastic Net Selection | Multivariate Adaptive Regression Splines | 0.737 (0.594–0.881) | 0.226 | 0.149 | 0.467 | 0.609 | 0.181 |
| Univariate AUC filter | Support Vector Machine (RBF) | 0.736 (0.570–0.902) | 0.286 | 0.195 | 0.533 | 0.524 | 0.160 |
| Minimum Redundancy–Maximum Relevance | Random Forest (ranger) | 0.735 (0.572–0.898) | 0.500 | 0.538 | 0.467 | 0.253 | 0.065 |
| Recursive Feature Elimination (RF) | Multivariate Adaptive Regression Splines | 0.729 (0.563–0.895) | 0.277 | 0.180 | 0.600 | 0.536 | 0.162 |
| LASSO (L1) Selection | Random Forest (ranger) | 0.727 (0.556–0.897) | 0.500 | 0.471 | 0.533 | 0.275 | 0.072 |
| Recursive Feature Elimination (RF) | Extreme Gradient Boosting (XGBoost) | 0.726 (0.548–0.904) | 0.432 | 0.364 | 0.533 | 0.693 | 0.250 |
| Boruta | Multivariate Adaptive Regression Splines | 0.724 (0.567–0.881) | 0.208 | 0.123 | 0.667 | 0.542 | 0.178 |
| LASSO (L1) Selection | Support Vector Machine (RBF) | 0.722 (0.555–0.889) | 0.286 | 0.206 | 0.467 | 0.441 | 0.128 |
| Univariate AUC filter | Logistic Regression (Elastic Net) | 0.715 (0.539–0.890) | 0.348 | 0.258 | 0.533 | 0.578 | 0.193 |
| Minimum Redundancy–Maximum Relevance | Extreme Gradient Boosting (XGBoost) | 0.713 (0.554–0.871) | 0.387 | 0.375 | 0.400 | 0.276 | 0.074 |
| Univariate AUC filter | Random Forest (ranger) | 0.706 (0.528–0.883) | 0.467 | 0.467 | 0.467 | 0.281 | 0.074 |
| Boruta | k-Nearest Neighbors | 0.689 (0.540–0.837) | 0.200 | 0.120 | 0.600 | 1.041 | 0.214 |
| Minimum Redundancy–Maximum Relevance | Logistic Regression (Elastic Net) | 0.679 (0.481–0.878) | 0.286 | 0.195 | 0.533 | 0.548 | 0.178 |
| LASSO (L1) Selection | Logistic Regression (Elastic Net) | 0.673 (0.489–0.857) | 0.314 | 0.222 | 0.533 | 0.612 | 0.210 |
| Minimum Redundancy–Maximum Relevance | Multivariate Adaptive Regression Splines | 0.670 (0.511–0.828) | 0.238 | 0.185 | 0.333 | 0.349 | 0.102 |
| Minimum Redundancy–Maximum Relevance | Support Vector Machine (RBF) | 0.666 (0.467–0.866) | 0.291 | 0.200 | 0.533 | 0.525 | 0.163 |
| Univariate AUC filter | Multivariate Adaptive Regression Splines | 0.664 (0.516–0.813) | 0.179 | 0.122 | 0.333 | 0.564 | 0.153 |
| Minimum Redundancy–Maximum Relevance | k-Nearest Neighbors | 0.664 (0.476–0.852) | 0.207 | 0.125 | 0.600 | 1.355 | 0.214 |
| Univariate AUC filter | Extreme Gradient Boosting (XGBoost) | 0.659 (0.484–0.834) | 0.364 | 0.333 | 0.400 | 0.322 | 0.076 |
| LASSO (L1) Selection | Multivariate Adaptive Regression Splines | 0.640 (0.474–0.806) | 0.203 | 0.136 | 0.400 | 0.550 | 0.169 |
| Univariate AUC filter | k-Nearest Neighbors | 0.595 (0.403–0.787) | 0.128 | 0.071 | 0.600 | 0.797 | 0.292 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustaalioğlu, İ.; Ak, R. Integrating Feature Selection, Machine Learning, and SHAP Explainability to Predict Severe Acute Pancreatitis. Diagnostics 2025, 15, 2473. https://doi.org/10.3390/diagnostics15192473
Ustaalioğlu İ, Ak R. Integrating Feature Selection, Machine Learning, and SHAP Explainability to Predict Severe Acute Pancreatitis. Diagnostics. 2025; 15(19):2473. https://doi.org/10.3390/diagnostics15192473
Chicago/Turabian StyleUstaalioğlu, İzzet, and Rohat Ak. 2025. "Integrating Feature Selection, Machine Learning, and SHAP Explainability to Predict Severe Acute Pancreatitis" Diagnostics 15, no. 19: 2473. https://doi.org/10.3390/diagnostics15192473
APA StyleUstaalioğlu, İ., & Ak, R. (2025). Integrating Feature Selection, Machine Learning, and SHAP Explainability to Predict Severe Acute Pancreatitis. Diagnostics, 15(19), 2473. https://doi.org/10.3390/diagnostics15192473






