Bug Wars: Artificial Intelligence Strikes Back in Sepsis Management
Abstract
1. Introduction
2. Methods
2.1. Study Design and Eligibility Criteria
2.2. Information Sources and Search Strategy
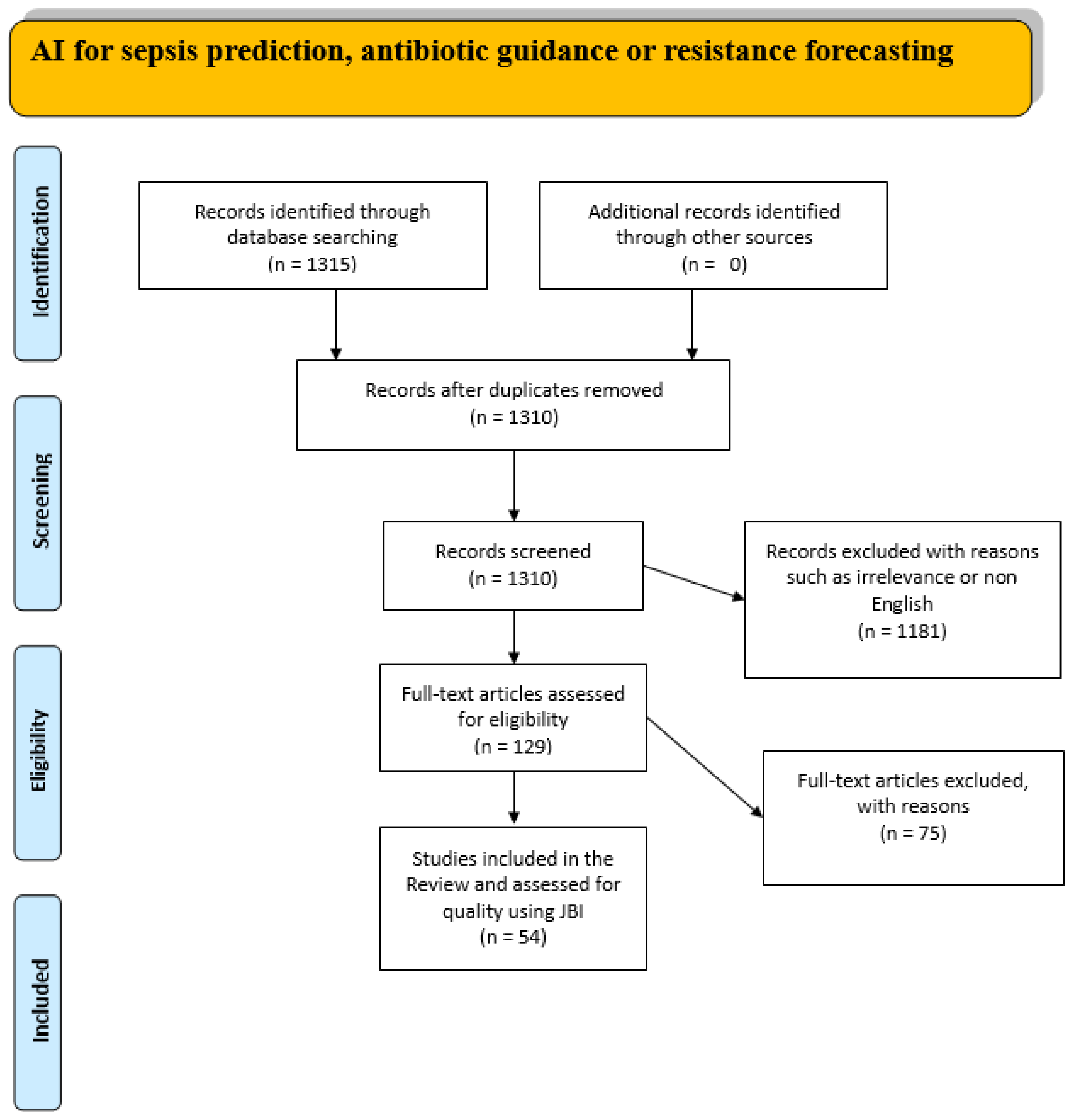
2.3. Study Selection Process
2.4. Quality Assessment
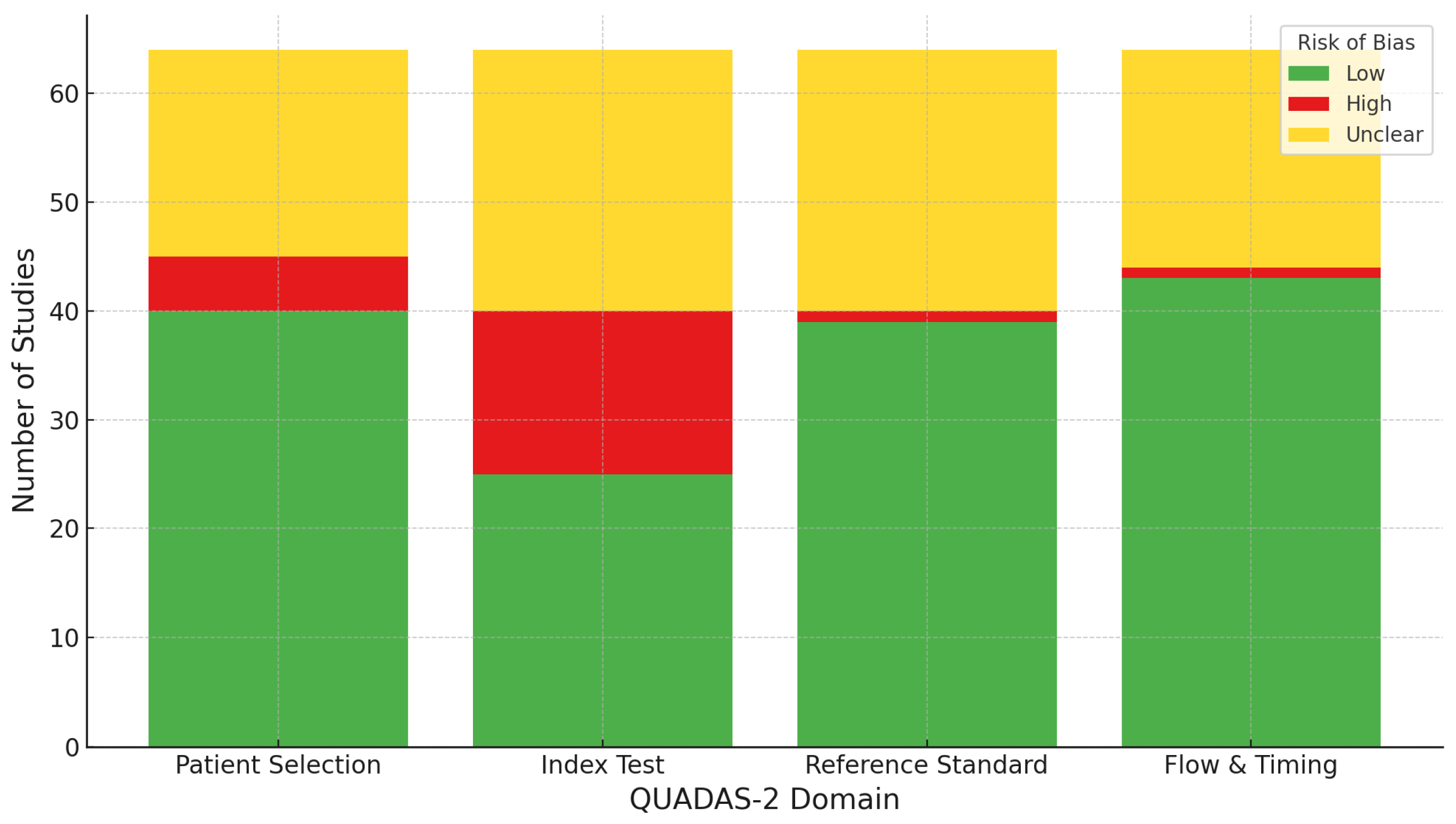
2.5. Risk of Bias Assessment
3. Results
3.1. Sepsis Detection in Daily Clinical Practice
3.2. AI in Sepsis Prediction
3.3. Limitations in Current Antibiotic Management of Sepsis
3.4. Antibiotic Optimization Through AI: Dosing and De-Escalation Strategies
3.5. Resistance Forecasting
3.6. Barriers to Clinical Adoption of AI in Sepsis Care: Challenges in Data Integration, Interpretability, and Ethical Implementation
4. Future Directions and Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cassini, A.; Allegranzi, B.; Fleischmann-Struzek, C.; Kortz, T.; Markwart, R.; Saito, H.; Bonet, M.; Brizuela, V.; Mehrtash, H.; Mingard, Ö.T.; et al. Global Report on the Epidemiology and Burden on Sepsis: Current Evidence, Identifying Gaps and Future Directions; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Global burden of disease and sepsis. Arch. Dis. Child. 2020, 105, 210. [CrossRef]
- Dantes, R.B.; Kaur, H.; Bouwkamp, B.A.; Haass, K.A.; Patel, P.; Dudeck, M.A.; Srinivasan, A.; Magill, S.S.; Wilson, W.W.; Whitaker, M.; et al. Sepsis Program Activities in Acute Care Hospitals—National Healthcare Safety Network, United States, 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 907–911. [Google Scholar] [CrossRef]
- Via, L.L.; Sangiorgio, G.; Stefani, S.; Marino, A.; Nunnari, G.; Cocuzza, S.; La Mantia, I.; Cacopardo, B.; Stracquadanio, S.; Spampinato, S.; et al. The Global Burden of Sepsis and Septic Shock. Epidemiologia 2024, 5, 456–478. [Google Scholar] [CrossRef]
- Ellaithy, I.; Elshiekh, H.; Elshennawy, S.; Elshenawy, S.; Al-Shaikh, B.; Ellaithy, A. Sepsis as a cause of death among elderly cancer patients: An updated SEER database analysis 2000–2021. Ann. Med. Surg. 2025, 87, 1838–1845. [Google Scholar] [CrossRef]
- Martín, S.; Pérez, A.; Aldecoa, C. Sepsis and immunosenescence in the elderly patient: A review. Front. Med. 2017, 4, 243980. [Google Scholar] [CrossRef]
- Andersson, M.; Östholm-Balkhed, Å.; Fredrikson, M.; Holmbom, M.; Hällgren, A.; Berg, S.; Hanberger, H. Delay of appropriate antibiotic treatment is associated with high mortality in patients with community-onset sepsis in a Swedish setting. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1223. [Google Scholar] [CrossRef]
- King, J.; Chenoweth, C.E.; England, P.C.; Heiler, A.; Kenes, M.T.; Raghavendran, K.; Wood, W.; Zhou, S.; Mack, M.; Wesorick, D.; et al. Early Recognition and Initial Management of Sepsis in Adult Patients. Ann Arbor (MI): Michigan Medicine University of Michigan. 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK598311/ (accessed on 25 April 2025).
- Jones Stephen, L.; Ashton, C.M.; Kiehne, L.; Gigliotti, E.; Bell-Gordon, C.; Disbot, M.; Masud, F.; Shirkey, B.A.; Wray, N.P. Reductions in Sepsis Mortality and Costs After Design and Implementation of a Nurse-Based Early Recognition and Response Program. Jt. Comm. J. Qual. Patient Saf. 2015, 41, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Toker, A.K.; Kose, S.; Turken, M. Comparison of sofa score, sirs, qsofa, and qsofa + l criteria in the diagnosis and prognosis of sepsis. Eur. J. Med. 2021, 53, 40–47. [Google Scholar] [CrossRef]
- Li, Y.; Yan, C.; Gan, Z.; Xi, X.; Tan, Z.; Li, J.; Li, G. Prognostic values of SOFA score, qSOFA score, and LODS score for patients with sepsis. Ann. Palliat. Med. 2020, 9, 1037044. [Google Scholar] [CrossRef] [PubMed]
- Raith, E.P.; Udy, A.A.; Bailey, M.; McGloughlin, S.; MacIsaac, C.; Bellomo, R.; Pilcher, D.V.; Australian and New Zealand Intensive Care Society (ANZICS) Centre for Outcomes and Resource Evaluation (CORE). Prognostic Accuracy of the SOFA Score, SIRS Criteria, and qSOFA Score for In-Hospital Mortality Among Adults With Suspected Infection Admitted to the Intensive Care Unit. JAMA 2017, 317, 290–300. [Google Scholar] [CrossRef]
- Shukla, P.; Rao, G.M.; Pandey, G.; Sharma, S.; Mittapelly, N.; Shegokar, R.; Mishra, P.R. Therapeutic interventions in sepsis: Current and anticipated pharmacological agents. Br. J. Pharmacol. 2014, 171, 5011. [Google Scholar] [CrossRef]
- Polat, G.; Ugan, R.A.; Cadirci, E.; Halici, Z. Sepsis and Septic Shock: Current Treatment Strategies and New Approaches. Eur. J. Med. 2017, 49, 53. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; van der Poll, T.; Marshall, J.C. The End of “One Size Fits All” Sepsis Therapies: Toward an Individualized Approach. Biomedicines 2022, 10, 2260. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, D.; Arina, P.; Edgeworth, J.; Curcin, V.; Guidetti, V.; Mandreoli, F.; Wang, Y.; Chaurasia, A. Using interpretable machine learning to predict bloodstream infection and antimicrobial resistance in patients admitted to ICU: Early alert predictors based on EHR data to guide antimicrobial stewardship. PLoS Digit. Health 2024, 3, e0000641. [Google Scholar] [CrossRef] [PubMed]
- Jarczak, D.; Kluge, S.; Nierhaus, A. Sepsis—Pathophysiology and Therapeutic Concepts. Front. Med. 2021, 8, 628302. [Google Scholar] [CrossRef]
- Wendland, P.; Schenkel-Häger, C.; Wenningmann, I.; Kschischo, M. An optimal antibiotic selection framework for Sepsis patients using Artificial Intelligence. NPJ Digit. Med. 2024, 7, 343. [Google Scholar] [CrossRef]
- Cavallaro, M.; Moran, E.; Collyer, B.; McCarthy, N.D.; Green, C.; Keeling, M.J. Informing antimicrobial stewardship with explainable AI. PLoS Digit. Health 2023, 2, e0000162. [Google Scholar] [CrossRef]
- Haas, R.; McGill, S.C. Artificial Intelligence for the Prediction of Sepsis in Adults: CADTH Horizon Scan. Ottawa (ON): Canadian Agency for Drugs and Technologies in Health. 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK596676/ (accessed on 27 April 2025).
- Huat, G.K.; Wang, L.; Yeow, A.Y.K.; Poh, H.; Li, K.; Yeow, J.J.L.; Tan, G.Y.H. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat. Commun. 2021, 12, 711. [Google Scholar] [CrossRef]
- Goldschmidt, E.; Rannon, E.; Bernstein, D.; Wasserman, A.; Roimi, M.; Shrot, A.; Coster, D.; Shamir, R. Predicting appropriateness of antibiotic treatment among ICU patients with hospital-acquired infection. NPJ Digit. Med. 2025, 8, 1–16. [Google Scholar] [CrossRef]
- Robert, G.; Forbes, D.; Boyer, N. Sepsis: Diagnosis and Management. Am. Fam. Physician 2020, 101, 409–418. [Google Scholar]
- Düvel, J.A.; Lampe, D.; Kirchner, M.; Elkenkamp, S.; Cimiano, P.; Düsing, C.; Marchi, H.; Schmiegel, S.; Fuchs, C.; Claßen, S.; et al. An AI-Based Clinical Decision Support System for Antibiotic Therapy in Sepsis (KINBIOTICS): Use Case Analysis. JMIR Hum. Factors 2025, 12, e66699. [Google Scholar] [CrossRef] [PubMed]
- Marwaha, J.S.; Landman, A.B.; Brat, G.A.; Dunn, T.; Gordon, W.J. Deploying digital health tools within large, complex health systems: Key considerations for adoption and implementation. NPJ Digit. Med. 2022, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pepper, D.J.; Sun, J.; Rhee, C.; Welsh, J.; Powers, J.H.; Danner, R.L.; Kadri, S.S. Procalcitonin-Guided Antibiotic Discontinuation and Mortality in Critically Ill Adults: A Systematic Review and Meta-analysis. Chest 2019, 155, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Nobre, V.; Harbarth, S.; Graf, J.D.; Rohner, P.; Pugin, J. Use of Procalcitonin to Shorten Antibiotic Treatment Duration in Septic Patients. Am. J. Respir. Crit. Care Med. 2012, 177, 498–505. [Google Scholar] [CrossRef]
- Kopterides, P.; Siempos, I.I.; Tsangaris, I.; Tsantes, A.; Armaganidis, A. Procalcitonin-guided algorithms of antibiotic therapy in the intensive care unit: A systematic review and meta-analysis of randomized controlled trials. Crit. Care Med. 2010, 38, 2229–2241. [Google Scholar] [CrossRef]
- Rhee, C.; Murphy, M.V.; Li, L.; Platt, R.; Klompas, M. Lactate Testing in Suspected Sepsis: Trends and Predictors of Failure to Measure Levels. Crit. Care Med. 2015, 43, 1669–1676. [Google Scholar] [CrossRef]
- Yang, J.; Hao, S.; Huang, J.; Chen, T.; Liu, R.; Zhang, P.; Feng, M.; He, Y.; Xiao, W.; Hong, Y.; et al. The application of artificial intelligence in the management of sepsis. Med. Rev. 2023, 3, 369–380. [Google Scholar] [CrossRef]
- Yang, M.B.; Liu, C.; Wang, X.; Li, Y.; Gao, H.B.; Liu, X.; Li, J. An Explainable Artificial Intelligence Predictor for Early Detection of Sepsis. Crit. Care Med. 2020, 48, e1091–e1096. [Google Scholar] [CrossRef]
- Li, X.; Xu, X.; Xie, F.; Xu, X.M.; Sun, Y.M.; Liu, X.M.; Jia, X.B.; Kang, Y.M.; Xie, L.; Wang, F.; et al. A Time-Phased Machine Learning Model for Real-Time Prediction of Sepsis in Critical Care. Crit. Care Med. 2020, 48, e884–e888. [Google Scholar] [CrossRef]
- Shashikumar, S.P.; Josef, C.S.; Sharma, A.; Nemati, S. DeepAISE—An interpretable and recurrent neural survival model for early prediction of sepsis. Artif. Intell. Med. 2021, 113, 102036. [Google Scholar] [CrossRef]
- Desautels, T.; Calvert, J.; Hoffman, J.; Jay, M.; Kerem, Y.; Shieh, L.; Shimabukuro, D.; Chettipally, U.; Feldman, M.D.; Barton, C.; et al. Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR Med. Inform. 2016, 4, e5909. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Lee, K.; Kim, K.J.; Ha, E.Y.; Kim, I.-C.; Park, S.H.; Cho, C.-H.; Yu, G.I.; Ahn, B.E.; Jeong, Y.; et al. Validation of an artificial intelligence-based algorithm for predictive performance and risk stratification of sepsis using real-world data from hospitalised patients: A prospective observational study. BMJ Health Care Inf. 2025, 32, e101353. [Google Scholar] [CrossRef]
- Zhao, C.-C.; Nan, Z.-H.; Li, B.; Yin, Y.-L.; Zhang, K.; Liu, L.-X.; Hu, Z.-J. Development and validation of a novel risk-predicted model for early sepsis-associated acute kidney injury in critically ill patients: A retrospective cohort study. BMJ Open 2025, 15, e088404. [Google Scholar] [CrossRef] [PubMed]
- Campagner, A.; Agnello, L.; Carobene, A.; Padoan, A.; Del Ben, F.; Locatelli, M.; Plebani, M.; Ognibene, A.; Lorubbio, M.; De Vecchi, E. Complete Blood Count and Monocyte Distribution Width–Based Machine Learning Algorithms for Sepsis Detection: Multicentric Development and External Validation Study. J. Med. Internet Res. 2025, 27, e55492. [Google Scholar] [CrossRef]
- Persson, I.; Macura, A.; Becedas, D.; Sjövall, F. Early prediction of sepsis in intensive care patients using the machine learning algorithm NAVOY® Sepsis, a prospective randomized clinical validation study. J. Crit. Care 2023, 80, 154400. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zhang, L.; Zhang, S.; Shi, J.; Hong, D.; Wu, S.; Pan, X.; Lin, W. Developing a rapid screening tool for high-risk ICU patients of sepsis: Integrating electronic medical records with machine learning methods for mortality prediction in hospitalized patients—Model establishment, internal and external validation, and visualization. J. Transl. Med. 2025, 23, 97. [Google Scholar] [CrossRef]
- Nemati, S.; Holder, A.; Razmi, F.; Stanley, M.D.; Clifford, G.D.; Buchman, T.G. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit. Care Med. 2018, 46, 547–553. [Google Scholar] [CrossRef]
- Taneja, I.; Reddy, B.; Damhorst, G.; Zhao, S.D.; Hassan, U.; Price, Z.; Jensen, T.; Ghonge, T.; Patel, M.; Wachspress, S.; et al. Combining Biomarkers with EMR Data to Identify Patients in Different Phases of Sepsis. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Fleuren, L.M.; Klausch, T.L.T.; Zwager, C.L.; Schoonmade, L.J.; Guo, T.; Roggeveen, L.F.; Swart, E.L.; Girbes, A.R.J.; Thoral, P.; Ercole, A.; et al. Machine learning for the prediction of sepsis: A systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020, 46, 383–400. [Google Scholar] [CrossRef]
- Zhang, G.; Shao, F.; Yuan, W.; Wu, J.; Qi, X.; Gao, J.; Shao, R.; Tang, Z.; Wang, T. Predicting sepsis in-hospital mortality with machine learning: A multi-center study using clinical and inflammatory biomarkers. Eur. J. Med. Res. 2024, 29, 156. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.; Bennett, N.; Plečko, D.; Horn, M.; Rieck, B.; Meinshausen, N.; Bühlmann, P.; Borgwardt, K. Predicting sepsis using deep learning across international sites: A retrospective development and validation study. EClinicalMedicine 2023, 62, 102124. [Google Scholar] [CrossRef]
- Ishan, T.; Damhorst, G.L.; Lopez‐Espina, C.; Zhao, S.D.; Zhu, R.; Khan, S.; White, K.; Kumar, J.; Vincent, A.; Yeh, L.; et al. Diagnostic and prognostic capabilities of a biomarker and EMR-based machine learning algorithm for sepsis. Clin. Transl. Sci. 2021, 14, 1578–1589. [Google Scholar] [CrossRef]
- Shimabukuro, D.W.; Barton, C.W.; Feldman, M.D.; Mataraso, S.J.; Das, R. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: A randomised clinical trial. BMJ Open Respir. Res. 2017, 4, e000234. [Google Scholar] [CrossRef]
- Zheng, L.; Lin, F.; Zhu, C.; Liu, G.; Wu, X.; Wu, Z.; Zheng, J.; Xia, H.; Cai, Y.; Liang, H.; et al. Machine Learning Algorithms Identify Pathogen-Specific Biomarkers of Clinical and Metabolomic Characteristics in Septic Patients with Bacterial Infections. Biomed. Res. Int. 2020, 2020, 6950576. [Google Scholar] [CrossRef] [PubMed]
- Wernly, B.; Mamandipoor, B.; Baldia, P.; Jung, C.; Osmani, V. Machine learning predicts mortality in septic patients using only routinely available ABG variables: A multi-centre evaluation. Int. J. Med. Inform. 2021, 145, 104312. [Google Scholar] [CrossRef] [PubMed]
- Pappada, S.M.; Owais, M.H.; Feeney, J.J.; Salinas, J.; Chaney, B.; Duggan, J.; Sparkle, T.; Aouthmany, S.; Hinch, B.; Papadimos, T.J. Development and validation of a sepsis risk index supporting early identification of ICU-acquired sepsis: An observational study. Anaesth. Crit. Care Pain. Med. 2024, 43, 101430. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Lei, M.; Wang, L.; Wang, X.; Li, X.; Mao, Z.; Kang, H.; Liu, H.; Sun, S.; Zhou, F. Prediction of sepsis among patients with major trauma using artificial intelligence: A multicenter validated cohort study. Int. J. Surg. 2025, 111, 467–480. [Google Scholar] [CrossRef]
- Li, K.; Shi, Q.; Liu, S.; Xie, Y.; Liu, J. Predicting in-hospital mortality in ICU patients with sepsis using gradient boosting decision tree. Medicine 2021, 100, e25813. [Google Scholar] [CrossRef]
- Li, J.; Xi, F.; Yu, W.; Sun, C.; Wang, X. Real-Time Prediction of Sepsis in Critical Trauma Patients: Machine Learning–Based Modeling Study. JMIR Form Res. 2023, 7, e42452. [Google Scholar] [CrossRef]
- Boussina, A.; Shashikumar, S.P.; Malhotra, A.; Owens, R.L.; El-Kareh, R.; Longhurst, C.A.; Quintero, K.; Donahue, A.; Chan, T.C.; Nemati, S.; et al. Impact of a deep learning sepsis prediction model on quality of care and survival. NPJ Digit. Med. 2024, 7, 14. [Google Scholar] [CrossRef]
- Sankavi, M.; Nelson, W.; Di, S.; McGillion, M.; Devereaux, P.J.; Barr, N.G.; Petch, J. Machine Learning-Based Early Warning Systems for Clinical Deterioration: Systematic Scoping Review. J. Med. Internet Res. 2021, 23, e25187. [Google Scholar] [CrossRef]
- Langley, R.J.; Tsalik, E.L.; van Velkinburgh, J.C.; Glickman, S.W.; Rice, B.J.; Wang, C.; Chen, B.; Carin, L.; Suarez, A.; Mohney, R.P.; et al. Sepsis: An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci. Transl. Med. 2013, 5, 195ra95. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S. Sepsis, Management & Advances in Metabolomics. Nanotheranostics 2024, 8, 270–284. [Google Scholar] [CrossRef]
- She, H.; Du, Y.; Du, Y.; Tan, L.; Yang, S.; Luo, X.; Li, Q.; Xiang, X.; Lu, H.; Hu, Y.; et al. Metabolomics and machine learning approaches for diagnostic and prognostic biomarkers screening in sepsis. BMC Anesthesiol. 2023, 23, 367. [Google Scholar] [CrossRef]
- D’Onofrio, V.; Salimans, L.; Bedenić, B.; Cartuyvels, R.; Barišić, I.; Gyssens, I.C. The Clinical Impact of Rapid Molecular Microbiological Diagnostics for Pathogen and Resistance Gene Identification in Patients With Sepsis: A Systematic Review. Open Forum Infect. Dis. 2020, 7, ofaa352. [Google Scholar] [CrossRef]
- Eubank, T.A.; Long, S.W.; Perez, K.K. Role of Rapid Diagnostics in Diagnosis and Management of Patients with Sepsis. J. Infect. Dis. 2020, 222, S103–S109. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S.; Carroll, K.C. Early Identification and Treatment of Pathogens in Sepsis: Molecular Diagnostics and Antibiotic Choice. Clin. Chest Med. 2016, 37, 191–207. [Google Scholar] [CrossRef]
- Sinha, M.; Jupe, J.; Mack, H.; Coleman, T.P.; Lawrence, S.M.; Fraley, S.I. Emerging technologies for molecular diagnosis of sepsis. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef] [PubMed]
- Plata-Menchaca, E.P.; Ferrer, R. Life-support tools for improving performance of the Surviving Sepsis Campaign Hour-1 bundle. Med. Intensiv. 2018, 42, 547–550. [Google Scholar] [CrossRef]
- Ranieri, V.M.; Thompson, B.T.; Barie, P.S.; Dhainaut, J.-F.; Douglas, I.S.; Finfer, S.; Gårdlund, B.; Marshall, J.C.; Rhodes, A.; Artigas, A.; et al. Drotrecogin alfa (activated) in adults with septic shock. N. Engl. J. Med. 2012, 366, 2055–2064. [Google Scholar] [CrossRef]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensive Care Med. 2018, 44, 925–928. [Google Scholar] [CrossRef]
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef]
- Martínez, M.L.; Plata-Menchaca, E.P.; Ruiz-Rodríguez, J.C.; Ferrer, R. An approach to antibiotic treatment in patients with sepsis. J. Thorac. Dis. 2020, 12, 1007. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Xia, Y.; Zhu, Z.; Zhang, Y. Knowledge, attitudes, and practices among physicians and pharmacists toward antibiotic use in sepsis. Front. Med. 2024, 11, 1454521. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.; Jeon, J.H.; Park, D.W. Antimicrobial Therapy and Antimicrobial Stewardship in Sepsis. Infect. Chemother. 2020, 52, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Donkor, E.S.; Codjoe, F.S. Methicillin Resistant Staphylococcus aureus and Extended Spectrum Beta-lactamase Producing Enterobacteriaceae: A Therapeutic Challenge in the 21st Century. Open Microbiol. J. 2019, 13, 94–100. [Google Scholar] [CrossRef]
- Department of Health Antimicrobial Resistance Strategy Analytical Working Group. Antimicrobial Resistance: Empirical and Statistical Evidence-Base. 2016. Available online: https://assets.publishing.service.gov.uk/media/5a7f417ce5274a2e87db4be3/AMR_EBO_2016.pdf (accessed on 26 May 2025).
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229. [Google Scholar] [CrossRef]
- Liu, V.X.; Fielding-Singh, V.; Greene, J.D.; Baker, J.M.; Iwashyna, T.J.; Bhattacharya, J.; Escobar, G.J. The timing of early antibiotics and hospital mortality in sepsis. Am. J. Respir. Crit. Care Med. 2017, 196, 856–863. [Google Scholar] [CrossRef]
- Ha, D.R.; Haste, N.M.; Gluckstein, D.P. The Role of Antibiotic Stewardship in Promoting Appropriate Antibiotic Use. Am. J. Lifestyle Med. 2017, 13, 376. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Kraft, W.K.; Farber, J.L. The Nephrotoxicity of Vancomycin. Clin. Pharmacol. Ther. 2017, 102, 459. [Google Scholar] [CrossRef]
- Wong-Beringer, A.; Joo, J.; Tse, E.; Beringer, P. Vancomycin-associated nephrotoxicity: A critical appraisal of risk with high-dose therapy. Int. J. Antimicrob. Agents 2011, 37, 95–101. [Google Scholar] [CrossRef]
- Song, J.H.; Kim, Y.S. Recurrent Clostridium difficile Infection: Risk Factors, Treatment, and Prevention. Gut Liver 2019, 13, 16–24. [Google Scholar] [CrossRef]
- Saleem, M.; Deters, B.; de la Bastide, A.; Korzen, M. Antibiotics Overuse and Bacterial Resistance. Ann. Microbiol. Res. 2019, 3, 93–99. [Google Scholar] [CrossRef]
- Husabø, G.; Nilsen, R.M.; Flaatten, H.; Solligård, E.; Frich, J.C.; Bondevik, G.T.; Braut, G.S.; Walshe, K.; Harthug, S.; Hovlid, E.; et al. Early diagnosis of sepsis in emergency departments, time to treatment, and association with mortality: An observational study. PLoS ONE 2020, 15, e0227652. [Google Scholar] [CrossRef]
- Tang, F.; Yuan, H.; Li, X.; Qiao, L. Effect of delayed antibiotic use on mortality outcomes in patients with sepsis or septic shock: A systematic review and meta-analysis. Int. Immunopharmacol. 2024, 129, 111616. [Google Scholar] [CrossRef]
- Neilson, H.K.M.; Fortier, J.H.M.; Finestone, P.R.; Ogilby, C.M.R.; Liu, R.M.; Bridges, E.J.M.; Garber, G.E.M. Diagnostic Delays in Sepsis: Lessons Learned From a Retrospective Study of Canadian Medico-Legal Claims. Crit Care Explor. 2023, 5, e0841. [Google Scholar] [CrossRef]
- Lesprit, P.; Landelle, C.; Brun-Buisson, C. Clinical impact of unsolicited post-prescription antibiotic review in surgical and medical wards: A randomized controlled trial. Clin. Microbiol. Infect. 2013, 19, E91–E97. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kiya, G.T.; Asefa, E.T.; Abebe, G.; Mekonnen, Z. Procalcitonin Guided Antibiotic Stewardship. Biomark. Insights 2024, 19, 11772719241298197. [Google Scholar] [CrossRef] [PubMed]
- Willmon, J.; Subedi, B.; Girgis, R.; Noe, M. Impact of Pharmacist-Directed Simplified Procalcitonin Algorithm on Antibiotic Therapy in Critically Ill Patients With Sepsis. Hosp. Pharm. 2021, 56, 501–506. [Google Scholar] [CrossRef]
- Moehring, R.W.; E Yarrington, M.; Warren, B.G.; Lokhnygina, Y.; Atkinson, E.; Bankston, A.; Coluccio, J.; David, M.Z.; Davis, A.; Davis, J.; et al. 14. Effects of an Opt-Out Protocol for Antibiotic De-escalation among Selected Patients with Suspected Sepsis: The DETOURS Trial. Open Forum Infect. Dis. 2021, 8, S10–S11. [Google Scholar] [CrossRef]
- Moehring, R.W.; E Yarrington, M.; Warren, B.G.; Lokhnygina, Y.; Atkinson, E.; Bankston, A.; Collucio, J.; David, M.Z.; E Davis, A.; Davis, J.; et al. Evaluation of an Opt-Out Protocol for Antibiotic De-Escalation in Patients With Suspected Sepsis: A Multicenter, Randomized, Controlled Trial. Clin. Infect. Dis. 2023, 76, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, I.; Tafelski, S.; Deja, M.; Halle, E.; Grebe, M.C.; Tamarkin, A.; Rothbart, A.; Uhrig, A.; Meyer, E.; Musial-Bright, L.; et al. Long-term effect of computer-assisted decision support for antibiotic treatment in critically ill patients: A prospective ‘before/after’ cohort study. BMJ Open 2014, 4, e005370. [Google Scholar] [CrossRef]
- Niederman, M.S.; Baron, R.M.; Bouadma, L.; Calandra, T.; Daneman, N.; DeWaele, J.; Kollef, M.H.; Lipman, J.; Nair, G.B. Initial antimicrobial management of sepsis. Crit. Care 2021, 25, 307. [Google Scholar] [CrossRef]
- Li, F.; Wang, S.; Gao, Z.; Qing, M.; Pan, S.; Liu, Y.; Hu, C. Harnessing artificial intelligence in sepsis care: Advances in early detection, personalized treatment, and real-time monitoring. Front. Med. 2024, 11, 1510792. [Google Scholar] [CrossRef] [PubMed]
- Marko, B.; Palmowski, L.; Nowak, H.; Witowski, A.; Koos, B.; Rump, K.; Bergmann, L.; Bandow, J.; Eisenacher, M.; Günther, P.; et al. Employing artificial intelligence for optimising antibiotic dosages in sepsis on intensive care unit: A study protocol for a prospective observational study (KI.SEP). BMJ Open 2024, 14, e086094. [Google Scholar] [CrossRef] [PubMed]
- Kijpaisalratana, N.; Saoraya, J.; Nhuboonkaew, P.; Vongkulbhisan, K.; Musikatavorn, K. Real-time machine learning-assisted sepsis alert enhances the timeliness of antibiotic administration and diagnostic accuracy in emergency department patients with sepsis: A cluster-randomized trial. Intern. Emerg. Med. 2024, 19, 1415–1424. [Google Scholar] [CrossRef]
- Conwill, A.; Sater, M.; Worley, N.; Wittenbach, J.; Huntley, M.H. 966. Large-Scale Evaluation of AST Prediction using Resistance Marker Presence/Absence vs. Machine Learning on WGS Data. Open Forum Infect. Dis. 2023, 10, ofad500.2461. [Google Scholar] [CrossRef]
- Jana, T.; Sarkar, D.; Ganguli, D.; Mukherjee, S.K.; Mandal, R.S.; Das, S. ABDpred: Prediction of active antimicrobial compounds using supervised machine learning techniques. Indian. J. Med. Res. 2024, 159, 78–90. [Google Scholar] [CrossRef]
- Lewin-Epstein, O.; Baruch, S.; Hadany, L.; Stein, G.Y.; Obolski, U. Predicting Antibiotic Resistance in Hospitalized Patients by Applying Machine Learning to Electronic Medical Records. Clin. Infect. Dis. 2021, 72, e848–e855. [Google Scholar] [CrossRef]
- Mooney, C.; Eogan, M.; Áinle, F.N.; Cleary, B.; Gallagher, J.J.; O’LOughlin, J.; Drew, R.J. Predicting bacteraemia in maternity patients using full blood count parameters: A supervised machine learning algorithm approach. Int. J. Lab. Hematol. 2021, 43, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Tang, J.-W.; Chen, J.; Liao, Y.-W.; Zhang, W.-W.; Wen, X.-R.; Liu, X.; Chen, H.-J.; Wang, L. SERS-ATB: A comprehensive database server for antibiotic SERS spectral visualization and deep-learning identification. Environ. Pollut. 2025, 373, 126083. [Google Scholar] [CrossRef]
- Tan, R.; Ge, C.; Wang, J.; Yang, Z.; Guo, H.; Yan, Y.; Du, Q. Interpretable machine learning model for early morbidity risk prediction in patients with sepsis-induced coagulopathy: A multi-center study. Front. Immunol. 2025, 16, 1552265. [Google Scholar] [CrossRef]
- Shi, J.; Han, H.; Chen, S.; Liu, W.; Li, Y. Machine learning for prediction of acute kidney injury in patients diagnosed with sepsis in critical care. PLoS ONE. 2024, 19, e0301014. [Google Scholar] [CrossRef]
- Pan, S.; Shi, T.; Ji, J.; Wang, K.; Jiang, K.; Yu, Y.; Li, C. Developing and validating a machine learning model to predict multidrug-resistant Klebsiella pneumoniae-related septic shock. Front. Immunol. 2025, 15, 1539465. [Google Scholar] [CrossRef]
- Özdede, M.; Zarakolu, P.; Metan, G.; Eser, Ö.K.; Selimova, C.; Kızılkaya, C.; Elmalı, F.; Akova, M. Predictive modeling of mortality in carbapenem-resistant Acinetobacter baumannii bloodstream infections using machine learning. J Investig. Med. 2024, 72, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Nielsen, A.D.; Goldberg, E.; Andreassen, S.; Tacconelli, E.; Almanasreh, N.; Frank, U.; Cauda, R.; Leibovici, L.; TREAT Study Group. Prediction of specific pathogens in patients with sepsis: Evaluation of TREAT, a computerized decision support system. J. Antimicrob. Chemother. 2007, 59, 1204–1207. [Google Scholar] [CrossRef]
- Paul, M.; Andreassen, S.; Nielsen, A.D.; Tacconelli, E.; Almanasreh, N.; Fraser, A.; Yahav, D.; Ram, R.; Leibovici, L.; TREAT Study Group. Prediction of bacteremia using TREAT, a computerized decision-support system. Clin. Infect. Dis. 2006, 42, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zheng, S.; He, J.; Zhang, Z.-M.; Wu, R.; Yu, Y.; Fu, H.; Han, L.; Zhu, H.; Xu, Y.; et al. An Easy and Quick Risk-Stratified Early Forewarning Model for Septic Shock in the Intensive Care Unit: Development, Validation, and Interpretation Study. J. Med. Internet Res. 2025, 27, e58779. [Google Scholar] [CrossRef]
- Park, S.W.; Yeo, N.Y.; Kang, S.; Ha, T.; Kim, T.-H.; Lee, D.; Kim, D.; Choi, S.; Kim, M.; Lee, D.; et al. Early Prediction of Mortality for Septic Patients Visiting Emergency Room Based on Explainable Machine Learning: A Real-World Multicenter Study. J. Korean Med. Sci. 2024, 39, e53. [Google Scholar] [CrossRef]
- Chen, X.; Hu, L.; Yu, R. Development and external validation of machine learning-based models to predict patients with cellulitis developing sepsis during hospitalisation. BMJ Open. 2024, 14, e084183. [Google Scholar] [CrossRef]
- Pan, X.; Xie, J.; Zhang, L.; Wang, X.; Zhang, S.; Zhuang, Y.; Lin, X.; Shi, S.; Shi, S.; Lin, W. Evaluate prognostic accuracy of SOFA component score for mortality among adults with sepsis by machine learning method. BMC Infect. Dis. 2023, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Ripoli, A.; Sozio, E.; Sbrana, F.; Bertolino, G.; Pallotto, C.; Cardinali, G.; Meini, S.; Pieralli, F.; Azzini, A.M.; Concia, E.; et al. Personalized machine learning approach to predict candidemia in medical wards. Infection 2020, 48, 749–759. [Google Scholar] [CrossRef]
- Madden, G.R.; Boone, R.H.; Lee, E.; Sifri, C.D.; Petri, W.A. Predicting Clostridioides difficile infection outcomes with explainable machine learning. EBioMedicine 2024, 106, 105244. [Google Scholar] [CrossRef]
- Islam, K.R.; Prithula, J.; Kumar, J.; Tan, T.L.; Reaz, M.B.I.; Sumon, S.I.; Chowdhury, M.E.H. Machine Learning-Based Early Prediction of Sepsis Using Electronic Health Records: A Systematic Review. J. Clin. Med. 2023, 12, 5658. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Ali, I.; van der Veer, S.; Martin, G.; Ainsworth, J.; Augustine, T. Adoption of clinical risk prediction tools is limited by a lack of integration with electronic health records. BMJ Health Care Inform. 2021, 28, e100253. [Google Scholar] [CrossRef] [PubMed]
- Barton, C.; Chettipally, U.; Zhou, Y.; Jiang, Z.; Lynn-Palevsky, A.; Le, S.; Calvert, J.; Das, R. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput. Biol. Med. 2019, 109, 79–84. [Google Scholar] [CrossRef]
- Sadasivuni, S.; Saha, M.; Bhatia, N.; Banerjee, I.; Sanyal, A. Fusion of fully integrated analog machine learning classifier with electronic medical records for real-time prediction of sepsis onset. Sci. Rep. 2022, 12, 5711. [Google Scholar] [CrossRef]
- Gao, W.; Pei, Y.; Liang, H.; Lv, J.; Chen, J.; Zhong, W. Multimodal AI system for the rapid diagnosis and surgical prediction of necrotizing enterocolitis. IEEE Access 2021, 9, 51050–51064. [Google Scholar] [CrossRef]
- Jacobs, L.; Wong, H.R. Emerging infection and sepsis biomarkers: Will they change current therapies? Expert Rev. Anti Infect. Ther. 2016, 14, 929–941. [Google Scholar] [CrossRef]
- Sakagianni, A.; Koufopoulou, C.; Feretzakis, G.; Kalles, D.; Verykios, V.S.; Myrianthefs, P.; Fildisis, G. Using Machine Learning to Predict Antimicrobial Resistance-A Literature Review. Antibiotics 2023, 12, 452. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.R.; Bonomo, R.A.; Rello, J. Managing sepsis in the era of precision medicine: Challenges and opportunities. Expert Rev. Anti Infect. Ther. 2022, 20, 871–880. [Google Scholar] [CrossRef] [PubMed]
| Study/Year | Model Type | Dataset Source | Sample Size | Validation Strategy | Overfitting Mitigation | Interpretability/Ethics | Performance |
|---|---|---|---|---|---|---|---|
| Li et al., 2020 (TASP) [33] | LightGBM, time-phased ML | 3 US hospitals, retrospective EHR | Not specified | 5-fold CV, internal/external | Time-phased strategy, cross-validation | Improved interpretability via time-phased cutoffs; focus on early prediction | AUROC 0.845 (internal), utility score 0.430 (internal) |
| DeepAISE (2020) [34] | Deep learning (time-phased) | Multicenter EHR, ICU | Not specified | Multicenter validation | Implied, not reported | Not specified (focus on accuracy and robustness) | AUROC >0.90 |
| InSight (2016) [35] | Machine learning (vital signs) | Single-center EHR | Not specified | External validation | Minimal feature set, cross-validation | Not interpretable, not reported | AUROC 0.88 (onset), 0.74 (4 h prior); APR 0.59 (onset), 0.27 (4 h prior) |
| VC-SEPS (2022) [36] | Deep learning | S. Korean hospital, real-world EMR | Not specified | Prospective, external | Not reported | Outperformed SOFA/qSOFA; robust real-world validation | AUROC 0.88, accuracy 87.3%, NPV 0.997, prediction 68 min before diagnosis |
| Zhao et al., 2022 [37] | Gradient boosting, ML | ICU SA-AKI, multi-institutional | Not specified | Split dev/test cohort | Not specified | SHAP analysis for feature importance, web-based app | AUROC 0.794, accuracy 78.3%, Sens 94.2%, Spec 32.1% |
| Cimenti et al., 2023 [38] | Extreme gradient boosting | 6 Italian hospitals, CBC data | 5344 | Internal and external | Cross-validation, cautious classification | Excellent performance, focus on blood count/MDW | AUC 0.91–0.98 (internal), 0.75–0.95 (external) |
| NAVOY (2022) [39] | ML (not specified) | Multicenter ICU, RCT validation | Not specified | Prospective RCT | Not reported | Largest RCT, clinical validation vs. Sepsis-3 | AUROC 0.80, Sens 0.80, Spec 0.78, accuracy 0.79 |
| Shi et al., 2022 [40] | Gradient boosting, ML | MIMIC-IV (US), Chinese EHR | Not specified | Internal and external | Not reported | SHAP for global/individual interpretability | AUC > 0.8, accuracy 78% (internal), 63–77% (external) |
| Nemati et al., 2018 [41] | AISE Algorithm | MIMIC-III, eICU | Not specified | External validation | Regularization, cross-validation | Interpretable predictions, feature highlighting | AUROC 0.83–0.85, Spec/Acc 0.63–0.67, up to 12 h pre-onset |
| Taneja et al., 2017 [42] | Ensemble ML + biomarkers | Single-center EHR + biomarkers | Not specified | Cross-validation | Cross-validation | Not reported | AUROC 0.75–0.81 |
| Fleuren et al., 2020 [43] | Meta-analysis (various ML/DL) | International pooled, mixed datasets | Not specified | Meta-analytic, multicenter | Mixed, varies by model | Summarizes interpretability and fairness across models | AUROC 0.68–0.99 (varies by model and setting) |
| Study/Year | Model Type | Dataset Source | Sample Size | Validation Strategy | Overfitting Mitigation | Interpretability/Ethics | Performance/Outcomes |
|---|---|---|---|---|---|---|---|
| KINBIOTICS [25] | Clinical decision support system (CDSS) | Multicenter microbiology, resistance, clinical data | Not specified | Real-time pilot | Not specified | Supports de-escalation and antimicrobial stewardship | Reduces toxicity (e.g., vancomycin nephrotoxicity) and broad-spectrum overuse |
| KI.SEP (2023) [90] | Personalized dosing recommender (AI/ML) | ICU patients with sepsis (prospective observational, Germany) | Not specified | Prospective, real-world | Not specified | Transparent dosing logic; focus on safety | AI-assisted dosing achieved optimal drug levels in >50% vs. 30–40% with conventional monitoring |
| MLASA (2023, cluster RCT) [91] | Machine learning–assisted sepsis alert | ED patients, tertiary hospital (>16,500 visits) | 574 sepsis patients | Cluster-randomized trial (intervention vs. control) | Not specified | Integrated into clinical workflow; validated against ethics board | 68.4% vs. 60.1% received antibiotics in 1 h; AUROC 0.93; sensitivity 79.7%, specificity 88.2%, NPV 98.9% |
| Study/Year | Model Type | Dataset Source/Sample Size | Validation Strategy | Overfitting Mitigation | Interpretability/Ethics | Performance/Outcomes |
|---|---|---|---|---|---|---|
| Conwill et al. [92] | ML (WGS-based, Keynome gAST) | WGS + phenotype, 107 species/drug combinations | Internal split + cross-validation | Not detailed | Genomic explainability; bias addressed | Median balanced accuracy 92% vs. 80% (markers) |
| Lewin-Epstein et al. [94] | Ensemble ML | EMR data, antibiotic history, comorbidities, microbiology/large Israeli hospital | Internal/external split sample | Not detailed | Feature importance analysis; EMR transparency | AUROC 0.73–0.79 (no species); 0.8–0.88 (w/species) |
| Maternal sepsis study [95] | ML (FBC parameters) | Pregnant/postpartum, blood cultures, n = 13 (MLR > 20.3) | Retrospective cohort | Not specified | Focus on explainable clinical features | 100% NLR > 20.3 and neutrophil > 19.2 → bacteremia |
| SERS-ATB [96] | Deep learning, Raman spectra | SERS-ATB database, multicenter, spectra, n not stated | External, real-world, multicenter | Not specified | Global database, accessibility | 98.9% accuracy, results in seconds |
| Pan et al. [99] | ML nomogram | MDR Klebsiella pneumoniae septic shock/ICU, China, n not stated | Not detailed | Not specified | Model nomogram presented | AUROC > 0.90 |
| Özdede et al. [100] | Five ML algorithms | Carbapenem-resistant Acinetobacter baumannii BI, n not stated | Stratified 10-fold cross-validation | Not specified | Critical predictor analysis | NB: AUC 0.822 (14d), RF: AUC 0.854 (30d) |
| TREAT CDSS [101,102] | Clinical decision support | Regional hospital, Gram-negative pathogens | Internal, simulated | Not specified | Recommender logic, scenario analysis | AUC 0.70–0.80, outperforms physicians |
| Liu et al. (SORP) [103] | Early warning ML system | ICU, vital/lab data, n not stated | Internal + test set | Not specified | Forewarning time stratification, risk group stratification | AUC 0.946, 13 h median warning time |
| Park et al. (ER mortality) [104] | CatBoost, XGBoost, etc. | ER sepsis, 19 Korean hospitals, n = 5112 | Internal + external, multi-site | Not specified | SHAP, variable importance | CatBoost: AUC 0.800; XGB: AUC 0.678; Acc 0.769–0.773 |
| Cellulitis ANN study [105] | ANN, 10 ML algorithms | MIMIC-IV (n = 6695), Yidu-Cloud (n = 2506, ext. val.) | External, international | Not specified | Robust to missing data, compared to LR | ANN: AUC 0.830, odds ratio 9.375, Acc > 0.9 |
| Pan et al. (SOFA scores) [106] | LR, Gaussian NB | 23,889 sepsis patients, China | K-fold cross-validation | Not specified | Differential weighting, organ-specific explainability | LR, GNB: AUC 0.76, Acc. 0.851–0.844 |
| Ripoli et al. (Candida) [107] | Random forest | Internal medicine wards, candidemia, n not stated | Not stated | Not specified | Not specified | AUROC 0.874 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barkas, G.I.; Dimeas, I.E.; Kotsiou, O.S. Bug Wars: Artificial Intelligence Strikes Back in Sepsis Management. Diagnostics 2025, 15, 1890. https://doi.org/10.3390/diagnostics15151890
Barkas GI, Dimeas IE, Kotsiou OS. Bug Wars: Artificial Intelligence Strikes Back in Sepsis Management. Diagnostics. 2025; 15(15):1890. https://doi.org/10.3390/diagnostics15151890
Chicago/Turabian StyleBarkas, Georgios I., Ilias E. Dimeas, and Ourania S. Kotsiou. 2025. "Bug Wars: Artificial Intelligence Strikes Back in Sepsis Management" Diagnostics 15, no. 15: 1890. https://doi.org/10.3390/diagnostics15151890
APA StyleBarkas, G. I., Dimeas, I. E., & Kotsiou, O. S. (2025). Bug Wars: Artificial Intelligence Strikes Back in Sepsis Management. Diagnostics, 15(15), 1890. https://doi.org/10.3390/diagnostics15151890









