Comparison of Five Assays for the Detection of Anti-dsDNA Antibodies and Their Correlation with Complement Consumption
Abstract
1. Introduction
- Farr Radioimmunoassay (RIA): This utilizes radiolabeled DNA to detect anti-dsDNA antibodies. It is known for its high sensitivity and specificity, although its use is less common due to concerns about radiation exposure. It detects anti-dsDNA antibodies of all occurring immunoglobulin isotypes: IgG, IgM, and IgA.
- Crithidia luciliae immunofluorescence test (CLIFT): This uses a kinetoplastid parasite, Crithidia luciliae, as the substrate for detecting anti-dsDNA antibodies of the IgG isotype. It is known to be highly specific for SLE but offers low sensitivity [7].
- Enzyme-linked immunosorbent assay (ELISA): This is a widely used technique for measuring the presence and levels of anti-dsDNA antibodies, providing quantitative results that aid in assessing disease activity. Most anti-dsDNA ELISAs are highly sensitive but have limited specificity, as they detect both high- and low-avidity antibodies, even though only high-avidity antibodies are clinically relevant for SLE [8]. Only a few of the commercially available anti-dsDNA ELISAs are designed to compensate for this lack of specificity by using a high salt concentration in the buffers to remove low-avidity antibodies [9]. Most commercial ELISAs detect anti-dsDNA IgG.
- Fluorescent enzyme immunoassay (FEIA): This method is similar to ELISA but uses fluorescence for the detection of anti-dsDNA IgG antibodies. FEIA is fully automated, offering ease of use; however, it has similar limitations to those of the ELISAs.
- Addressable laser bead assay (ALBIA): This modern multiplex method enables the simultaneous detection of multiple autoantibodies, including anti-dsDNA IgG, in a single test, and is based on a laser detection method. It can offer a more comprehensive autoimmune profile in one reaction.
- Chemiluminescence immunoassay (CIA): This modern technology is based on antigen-coated paramagnetic beads and chemiluminescence as the detection method [10]. Due to the stringent washing procedure, made possible through the wash steps with magnetic fixation of the beads, this assay is highly specific for SLE, as most low-avidity antibodies are removed. The antibodies detected are of the IgG class.
- Particle-based multi-analyte technology (PMAT): This is a novel method for measuring IgG isotype autoantibodies to dsDNA, which has recently become available to laboratories [11]. Similarly to ALBIA, PMAT offers the simultaneous detection of multiple autoantibodies using an advanced and sensitive detection approach based on fluorescence.
2. Materials and Methods
2.1. Patient Samples
2.2. Measurement of Anti-dsDNA Antibodies
2.3. Measurement of Complement Consumption
2.4. Statistical Analysis
3. Results
3.1. Frequency and Intersection of Anti-dsDNA Positivity Across Platforms
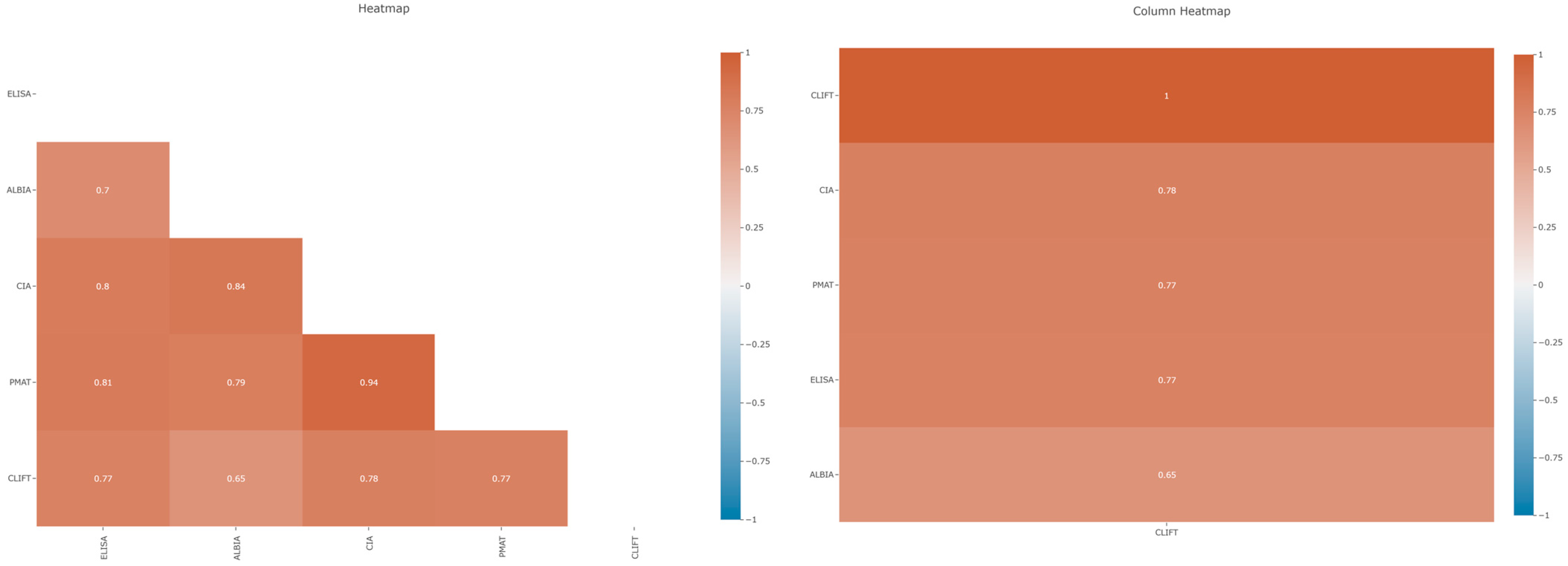
3.2. Correlation Between Anti-dsDNA Assays
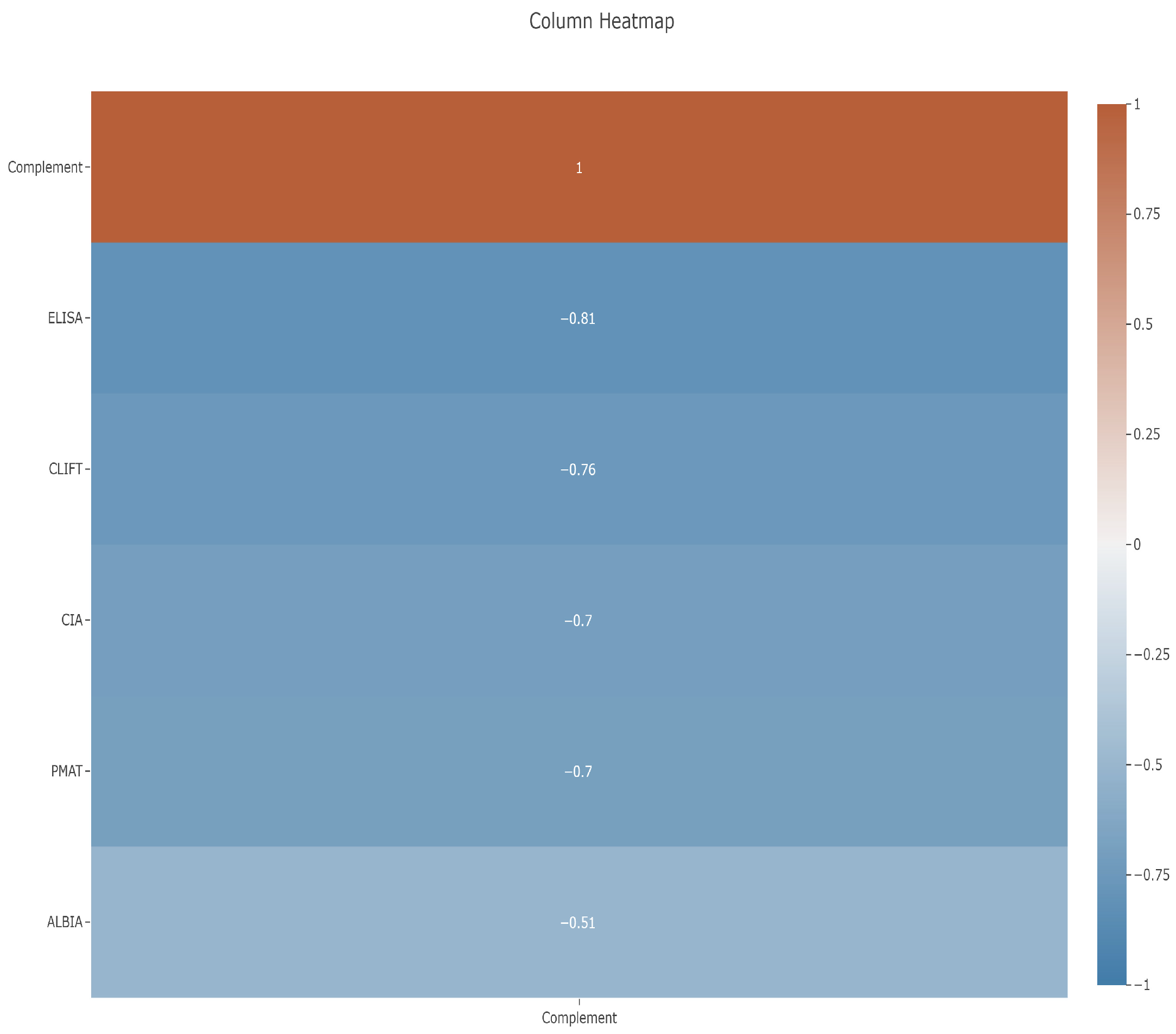
3.3. Correlation Between Anti-dsDNA Assays and Complements C3 and C4
4. Discussion
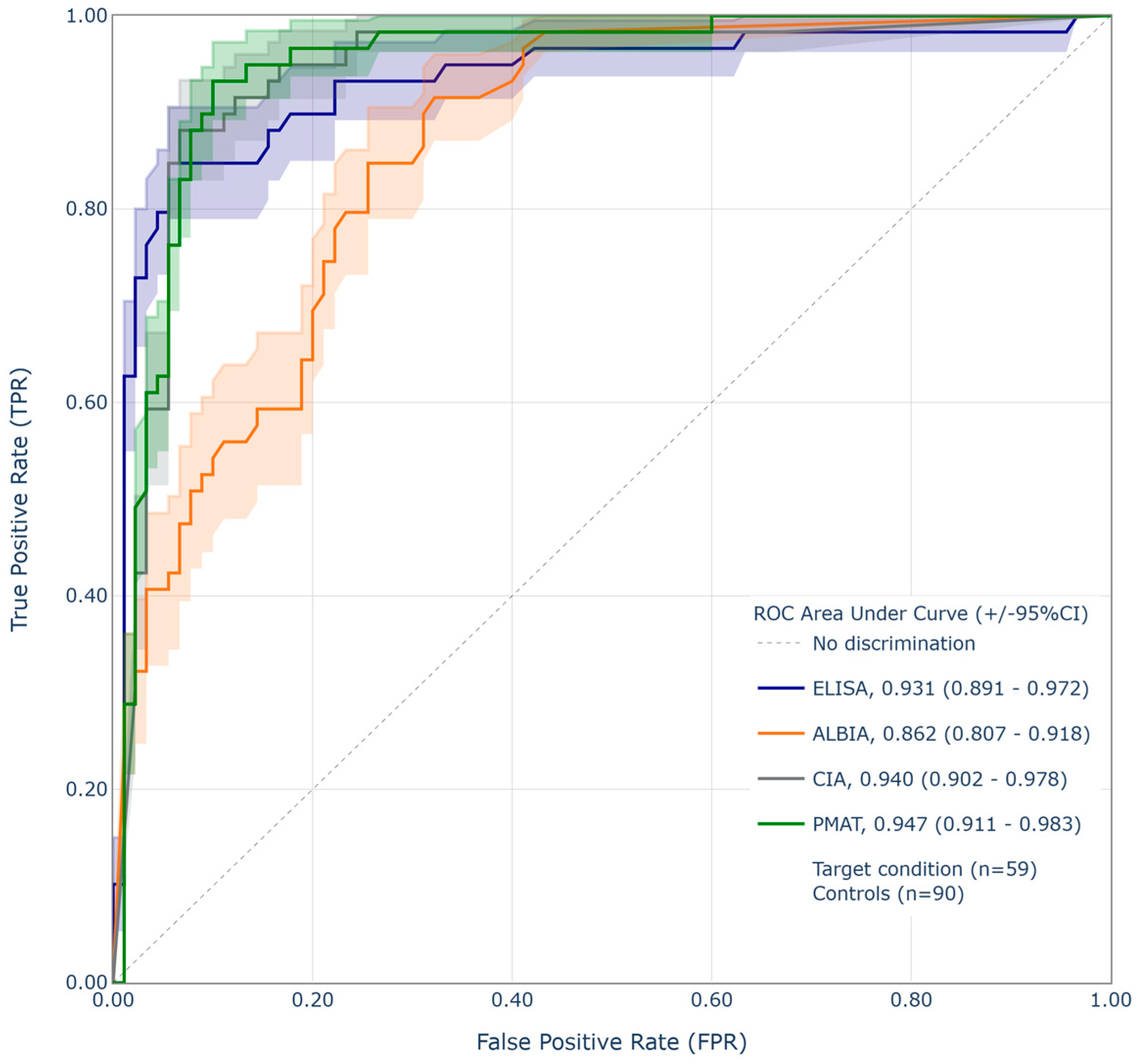
4.1. Correlation of Different Anti-dsDNA Assays
4.2. Performance Relative to CLIFT
4.3. Correlation Between Anti-dsDNA Assays and C3 as Proxy for Disease Activity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann. Rheum. Dis. 2019, 78, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Villalta, D.; Bizzaro, N.; Bassi, N.; Zen, M.; Gatto, M.; Ghirardello, A.; Iaccarino, L.; Punzi, L.; Doria, A.; Rieux-Laucat, F. Anti-dsDNA antibody isotypes in systemic lupus erythematosus: IgA in addition to IgG anti-dsDNA help to identify glomerulonephritis and active disease. PLoS ONE 2013, 8, e71458. [Google Scholar] [CrossRef]
- Witte, T. IgM antibodies against dsDNA in SLE. Clin. Rev. Allergy Immunol. 2008, 34, 345–347. [Google Scholar] [CrossRef]
- Lee, A.Y.S. IgA anti-dsDNA antibodies: A neglected serological parameter in systemic lupus erythematosus. Lupus 2022, 31, 137–142. [Google Scholar] [CrossRef]
- Mummert, E.; Fritzler, M.J.; Sjöwall, C.; Bentow, C.; Mahler, M. The clinical utility of anti-double-stranded DNA antibodies and the challenges of their determination. J. Immunol. Methods 2018, 459, 11–19. [Google Scholar] [CrossRef]
- Infantino, M.; Meacci, F.; Bentow, C.; Martis, P.; Benucci, M.; Afeltra, A.; Rigon, A.; Atzeni, F.; Sarzi-Puttini, P.; Manfredi, M.; et al. Clinical comparison of QUANTA Flash dsDNA chemiluminescent immunoassay with four current assays for the detection of anti-dsDNA autoantibodies. J. Immunol. Res. 2015, 2015, 902821. [Google Scholar] [CrossRef]
- Aguilera, A.T.; Serrano, R.B.; Navas, A.; Molina, J.A.; Romero, P.A.; Roger, A.J. Longitudinal study of patients with discrepant results in CLIFT and a solid-phase dsDNA antibody assay: Does a gold standard dsDNA assay exist? Lupus Sci. Med. 2023, 10, e000984. [Google Scholar] [CrossRef] [PubMed]
- Villalta, D.; Romelli, P.B.; Savina, C.; Bizzaro, N.; Tozzoli, R.; Tonutti, E.; Ghirardello, A.; Doria, A. Anti-dsDNA antibody avidity determination by a simple reliable ELISA method for SLE diagnosis and monitoring. Lupus 2003, 12, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Fiegel, F.; Buhl, A.; Jaekel, H.-P.; Werle, E.; Schmolke, M.; Ollert, M.; Luppa, P. Autoantibodies to double-stranded DNA—Intermethod comparison between four commercial immunoassays and a research biosensor-based device. Lupus 2010, 19, 957–964. [Google Scholar] [CrossRef]
- Mahler, M.; Bentow, C.; Serra, J.; Fritzler, M.J. Detection of autoantibodies using chemiluminescence technologies. Immunopharmacol. Immunotoxicol. 2016, 38, 14–20. [Google Scholar]
- Richards, M.; La Torre, I.G.-D.; González-Bello, Y.C.; Mercado, M.V.-D.; Andrade-Ortega, L.; Medrano-Ramírez, G.; Navarro-Zarza, J.E.; Maradiaga-Ceceña, M.; Loyo, E.; Rojo-Mejía, A.; et al. Autoantibodies to Mi-2 alpha and Mi-2 beta in patients with idiopathic inflammatory myopathy. Rheumatology 2019, 58, 1655–1661. [Google Scholar] [CrossRef]
- Ayano, M.; Horiuchi, T. Complement as a Biomarker for Systemic Lupus Erythematosus. Biomolecules 2023, 13, 367. [Google Scholar] [CrossRef]
- Yuan, W.; Cao, H.; Wan, P.; Shi, R.; Zhou, S.; Zheng, J. Clinical evaluation of total and high-avidity anti-dsDNA antibody assays for the diagnosis of systemic lupus erythematosus. Lupus 2019, 28, 1387–1396. [Google Scholar] [CrossRef]
- Jaekell, H.P.; Trabandt, A.; Grobe, N.; Werle, E. Anti-dsDNA antibody subtypes and anti-C1q antibodies: Toward a more reliable diagnosis and monitoring of systemic lupus erythematosus and lupus nephritis. Lupus 2006, 15, 335–345. [Google Scholar] [CrossRef]
- Infantino, M.; Nagy, E.; Bizzaro, N.; Fischer, K.; Bossuyt, X.; Damoiseaux, J. Anti-dsDNA antibodies in the classification criteria of systemic lupus erythematosus. J. Transl. Autoimmun. 2022, 5, 100139. [Google Scholar] [CrossRef]
- Bizzaro, N.; Villalta, D.; Bini, V.; Migliorini, P.; Franceschini, F.; Piantoni, S.; Garrafa, E.; Riccieri, V.; Fioravanti, A.; Bellisai, F.; et al. Multiparametric autoantibody analysis: A new paradigm for the diagnosis of connective tissue diseases. Arthritis Res. Ther. 2022, 24, 278. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Practice Surveys. Available online: https://www.cap.org/advocacy/latest-news-and-practice-data/practice-surveys (accessed on 27 June 2025).
- UK NEQAS for Antibodies to Nuclear and Related Antigens. UK NEQAS Immunology, Immunochemistry & Allergy. 2024. Available online: https://www.immqas.org.uk/ (accessed on 27 June 2025).
- Infantino, M.; Palterer, B.; Previtali, G.; Alessio, M.; Villalta, D.; Carbone, T.; Platzgummer, S.; Paura, G.; Castiglione, C.; Fabris, M.; et al. Comparison of current methods for anti-dsDNA antibody detection and reshaping diagnostic strategies. Scand. J. Immunol. 2022, 96, e13220. [Google Scholar] [CrossRef] [PubMed]
- Zaminski, D.; Saxena, A.; Izmirly, P.; Buyon, J.P.; Belmont, H.M. Clinical implications of discordance between anti-dsDNA antibodies by multiplex flow immunoassay and Crithidia luciliae assay in a multiethnic racial cohort of patients with SLE. Lupus Sci. Med. 2023, 10, e001012. [Google Scholar] [CrossRef]
- He, S.; Wu, X.; Li, L.; Jiang, K.; He, Q.; Xie, L. A comparison of the chemiluminescence immunoassay and Crithidia luciliae immunofluorescence test in detecting anti-dsDNA antibodies and assessing the activity of systemic lupus erythematosus. Lupus 2023, 32, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, K.; Wang, X.; Li, T.; Guo, L.; Gu, L.; Chen, Z.; Sun, F.; Wang, H.; Li, J.; et al. The performance of different anti-dsDNA autoantibodies assays in Chinese systemic lupus erythematosus patients. Clin. Rheumatol. 2018, 37, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.; Yang, J.; Ren, H.; Zhang, J.; Chen, S.; Ren, J.; Zhou, L. Potential biomarkers for diagnosis and assessment of disease activity in systemic lupus erythematosus. Int. Immunopharmacol. 2022, 111, 109155. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Ren, L.; Sun, G.; Yu, T.; Yao, Y.; Wang, L.; Liu, F.; Zhang, L.; He, X.; Liu, M. Anti-dsDNA, anti-nucleosome, anti-C1q, and anti-histone antibodies as markers of active lupus nephritis and systemic lupus erythematosus disease activity. Immun. Inflamm. Dis. 2021, 9, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Andrejevic, S.; Jeremic, I.; Sefik-Bukilica, M.; Nikolic, M.; Stojimirovic, B.; Bonaci-Nikolic, B. Immunoserological parameters in SLE: High-avidity anti-dsDNA detected by ELISA are the most closely associated with the disease activity. Clin. Rheumatol. 2013, 32, 1619–1626. [Google Scholar] [CrossRef]


| Technology | CLIFT | ELISA | ALBIA | CIA | PMAT |
|---|---|---|---|---|---|
| Platform | EUROPattern | QUANTA Lite HA | BioPlex 2200 | QUANTA Flash | Aptiva |
| Manufacturer | Euroimmun | Werfen | Bio-Rad | Werfen | Werfen |
| Assay time | 90 min | 90 min | 45 min | 30 min | 30 min |
| Detection | Semi-quantitative | Quantitative | |||
| Analytical measuring range | N/A | 12.3–1000 IU/mL | 1–300 IU/mL | 9.8–666.9 IU/mL | 2.30–814.10 IU/mL |
| Cut-off value | N/A | ≥30 IU/mL | ≥5 IU/mL | ≥35 IU/mL | >35.00 IU/mL |
| Interpretation | N/A | ≤30 negative >30 positive | 1–5 negative 5–9 indeterminate >9 positive | 9.8–35 negative 35–45 equivocal >45 positive | 2.3–27.00 IU/mL negative 27.00–35.00 IU/mL indeterminate >35.00 IU/mL positive |
| Solid phase | Slide | Well | Bead | ||
| Antigen source | Crithidia luciliae | Native calf thymus | Synthetic dsDNA | ||
| Spearman | CLIFT | ALBIA | ELISA | CIA | PMAT |
|---|---|---|---|---|---|
| CLIFT | |||||
| ALBIA | 0.65 (0.54–0.73) | ||||
| ELISA | 0.77 (0.69–0.83) | 0.70 (0.61–0.78) | |||
| CIA | 0.78 (0.71–0.84) | 0.84 (0.78–0.86) | 0.80 (0.74–0.86) | ||
| PMAT | 0.77 (0.70–0.83) | 0.79 (0.72–0.84) | 0.81 (0.75–0.86) | 0.94 (0.91–0.96) |
| Assay | Original Cut-Off | Threshold | Relative Specificity % | Relative Sensitivity % |
|---|---|---|---|---|
| PMAT | 35 IU/mL | 25.9 IU/mL | 94.4 | 76.3 |
| ALBIA | 10 IU/mL | 170 IU/mL | 94.4 | 42.4 |
| ELISA | ≤30 IU/mL | 70.7 IU/mL | 94.4 | 84.7 |
| CIA | ≥35 IU/mL | 144.8 IU/mL | 94.4 | 84.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricchiuti, V.; Obney, J.; Holloway, B.; Aure, M.A.; Shapiro, M.; Bentow, C.; Mahler, M. Comparison of Five Assays for the Detection of Anti-dsDNA Antibodies and Their Correlation with Complement Consumption. Diagnostics 2025, 15, 2430. https://doi.org/10.3390/diagnostics15192430
Ricchiuti V, Obney J, Holloway B, Aure MA, Shapiro M, Bentow C, Mahler M. Comparison of Five Assays for the Detection of Anti-dsDNA Antibodies and Their Correlation with Complement Consumption. Diagnostics. 2025; 15(19):2430. https://doi.org/10.3390/diagnostics15192430
Chicago/Turabian StyleRicchiuti, Vincent, Jacob Obney, Brooke Holloway, Mary Ann Aure, Marti Shapiro, Chelsea Bentow, and Michael Mahler. 2025. "Comparison of Five Assays for the Detection of Anti-dsDNA Antibodies and Their Correlation with Complement Consumption" Diagnostics 15, no. 19: 2430. https://doi.org/10.3390/diagnostics15192430
APA StyleRicchiuti, V., Obney, J., Holloway, B., Aure, M. A., Shapiro, M., Bentow, C., & Mahler, M. (2025). Comparison of Five Assays for the Detection of Anti-dsDNA Antibodies and Their Correlation with Complement Consumption. Diagnostics, 15(19), 2430. https://doi.org/10.3390/diagnostics15192430






