Deep Learning Network Selection and Optimized Information Fusion for Enhanced COVID-19 Detection: A Literature Review
Abstract
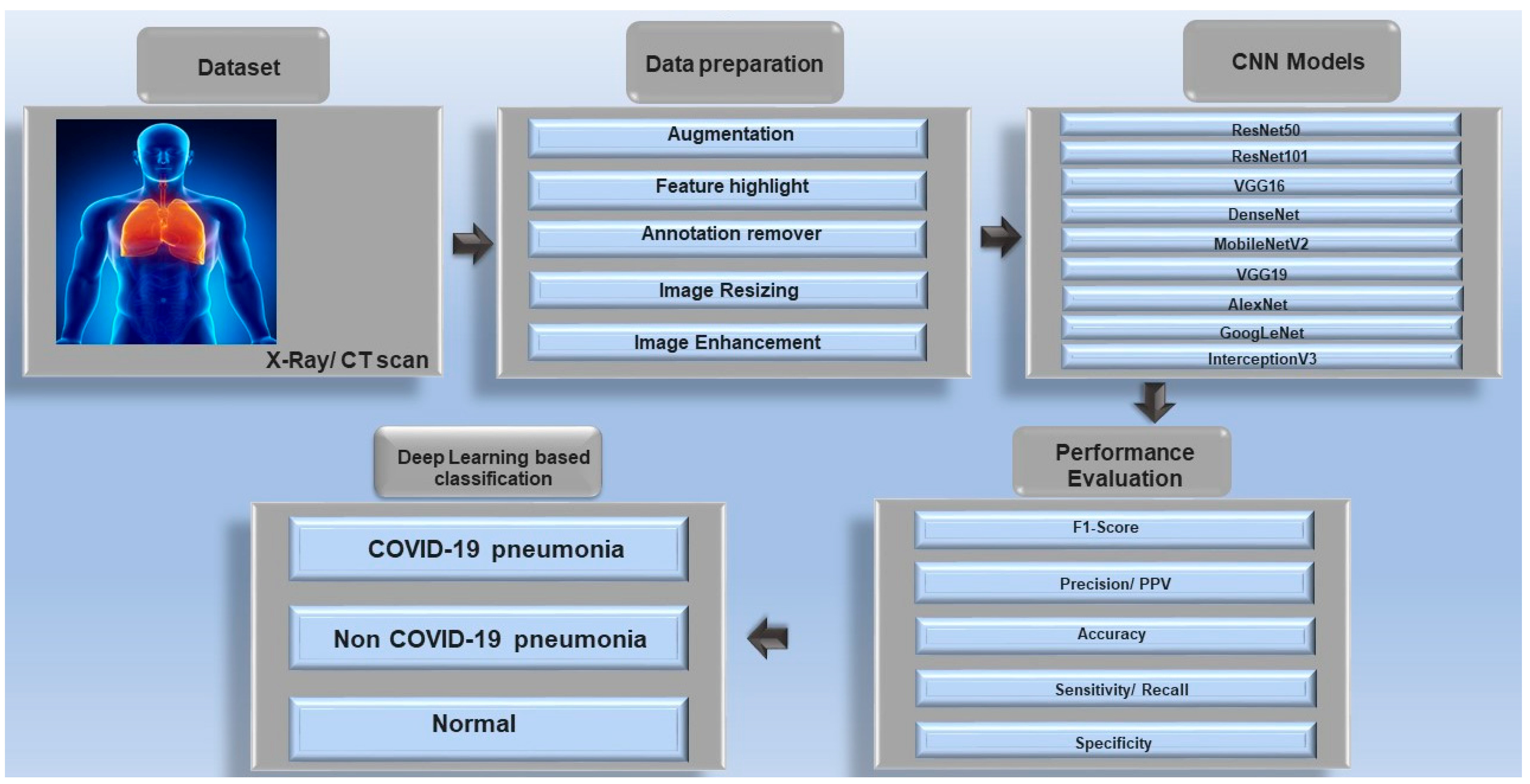
1. Introduction
2. Methods
3. Data Modalities for COVID-19 Diagnosis
3.1. Chest Radiography (X-Ray)
3.2. Computed Tomography (CT)
3.3. Other Modalities (Ultrasound, Audio, and Clinical Data)
4. Deep Learning Models and Architectures for COVID-19 Detection
4.1. Convolutional Neural Networks (CNNs)
4.2. Transformer-Based Models
4.3. Ensembles and Hybrid Models
5. Information Fusion Strategies for Enhanced Diagnosis
5.1. Data-Level (Early) Fusion
5.2. Feature-Level (Mid) Fusion
5.3. Decision-Level (Late) Fusion
6. Major Sources of Non-Uniformity in Chest Imaging Datasets for COVID-19 Detection and Their Impact on the Model
7. The Role of Federated Learning in Ensuring Data Privacy and Enhancing Model Robustness in Healthcare
8. Evaluating Model Resilience to Image Artifacts, Comorbidities, and COVID-19 Mimickers
9. The Role and Importance of Explainable AI (XAI) in Clinical Diagnosis
10. Key Challenges and Limitations
10.1. Data Quality and Noise
10.2. Overfitting and Generalization
10.3. Evaluation Metrics and Reporting
10.4. Clinical Integration and Trust
11. Recent Advancements and Future Directions
11.1. Improved Model Performance
11.2. Multimodal and Multi-Task Fusion
11.3. Hierarchical and Explainable Models
11.4. Generalization and Deployment
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| CNN | Convolutional neural network |
| COPD | Chronic obstructive pulmonary disease |
| CT | Computed tomography |
| CXR | Chest X-ray |
| DL | Deep Learning |
| FL | Federated learning |
| GDPR | General Data Protection Regulation |
| RT-PCR | Real-time polymerase chain reaction |
| ViT | Vision transformer |
| XAI | Explainable AI |
| X-Ray | Radiography |
References
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- DeGrave, A.J.; Janizek, J.D.; Lee, S.I. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 2021, 3, 610–619. [Google Scholar] [CrossRef]
- Hardy-Werbin, M.; Maiques, J.M.; Busto, M.; Cirera, I.; Aguirre, A.; Garcia-Gisbert, N.; Zuccarino, F.; Carbullanca, S.; Del Carpio, L.A.; Ramal, D.; et al. MultiCOVID: A multi modal deep learning approach for COVID-19 diagnosis. Sci. Rep. 2023, 13, 18761. [Google Scholar] [CrossRef] [PubMed]
- Cleverley, J.; Piper, J.; Jones, M.M. The role of chest radiography in confirming COVID-19 pneumonia. BMJ 2020, 370, m2426. [Google Scholar] [CrossRef] [PubMed]
- Archana, K.; Kaur, A.; Gulzar, Y.; Hamid, Y.; Mir, M.S.; Soomro, A.B. Deep learning models/techniques for COVID-19 detection: A survey. Front. Appl. Math. Stat. 2023, 9, 1303714. [Google Scholar] [CrossRef]
- Cruz, B.G.S.; Bossa, M.N.; Sölter, J.; Husch, A.D. Public COVID-19 X-ray datasets and their impact on model bias—A systematic review of a significant problem. Med. Image Anal. 2021, 74, 102225. [Google Scholar] [CrossRef]
- Hammoudi, K.; Benhabiles, H.; Melkemi, M.; Dornaika, F.; Arganda-Carreras, I.; Collard, D.; Scherpereel, A. Deep Learning on Chest X-ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19. J. Med. Syst. 2021, 45, 75. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Hemdan, E.E.-D.; Shouman, M.A.; Karar, M.E. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images. arXiv 2020, arXiv:2003.11055. Available online: https://arxiv.org/pdf/2003.11055 (accessed on 29 April 2025).
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Horry, M.J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. X-Ray Image based COVID-19 Detection using Pre-trained Deep Learning Models. Preprint 2020, 1–13. [Google Scholar] [CrossRef]
- Bukhari, S.U.K.; Safwan, S.; Bukhari, K.; Syed, A.; Sajid, S.; Shah, H. The diagnostic evaluation of Convolutional Neural Network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. MedRxiv 2020, 20044610. [Google Scholar] [CrossRef]
- Benmalek, E.; Elmhamdi, J.; Jilbab, A. Comparing CT scan and chest X-ray imaging for COVID-19 diagnosis. Biomed. Eng. Adv. 2021, 1, 100003. [Google Scholar] [CrossRef] [PubMed]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef]
- Gürsoy, E.; Kaya, Y. An overview of deep learning techniques for COVID-19 detection: Methods, challenges, and future works. Multimed. Syst. 2023, 29, 1603. [Google Scholar] [CrossRef]
- Ilhan, H.O.; Serbes, G.; Aydin, N. Decision and feature level fusion of deep features extracted from public COVID-19 data-sets. Appl. Intell. 2021, 52, 8551–8571. [Google Scholar] [CrossRef]
- Ye, Q.; Gao, Y.; Ding, W.; Niu, Z.; Wang, C.; Jiang, Y.; Wang, M.; Fang, E.F.; Menpes-Smith, W.; Xia, J.; et al. Robust weakly supervised learning for COVID-19 recognition using multi-center CT images. Appl. Soft Comput. 2022, 116, 108291. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020, 296, E65–E71. [Google Scholar] [CrossRef]
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19). Eur. Radiol. 2021, 31, 6096–6104. [Google Scholar] [CrossRef]
- Nabavi, S.; Ejmalian, A.; Moghaddam, M.E.; Abin, A.A.; Frangi, A.F.; Mohammadi, M.; Rad, H.S. Medical imaging and computational image analysis in COVID-19 diagnosis: A review. Comput. Biol. Med. 2021, 135, 104605. [Google Scholar] [CrossRef]
- Serte, S.; Demirel, H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 2021, 132, 104306. [Google Scholar] [CrossRef]
- Wu, X.; Hui, H.; Niu, M.; Li, L.; Wang, L.; He, B.; Yang, X.; Li, L.; Li, H.; Tian, J.; et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: A multicentre study. Eur. J. Radiol. 2020, 128, 109041. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Chen, Y.; Su, J.; Lang, G. A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering 2020, 6, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Naz, S.; Khan, A.; Zaib, A.; Razzak, I. Improving Coronavirus (COVID-19) Diagnosis Using Deep Transfer Learning. Lect. Notes Netw. Syst. 2022, 350, 23–37. [Google Scholar] [CrossRef]
- Jin, C.; Chen, W.; Cao, Y.; Xu, Z.; Tan, Z.; Zhang, X.; Deng, L.; Zheng, C.; Zhou, J.; Shi, H.; et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020, 11, 5088. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Rafiee, M.J.; Naderkhani, F.; Heidarian, S.; Enshaei, N.; Oikonomou, A.; Fard, F.B.; Anconina, R.; Farahani, K.; Plataniotis, K.N.; et al. Human-level COVID-19 diagnosis from low-dose CT scans using a two-stage time-distributed capsule network. Sci. Rep. 2022, 12, 4827. [Google Scholar] [CrossRef]
- Yousefzadeh, M.; Esfahanian, P.; Movahed, S.M.S.; Gorgin, S.; Rahmati, D.; Abedini, A.; Nadji, S.A.; Haseli, S.; Karam, M.B.; Kiani, A.; et al. ai-corona: Radiologist-assistant deep learning framework for COVID-19 diagnosis in chest CT scans. PLoS ONE 2021, 16, e0250952. [Google Scholar] [CrossRef]
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Chen, Q.; Huang, S.; Yang, M.; Yang, X.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020, 10, 19196. [Google Scholar] [CrossRef]
- Javaheri, T.; Homayounfar, M.; Amoozgar, Z.; Reiazi, R.; Homayounieh, F.; Abbas, E.; Laali, A.; Radmard, A.R.; Gharib, M.H.; Mousavi, S.A.J.; et al. CovidCTNet: An open-source deep learning approach to diagnose COVID-19 using small cohort of CT images. NPJ Digit. Med. 2021, 4, 29. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef]
- Xing, W.; He, C.; Li, J.; Qin, W.; Yang, M.; Li, G.; Li, Q.; Ta, D.; Wei, G.; Li, W.; et al. Automated lung ultrasound scoring for evaluation of coronavirus disease 2019 pneumonia using two-stage cascaded deep learning model. Biomed. Signal Process. Control 2022, 75, 103561. [Google Scholar] [CrossRef]
- Born, J.; Wiedemann, N.; Cossio, M.; Buhre, C.; Brändle, G.; Leidermann, K.; Goulet, J.; Aujayeb, A.; Moor, M.; Rieck, B.; et al. Accelerating Detection of Lung Pathologies with Explainable Ultrasound Image Analysis. Appl. Sci. 2021, 11, 672. [Google Scholar] [CrossRef]
- COVID-19 Cough Audio Classification. Available online: https://www.kaggle.com/datasets/andrewmvd/covid19-cough-audio-classification (accessed on 29 April 2025).
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Plante, T.B.; Blau, A.M.; Berg, A.N.; Weinberg, A.S.; Jun, I.C.; Tapson, V.F.; Kanigan, T.S.; Adib, A.B. Development and external validation of a machine learning tool to rule out COVID-19 among adults in the emergency department using routine blood tests: A large, multicenter, real-world study. J. Med. Internet. Res. 2020, 22, e24048. [Google Scholar] [CrossRef] [PubMed]
- Ahrabi, S.S.; Scarpiniti, M.; Baccarelli, E.; Momenzadeh, A. An Accuracy vs. Complexity Comparison of Deep Learning Architectures for the Detection of COVID-19 Disease. Computation 2021, 9, 3. [Google Scholar] [CrossRef]
- Loey, M.; Smarandache, F.; Khalifa, N.E.M. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef] [PubMed]
- Rahimzadeh, M.; Attar, A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform. Med. Unlocked 2020, 19, 100360. [Google Scholar] [CrossRef]
- Yildirim, O.; Talo, M.; Ay, B.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Automated detection of diabetic subject using pre-trained 2D-CNN models with frequency spectrum images extracted from heart rate signals. Comput. Biol. Med. 2019, 113, 103387. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.S.; Asari, V.K. A State-of-the-Art Survey on Deep Learning Theory and Architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef]
- Keidar, D.; Yaron, D.; Goldstein, E.; Shachar, Y.; Blass, A.; Charbinsky, L.; Aharony, I.; Lifshitz, L.; Lumelsky, D.; Neeman, Z.; et al. COVID-19 classification of X-ray images using deep neural networks. Eur. Radiol. 2021, 31, 9654–9663. [Google Scholar] [CrossRef] [PubMed]
- Bhuyan, H.K.; Chakraborty, C.; Shelke, Y.; Pani, S.K. COVID-19 diagnosis system by deep learning approaches. Expert Syst. 2022, 39, e12776. [Google Scholar] [CrossRef] [PubMed]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the ICLR 2021—9th International Conference on Learning Representations, Vienna, Austria, 4 May 2021; Available online: https://arxiv.org/pdf/2010.11929 (accessed on 29 April 2025).
- Gunraj, H.; Wang, L.; Wong, A. COVIDNet-CT: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest CT Images. Front. Med. 2020, 7, 608525. [Google Scholar] [CrossRef]
- Gunraj, H.; Tuinstra, T.; Wong, A. COVIDx CT-3: A Large-Scale, Multinational, Open-Source Benchmark Dataset for Computer-aided COVID-19 Screening from Chest CT Images. arXiv 2022, arXiv:2206.03043. Available online: https://arxiv.org/pdf/2206.03043 (accessed on 29 April 2025).
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A.P. xViTCOS: Explainable Vision Transformer Based COVID-19 Screening Using Radiography. IEEE. J. Transl. Eng. Health Med. 2021, 10, 1100110. [Google Scholar] [CrossRef]
- Owais, M.; Lee, Y.W.; Mahmood, T.; Haider, A.; Sultan, H.; Park, K.R. Multilevel Deep-Aggregated Boosted Network to Recognize COVID-19 Infection from Large-Scale Heterogeneous Radiographic Data. IEEE. J. Biomed. Health Inform. 2021, 25, 1881–1891. [Google Scholar] [CrossRef]
- Bai, H.X.; Wang, R.; Xiong, Z.; Hsieh, B.; Chang, K.; Halsey, K.; Tran, T.M.L.; Choi, J.W.; Wang, D.-C.; Shi, L.-B.; et al. Artificial Intelligence Augmentation of Radiologist Performance in Distinguishing COVID-19 from Pneumonia of Other Origin at Chest CT. Radiology 2020, 296, E156–E165. [Google Scholar] [CrossRef]
- Al Rahhal, M.M.; Bazi, Y.; Jomaa, R.M.; AlShibli, A.; Alajlan, N.; Mekhalfi, M.L.; Melgani, F. COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers. J. Pers. Med. 2022, 12, 310. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar] [CrossRef]
- Costa, G.S.S.; Paiva, A.C.; Junior, G.B.; Ferreira, M.M. COVID-19 Automatic Diagnosis with CT Images Using the Novel Transformer Architecture. In Proceedings of the Anais do XXI Simpósio Brasileiro de Computação Aplicada à Saúde, Virtual Event, 15–18 June 2021; pp. 293–301, References—Scientific Research Publishing. Available online: https://www.scirp.org/reference/referencespapers?referenceid=3278554 (accessed on 29 April 2025).
- van Tulder, G.; Tong, Y.; Marchiori, E. Multi-view analysis of unregistered medical images using cross-view transformers. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer International Publishing: Berlin/Heidelberg, Germany, 2021; Volume 12903 LNCS, pp. 104–113. [Google Scholar] [CrossRef]
- (PDF) MIA-COV19D: A Transformer-Based Framework for COVID19 Classification in Chest CTs. Available online: https://www.researchgate.net/publication/353105641_MIA-COV19D_A_transformer-based_framework_for_COVID19_classification_in_chest_CTs (accessed on 29 April 2025).
- Naidji, M.R.; Elberrichi, Z. A Novel Hybrid Vision Transformer CNN for COVID-19 Detection from ECG Images. Computers 2024, 13, 109. [Google Scholar] [CrossRef]
- Tehrani, S.S.M.; Zarvani, M.; Amiri, P.; Ghods, Z.; Raoufi, M.; Safavi-Naini, S.A.A.; Soheili, A.; Gharib, M.; Abbasi, H. Visual transformer and deep CNN prediction of high-risk COVID-19 infected patients using fusion of CT images and clinical data. BMC Med. Inform. Decis. Mak. 2023, 23, 265. [Google Scholar] [CrossRef]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef] [PubMed]
- Sethy, P.K.; Behera, S.K. Detection of Coronavirus Disease (COVID-19) Based on Deep Features. Preprint 2020, 1–9. Available online: https://www.preprints.org/manuscript/202003.0300/v1 (accessed on 14 July 2025).
- Vinod, D.N.; Prabaharan, S.R.S. Elucidation of infection asperity of CT scan images of COVID-19 positive cases: A Machine Learning perspective. Sci. Afr. 2023, 20, e01681. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jin, S.; Yan, Q.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system. Appl. Soft Comput. 2021, 98, 106897. [Google Scholar] [CrossRef]
- Hassan, H.; Ren, Z.; Zhao, H.; Huang, S.; Li, D.; Xiang, S.; Kang, Y.; Chen, S.; Huang, B. Review and classification of AI-enabled COVID-19 CT imaging models based on computer vision tasks. Comput. Biol. Med. 2022, 141, 105123. [Google Scholar] [CrossRef]
- Sadik, F.; Dastider, A.G.; Subah, M.R.; Mahmud, T.; Fattah, S.A. A dual-stage deep convolutional neural network for automatic diagnosis of COVID-19 and pneumonia from chest CT images. Comput. Biol. Med. 2022, 149, 105806. [Google Scholar] [CrossRef]
- Kordnoori, S.; Sabeti, M.; Mostafaei, H.; Banihashemi, S.S.A. Analysis of lung scan imaging using deep multi-task learning structure for COVID-19 disease. IET Image Process. 2023, 17, 1534–1545. [Google Scholar] [CrossRef]
- Nafisah, S.I.; Muhammad, G.; Hossain, M.S.; AlQahtani, S.A. A Comparative Evaluation between Convolutional Neural Networks and Vision Transformers for COVID-19 Detection. Mathematics 2023, 11, 1489. [Google Scholar] [CrossRef]
- Tahir, A.M.; Chowdhury, M.E.H.; Khandakar, A.; Rahman, T.; Qiblawey, Y.; Khurshid, U.; Kiranyaz, S.; Ibtehaz, N.; Rahman, M.S.; Al-Maadeed, S.; et al. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021, 139, 105002. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferraz, A.; Betini, R.C. Comparative Evaluation of Deep Learning Models for Diagnosis of COVID-19 Using X-ray Images and Computed Tomography. J. Braz. Comput. Soc. 2025, 31, 99–131. [Google Scholar] [CrossRef]
- Irkham, I.; Ibrahim, A.U.; Nwekwo, C.W.; Al-Turjman, F.; Hartati, Y.W. Current Technologies for Detection of COVID-19: Biosensors, Artificial Intelligence and Internet of Medical Things (IoMT): Review. Sensors 2022, 23, 426. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mishra, S.; Satapathy, S.K.; Cho, S.-B.; Mohanty, S.N.; Sah, S.; Sharma, S. Advancing COVID-19 poverty estimation with satellite imagery-based deep learning techniques: A systematic review. Spat. Inf. Res. 2024, 32, 583–592. [Google Scholar] [CrossRef]
- Ning, W.; Lei, S.; Yang, J.; Cao, Y.; Jiang, P.; Yang, Q.; Zhang, J.; Wang, X.; Chen, F.; Geng, Z.; et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020, 4, 1197–1207. [Google Scholar] [CrossRef]
- Padmavathi; Ganesan, K. Metaheuristic optimizers integrated with vision transformer model for severity detection and classification via multimodal COVID-19 images. Sci. Rep. 2025, 15, 13941. [Google Scholar] [CrossRef]
- Satapathy, S.K.; Saravanan, S.; Mishra, S.; Mohanty, S.N. A Comparative Analysis of Multidimensional COVID-19 Poverty Determinants: An Observational Machine Learning Approach. New Gener. Comput. 2023, 41, 155–184. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Z.; Lei, H.; Chen, G.; Li, H.; Li, C.; Gao, W.; Chen, Y.; Wang, Y.; Xu, H.; Ma, G.; et al. Multi-center sparse learning and decision fusion for automatic COVID-19 diagnosis. Appl. Soft Comput. 2022, 115, 108088. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, S.C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. NPJ Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef]
- Guarrasi, V.; Aksu, F.; Caruso, C.M.; Di Feola, F.; Rofena, A.; Ruffini, F.; Soda, P. A systematic review of intermediate fusion in multimodal deep learning for biomedical applications. Image Vis. Comput. 2025, 158, 105509. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Bin Mahbub, Z.; Hossain, S.A.; Alhatou, A.; Abdalla, E.; Muthiyal, S.; Islam, K.F.; Kashem, S.B.A.; et al. BIO-CXRNET: A robust multimodal stacking machine learning technique for mortality risk prediction of COVID-19 patients using chest X-ray images and clinical data. Neural. Comput. Appl. 2023, 35, 17461–17483. [Google Scholar] [CrossRef]
- Tur, K. Multi-Modal Machine Learning Approach for COVID-19 Detection Using Biomarkers and X-Ray Imaging. Diagnostics 2024, 14, 2800. [Google Scholar] [CrossRef] [PubMed]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief. Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.N.; Falcone, G.J.; Rajpurkar, P.; Topol, E.J. Multimodal biomedical AI. Nat. Med. 2022, 28, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Althenayan, A.S.; AlSalamah, S.A.; Aly, S.; Nouh, T.; Mahboub, B.; Salameh, L.; Alkubeyyer, M.; Mirza, A. COVID-19 Hierarchical Classification Using a Deep Learning Multi-Modal. Sensors 2024, 24, 2641. [Google Scholar] [CrossRef] [PubMed]
- Mukhi, S.E.; Varshini, R.T.; Sherley, S.E.F. Diagnosis of COVID-19 from Multimodal Imaging Data Using Optimized Deep Learning Techniques. SN Comput. Sci. 2023, 4, 1–9. [Google Scholar] [CrossRef]
- Ali, M.U.; Zafar, A.; Tanveer, J.; Khan, M.A.; Kim, S.H.; Alsulami, M.M.; Lee, S.W. Deep learning network selection and optimized information fusion for enhanced COVID-19 detection. Int. J. Imaging Syst. Technol. 2024, 34, e23001. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Bhaik, D.; Kumar, S.; Thakur, P.; Sharma, R.; Pachori, R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process. Control 2022, 71, 103182. [Google Scholar] [CrossRef]
- Nguyen-Tat, T.B.; Tran-Thi, V.T.; Ngo, V.M. Predicting the Severity of COVID-19 Pneumonia from Chest X-Ray Images: A Convolutional Neural Network Approach. EAI Endorsed Trans. Ind. Netw. Intell. Syst. 2025, 12, 1–19. [Google Scholar] [CrossRef]
- Junia, R.C.; Selvan, K. Resolute neuronet: Deep learning-based segmentation and classification COVID-19 using chest X-Ray images. Int. J. Syst. Assur. Eng. Manag. 2024, 1–13. [Google Scholar] [CrossRef]
- Chen, L.; Lin, X.; Ma, L.; Wang, C. A BiLSTM model enhanced with multi-objective arithmetic optimization for COVID-19 diagnosis from CT images. Sci. Rep. 2025, 15, 10841. [Google Scholar] [CrossRef]
- Veluchamy, S.; Sudharson, S.; Annamalai, R.; Bassfar, Z.; Aljaedi, A.; Jamal, S.S. Automated Detection of COVID-19 from Multimodal Imaging Data Using Optimized Convolutional Neural Network Model. J. Imaging Inform. Med. 2024, 37, 2074–2088. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Tartaglione, E.; Barbano, C.A.; Berzovini, C.; Calandri, M.; Grangetto, M. Unveiling COVID-19 from Chest X-ray with Deep Learning: A Hurdle Race with Small Data. arXiv 2020, arXiv:2004.05405. [Google Scholar]
- Li, T.; Sahu, A.K.; Talwalkar, A.; Smith, V. Federated Learning: Challenges, Methods, and Future Directions. IEEE Signal Process. Mag. 2020, 37, 50–60. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated Learning in Medicine: Facilitating Multi-Institutional Collaborations without Sharing Patient Data. Med. Image. Anal. 2020, 65, 101759. [Google Scholar] [CrossRef] [PubMed]
- Dayan, I.; Roth, H.R.; Zhong, A.; Harouni, A.; Gentili, A.; Abidin, A.Z.; Liu, A.; Costa, A.B.; Wood, B.J.; Tsai, C.-S.; et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021, 27, 1735–1743. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Makkar, A.; Santosh, K.C. SecureFed: Federated learning empowered medical imaging technique to analyze lung abnormalities in chest X-rays. Int. J. Mach. Learn Cybern. 2023, 14, 1–12. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sandhu, S.S.; Gorji, H.T.; Tavakolian, P.; Tavakolian, K.; Akhbardeh, A. Medical Imaging Applications of Federated Learning. Diagnostics 2023, 13, 3140. [Google Scholar] [CrossRef]
- Feki, I.; Ammar, S.; Kessentini, Y.; Muhammad, K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021, 106, 107330. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dou, Q.; So, T.Y.; Jiang, M.; Liu, Q.; Vardhanabhuti, V.; Kaissis, G.; Li, Z.; Si, W.; Lee, H.H.C.; Yu, K.; et al. Federated deep learning for detecting COVID-19 lung abnormalities in CT: A privacy-preserving multinational validation study. NPJ Digit. Med. 2021, 4, 60. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Shen, J.; Li, Z.; Sang, Y.; Wu, X.; Zha, Y.; Liang, W.; Wang, C.; Wang, K.; et al. Clinically Applicable AI System for Accurate Diagnosis, Quantitative Measurements, and Prognosis of COVID-19 Pneumonia Using Computed Tomography. Nat. Commun. 2020, 11, 5088. [Google Scholar]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.; Debray, T.P.A.; et al. Prediction Models for Diagnosis and Prognosis of COVID-19 Infection: Systematic Review and Critical Appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What Do We Need to Build Explainable AI Systems for the Medical Domain? arXiv 2017, arXiv:1712.09923. [Google Scholar] [CrossRef]
- Sadeghi, Z.; Alizadehsani, R.; Cifci, M.A.; Kausar, S.; Rehman, R.; Mahanta, P.; Bora, P.K.; Almasri, A.; Alkhawaldeh, R.S.; Hussain, S.; et al. A review of Explainable Artificial Intelligence in healthcare. Comput. Electr. Eng. 2024, 118, 109370. [Google Scholar] [CrossRef]
- Önder, M.; Şentürk, Ü.; Polat, K. Shallow Learning Versus Deep Learning in Biomedical Applications, In Shallow Learning vs. Deep Learning The Springer Series in Applied Machine Learning; Ertuğrul, Ö.F., Guerrero, J.M., Yilmaz, M., Eds.; Springer: Cham, Switzerland, 2024. [Google Scholar] [CrossRef]
- Rundo, L.; Militello, C. Image biomarkers and explainable AI: Handcrafted features versus deep learned features. Eur. Radiol. Exp. 2024, 8, 130. [Google Scholar] [CrossRef]
- Amann, J.; Vetter, D.; Blomberg, S.N.; Christensen, H.C.; Coffee, M.; Gerke, S.; Gilbert, T.K.; Hagendorff, T.; Holm, S.; Livne, M.; et al. Z-Inspection initiative. To explain or not to explain? Artificial intelligence explainability in clinical decision support systems. PLoS Digit. Health 2022, 17, e0000016. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saarela, M.; Podgorelec, V. Recent Applications of Explainable AI (XAI): A Systematic Literature Review. Appl. Sci. 2024, 14, 8884. [Google Scholar] [CrossRef]
- Ko, H.; Chung, H.; Kang, W.S.; Kim, K.W.; Shin, Y.; Kang, S.J.; Lee, J.H.; Kim, Y.J.; Kim, N.Y.; Jung, H.; et al. COVID-19 pneumonia diagnosis using a simple 2d deep learning framework with a single chest CT image: Model development and validation. J. Med. Internet. Res. 2020, 22, e19569. [Google Scholar] [CrossRef]
- Hasan, A.M.; Al-Jawad, M.M.; Jalab, H.A.; Shaiba, H.; Ibrahim, R.W.; Al-Shamasneh, A.R. Classification of COVID-19 Coronavirus, Pneumonia and Healthy Lungs in CT Scans Using Q-Deformed Entropy and Deep Learning Features. Entropy 2020, 22, 517. [Google Scholar] [CrossRef]
- Waheed, A.; Goyal, M.; Gupta, D.; Khanna, A.; Al-Turjman, F.; Pinheiro, P.R. CovidGAN: Data Augmentation Using Auxiliary Classifier GAN for Improved COVID-19 Detection. IEEE Access 2020, 8, 91916–91923. [Google Scholar] [CrossRef]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable Deep Learning for Pulmonary Disease and Coronavirus COVID-19 Detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Singh, V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos. Solitons Fractals 2020, 138, 109944. [Google Scholar] [CrossRef]
- Ni, Q.; Sun, Z.Y.; Qi, L.; Chen, W.; Yang, Y.; Wang, L.; Zhang, X.; Yang, L.; Fang, Y.; Xing, Z.; et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020, 30, 6517–6527. [Google Scholar] [CrossRef] [PubMed]
- Pathak, Y.; Shukla, P.K.; Arya, K.V. Deep Bidirectional Classification Model for COVID-19 Disease Infected Patients. IEEE./ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.M.; Bertolini, D.; Teixeira, L.O.; Silla, C.N.; Costa, Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020, 194, 105532. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zha, Y.; Li, W.; Wu, Q.; Li, X.; Niu, M.; Wang, M.; Qiu, X.; Li, H.; Yu, H.; et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020, 56, 2000775. [Google Scholar] [CrossRef]
- Lan, M.; Meng, M.; Yu, J.; Wu, J. Learning to Discover Knowledge: A Weakly-Supervised Partial Domain Adaptation Approach. IEEE Trans. Image. Process 2024, 33, 4090–4103. [Google Scholar] [CrossRef]
- Yang, J.; Dung, N.T.; Thach, P.N.; Phong, N.T.; Phu, V.D.; Phu, K.D.; Yen, L.M.; Thy, D.B.X.; Soltan, A.A.S.; Thwaites, L.; et al. Generalizability assessment of AI models across hospitals in a low-middle and high income country. Nat. Commun. 2024, 15, 8270. [Google Scholar] [CrossRef]
- Haynes, S.C.; Johnston, P.; Elyan, E. Generalisation challenges in deep learning models for medical imagery: Insights from external validation of COVID-19 classifiers. Multimed. Tools Appl. 2024, 83, 76753–76772. [Google Scholar] [CrossRef]
- Du, K.L.; Zhang, R.; Jiang, B.; Zeng, J.; Lu, J. Foundations and Innovations in Data Fusion and Ensemble Learning for Effective Consensus. Mathematics 2025, 13, 587. [Google Scholar] [CrossRef]
- Kim, T.; Lee, H. Evaluating AI Models and Predictors for COVID-19 Infection Dependent on Data from Patients with Cancer or Not: A Systematic Review. Korean J. Clin. Pharm. 2024, 34, 141–154. [Google Scholar] [CrossRef]
- Nishio, M.; Kobayashi, D.; Nishioka, E.; Matsuo, H.; Urase, Y.; Onoue, K.; Ishikura, R.; Kitamura, Y.; Sakai, E.; Tomita, M.; et al. Deep learning model for the automatic classification of COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy: A multi-center retrospective study. Sci. Rep. 2022, 12, 8214. [Google Scholar] [CrossRef]
- Komolafe, T.E.; Cao, Y.; Nguchu, B.A.; Monkam, P.; Olaniyi, E.O.; Sun, H.; Zheng, J.; Yang, X. Diagnostic Test Accuracy of Deep Learning Detection of COVID-19: A Systematic Review and Meta-Analysis. Acad. Radiol. 2021, 28, 1507–1523. [Google Scholar] [CrossRef]
- Moulaei, K.; Shanbehzadeh, M.; Mohammadi-Taghiabad, Z.; Kazemi-Arpanahi, H. Comparing machine learning algorithms for predicting COVID-19 mortality. BMC Med. Inform. Decis. Mak. 2022, 22, 2. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Peng, L.; Li, T.; Adila, D.; Zaiman, Z.; Melton-Meaux, G.B.; Ingraham, N.E.; Murray, E.; Boley, D.; Switzer, S.; et al. Performance of a Chest Radiograph AI Diagnostic Tool for COVID-19: A Prospective Observational Study. Radiol. Artif. Intell. 2022, 4, e210217. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, A.H.; Saba, A.I.; Panchal, H.; Shanmugan, S.; Alsaleh, N.A.; Ahmadein, M. Artificial Intelligence for Forecasting the Prevalence of COVID-19 Pandemic: An Overview. Healthcare 2021, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, Y.; Rios, I.; Araújo, P.; Macambira, A.; Guimarães, M.; Sales, L.; Júnior, M.R.; Nicola, A.; Nakayama, M.; Paschoalick, H.; et al. Artificial intelligence in triage of COVID-19 patients. Front. Artif. Intell. 2024, 7, 1495074. [Google Scholar] [CrossRef]
- Al-Azzawi, A.; Al-Jumaili, S.; Duru, A.D.; Duru, D.G.; Uçan, O.N. Evaluation of Deep Transfer Learning Methodologies on the COVID-19 Radiographic Chest Images. Trait. Du Signal 2023, 40, 407–420. [Google Scholar] [CrossRef]
- Talib, M.A.; Moufti, M.A.; Nasir, Q.; Kabbani, Y.; Aljaghber, D.; Afadar, Y. Transfer Learning-Based Classifier to Automate the Extraction of False X-Ray Images from Hospital’s Database. Int. Dent. J. 2024, 74, 1471–1482. [Google Scholar] [CrossRef]
- Amyar, A.; Modzelewski, R.; Li, H.; Ruan, S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Comput. Biol. Med. 2020, 126, 104037. [Google Scholar] [CrossRef]
- Gorriz, J.M.; Dong, Z.; Chetoui, M.; Akhloufi, M.A. Explainable Vision Transformers and Radiomics for COVID-19 Detection in Chest X-rays. J. Clin. Med. 2022, 11, 3013. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.H.; Sohail, A.; Zafar, M.M.; Khan, A. Coronavirus disease analysis using chest X-ray images and a novel deep convolutional neural network. Photodiagnosis Photodyn. Ther. 2021, 35, 102473. [Google Scholar] [CrossRef] [PubMed]
- Sharmila, V.J. Deep Learning Algorithm for COVID-19 Classification Using Chest X-Ray Images. Comput. Math. Methods Med. 2021, 2021, 9269173. [Google Scholar] [CrossRef] [PubMed]
- Gouda, W.; Almurafeh, M.; Humayun, M.; Jhanjhi, N.Z. Detection of COVID-19 Based on Chest X-rays Using Deep Learning. Healthcare 2022, 10, 343. [Google Scholar] [CrossRef]
- Assessment of a Deep Learning Model for COVID-19 Classification on Chest Radiographs: A Comparison Across Image Acquisition Techniques and Clinical Factors. Available online: https://www.spiedigitallibrary.org/journals/journal-of-medical-imaging/volume-10/issue-06/064504/Assessment-of-a-deep-learning-model-for-COVID-19-classification/10.1117/1.JMI.10.6.064504.full (accessed on 29 April 2025).
- Krones, F.; Marikkar, U.; Parsons, G.; Szmul, A.; Mahdi, A. Review of multimodal machine learning approaches in healthcare. Inf. Fusion. 2025, 114, 102690. [Google Scholar] [CrossRef]
- Sun, C.; Hong, S.; Song, M.; Li, H.; Wang, Z. Predicting COVID-19 disease progression and patient outcomes based on temporal deep learning. BMC Med. Inform. Decis. Mak. 2021, 21, 45. [Google Scholar] [CrossRef]
- Leitner, J.; Behnke, A.; Chiang, P.H.; Ritter, M.; Millen, M.; Dey, S. Classification of Patient Recovery From COVID-19 Symptoms Using Consumer Wearables and Machine Learning. IEEE J. Biomed. Health Inform. 2023, 27, 1271–1282. [Google Scholar] [CrossRef]
- Saini, K.; Devi, R. A systematic scoping review of the analysis of COVID-19 disease using chest X-ray images with deep learning models. J. Auton. Intell. 2024, 7, 1–19. [Google Scholar] [CrossRef]
- Andrade-Girón, D.C.; Marín-Rodriguez, W.J.; Lioo-Jordán, F.de.M.; Villanueva-Cadenas, G.J.; Garivay-Torres, d.M.F. Neural Networks for the Diagnosis of COVID-19 in Chest X-ray Images: A Systematic Review and Meta-Analysis. EAI Endorsed. Trans. Pervasive. Health Technol. 2023, 9, 2023. [Google Scholar] [CrossRef]
- Park, S.; Kim, G.; Oh, Y.; Seo, J.B.; Lee, S.M.; Kim, J.H.; Moon, S.; Lim, J.-K.; Ye, J.C. Multi-task vision transformer using low-level chest X-ray feature corpus for COVID-19 diagnosis and severity quantification. Med. Image. Anal. 2022, 75, 102299. [Google Scholar] [CrossRef]
- An, P.; Li, X.; Qin, P.; Ye, Y.; Zhang, J.; Guo, H.; Duan, P.; He, Z.; Song, P.; Li, M.; et al. Predicting model of mild and severe types of COVID-19 patients using Thymus CT radiomics model: A preliminary study. Math. Biosci. Eng. 2023, 4, 6612–6629. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Liu, G.; Bai, Q. Deep Learning in COVID-19 Diagnosis, Prognosis and Treatment Selection. Mathematics 2023, 11, 1279. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, Y.; Sebe, N.; Yan, Y. Vision + X: A Survey on Multimodal Learning in the Light of Data. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 9102–9122. [Google Scholar] [CrossRef] [PubMed]
- Ajagbe, S.A.; Adigun, M.O. Deep learning techniques for detection and prediction of pandemic diseases: A systematic literature review. Multimed. Tools Appl. 2024, 83, 5893–5927. [Google Scholar] [CrossRef]
- El-Bouzaidi, Y.E.I.; Abdoun, O. Advances in artificial intelligence for accurate and timely diagnosis of COVID-19: A comprehensive review of medical imaging analysis. Sci. Afr. 2023, 22, e01961. [Google Scholar] [CrossRef]
- Yurdem, B.; Kuzlu, M.; Gullu, M.K.; Catak, F.; Tabassum, M. Federated learning: Overview, strategies, applications, tools and future directions. Helyion 2024, 10, e38137. [Google Scholar] [CrossRef]
- Subramanian, N.; Elharrouss, O.; Al-Maadeed, S.; Chowdhury, M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med 2022, 13, 105233. [Google Scholar] [CrossRef]
| Study (Year) | Data (Chest X-Ray) | Classes/Task | Model(s) and Approach | Performance |
|---|---|---|---|---|
| Narin et al. (2021) [8] | 100 images (50 COVID and 50 normal) | COVID vs. Normal (binary) | ResNet50 (TL fine-tuned) | Acc 98.0% |
| Hemdan et al. (2020) [9] | 50 images (25 COVID and 25 normal) | COVID vs. Normal (binary) | VGG19 and DenseNet201 (TL; ensemble reported) | Acc~90%; F1 91% |
| Apostolopoulos et al. (2020) [10] | ~2900 images (mixed sources) | COVID vs. Pneumonia vs. Normal | MobileNetV2 (TL) | Acc 96–98% (3-class) |
| Horry et al. (2020) [11] | 400 images (100 COVID, 100 pneumonia, and 200 healthy) | COVID vs. Pneumonia vs. Normal | Inception, Xception, ResNet, and VGG (ensemble) | Prec 83%; Sens 80%; F1 80% |
| Bukhari et al. (2020) [12] | 278 images (3 classes: COVID, normal, and pneumonia) | COVID vs. Pneumonia vs. Normal | ResNet50 (TL fine-tuned) | Acc 98.2%; Sens 98.2%; F1 98.2% |
| Benmalek et al. (2021 [13] | 158 images (COVID vs. others) | COVID vs. Other diseases (binary) | ResNet50 + SVM (a CNN for features and an SVM classifier) | Acc 95.4% |
| Minaee et al. (2020) [14] | 5071 images (many sources; 100 COVID) | COVID vs. Normal (binary) | Ensemble of ResNet18, ResNet50 and SqueezeNet (TL) | Acc 98.0%; Sens 100% |
| Kaya & Gürsoy (2023 [15] | 9457 images (3-class dataset) | COVID vs. Pneumonia vs. Normal | MobileNetV2 (TL; 5-fold cross-val) | Acc 97.6% (3-class) |
| Ilhan et al. (2021) [16] | ~23,000 images (from 3 public sets combined) | COVID vs. Pneumonia vs. Normal | Feature fusion of 7 CNNs + ensemble classifiers | Acc 90.7%; Prec 93%; Sens 91% (3-class) |
| Study (Year) | Data (Chest CT) | Classes/Task | Model(s) and Approach | Performance |
|---|---|---|---|---|
| Wu et al. (2020) [22] | 495 CT images (368 COVID and 127 other infections) | COVID vs. Other infections | ResNet50 (TL and multi-view fusion of slices) | Acc 76%; Sens 81%; Spec 62% |
| Xu et al. (2020) [23] | – (CT images and a small dataset) | COVID vs. Non-COVID (binary) | ResNet18 (TL, with lung segmentation) | Acc 86.7% |
| Rehman et al. (2022 [24] | – (CT and a three-class problem) | COVID vs. Viral vs. Bacterial Pneu. | ResNet101 (TL fine-tuned) | Acc 98.75% (on the COVID class) |
| Jin et al. (2020) [25] | 1881 images (496 COVID and 1385 healthy) | COVID vs. Normal (binary) | ResNet152 (TL fine-tuned) | Acc 94.98% |
| Afshar et al. (2022 [26] | 1020 CT scans (COVID vs. healthy) | COVID vs. Normal (binary) | ResNet101 (TL; evaluated on the hold-out set) | Acc 99.5% |
| Yousefzadeh et al. (2021) [27] | 2124 CT scans (706 COVID and 1418 normal) | COVID vs. Normal (binary) | Ensemble of ResNet, EfficientNet, DenseNet, etc. (COVID-AI) | Acc 96.4% |
| Chen et al. (2020) [28] | 46,096 CT images (from hospital) | COVID vs. Normal (binary) | UNet++ + ResNet50 (segmentation + classification) | Acc 98.85% |
| Javaheri et al. (2021) [29] | 89,145 CT images (32 k COVID, 25 k CAP, and 31 k healthy) | COVID vs. CAP vs. Normal (3-class) | CovidCTNet (3D U-Net for localization + CNN) | Acc 91.66%; Sens 87.5%; Spec 94.0%; AUC 0.95 |
| Study (Year) | Datasets (Modality) | Public Availability | Dataset Diversity | Key Findings (CNN vs. ViT) |
|---|---|---|---|---|
| Nafisah et al., 2023 [66] MDPI Mathematics | COVID-QU-Ex (CXR): ~21,165 images (10,192 normal, 7357 non-COVID-19 pneumonia, and 3616 COVID-19) with lung masks [66,67]. | Yes—open dataset (COVID-QU-Ex) available via Kaggle | Multi-source and large: Compiled from multiple public repositories (the largest COVID-19 CXR dataset). Images vary in quality, resolution, and source (frontal CXRs from diverse hospitals), enhancing generalizability [67]. | CNN vs. ViT: Achieved comparably high accuracy. The best CNN (EfficientNet-B7) achieved 99.82% accuracy, slightly outperforming the best ViT model (SegFormer, 99.7% range) [66]. Both model types showed near-ceiling performance on this dataset, particularly after lung-region segmentation and augmentation, indicating no clear advantage of the ViT over the CNN in this setting. |
| Ferraz & Betini, 2025 [68] J. Brazilian Comp. Society | COVID-QU-Ex (CXR), HCV-UFPR-COVID-19 (CXR), HUST-19 (CT), and SARS-CoV-2 CT-Scan (CT). These four benchmark datasets cover both X-rays and CTs [67,68]. | Mixed: COVID-QU-Ex, HUST-19, and SARS-CoV-2 CT are public; HCV-UFPR is private (available upon request). COVID-QU-Ex and HUST-19 are open-access datasets [68]; the HCV-UFPR X-rays are provided on a case-by-case basis by Hospital da Cruz Vermelha (Brazil) [69]; the SARS-CoV-2 CT set is from Kaggle (Al Rahhal et al., 2022) [52]. | COVID-QU-Ex: Multi-institution global CXR collection (highly diverse in source institutions and imaging conditions) [70]. HCV-UFPR: Single-hospital in Brazil (281 COVID-19 and 232 normal CXRs [69]; limited demographic variety). HUST-19: Large CT dataset (≈13,980 images from ~1521 patients) from Huazhong Univ. hospitals in Wuhan [65]—sizable but geographically localized. SARS-CoV-2 CT: Collected from multiple hospitals in São Paulo (2482 CT images) [71]; some diversity within one region. | CNN vs. ViT: All models achieved strong classification results, but the Swin Transformer (ViT) consistently outperformed the CNN (ResNet-50) on both CXR and CT tasks [67]. Notably, Swin demonstrated greater generalization in cross-dataset experiments (training on one dataset and testing on another), achieving an AUC/accuracy of up to 1.0 on some test sets [67]. This suggests that ViT-based models (especially Swin) handle distribution shifts between institutions better than the CNN in this study. |
| Padmavathi & Ganesan, 2025 [72] Scientific Reports (Nature) | COVID-QU-Ex (CXR, 33,920 images across COVID-19/non-COVID-19/normal classes, with ground-truth lung masks) and a Wuhan CT Collection (CT images from Union and Liyuan Hospitals, Wuhan; 8000 CT slices balanced between COVID-19-positive and normal) [72]. | Yes—both datasets were made public by the authors on Kaggle (the CXR set is the COVID-QU-Ex Kaggle version, and the CT set contained preprocessed data from the two hospitals) [72]. | COVID-QU-Ex: Very diverse; drawn from multiple international CXR sources; covers varied patient demographics, imaging devices, and conditions [67]. Wuhan CT dataset: Collected from two large hospitals (common imaging protocols); less geographic diversity (all patients from the same region), but a substantial sample size, ensuring statistical power [72]. | CNN vs. ViT: A ViT-based approach with optimization techniques significantly outperformed classic CNNs in COVID detection. The proposed hybrid ViT model achieved ~99.1% accuracy in binary CXR classification, compared with 84–93% for standard CNN baselines (ResNet34, ~84.2%; VGG19, ~93.2%) [72]. Similar trends were observed with the CT data (~98.9% vs. mid-80 s%). The ViT’s attention mechanism (enhanced by Gray Wolf and PSO optimizers) captured global lung features, yielding superior performance and robustness [72]. This highlights significant performance gains for ViTs over CNNs in both modalities when advanced tuning is applied. |
| Tehrani et al., 2023 [58] BMC Med. Inf. and Dec. Making | Private Iran COVID-CT Dataset: 380 COVID-19 patients’ CT scans (each with 50–70 slice images) plus corresponding clinical data (demographics, vitals, labs) [58]. After preprocessing, 321 patients’ data were used for outcome prediction (e.g., survival vs. deterioration) [58]. | No (Private)—Data were collected in-house and are not publicly available (only shared by authors upon reasonable request) [58]. | Locally collected: All CT scans and patient records come from a limited number of hospitals (single country); thus, patient demographics and scan conditions are relatively homogeneous. The authors note the challenge of obtaining large multi-center clinical datasets [58]. (They mitigated data scarcity using data fusion and augmentation, but the dataset lacks the multi-national diversity of open datasets.) | CNN vs. ViT: This study fused 3D chest CT images with clinical features to predict disease outcomes. A 3D video Swin transformer (ViT) model outperformed several CNN models, given the same input data [58]. In predicting high-risk patients, the Swin transformer achieved the highest true-positive rate (~0.95) and best overall AUC (0.77) among the tested models [58]. In contrast, conventional 3D CNNs on the same task showed lower accuracy (The TPR for CNNs was lower, not reaching 0.95). This indicates the ViT’s stronger ability to leverage 3D imaging + clinical information for COVID-19 severity prediction. |
| Fusion Type | Description | Performance Summary | Strengths | Limitations |
|---|---|---|---|---|
| Data-Level Fusion | Merges aggregate data (or images) across several modalities (e.g., CT + X-ray images or image + clinical data) into a common input space. | Imaging + clinical data are rarely used. They are used in imaging (e.g., multi-view X-rays). Middling gains are easily susceptible to noise gain. | Stores the largest quantity of data; easy to apply to data of the same type (e.g., multi-view X-rays). | Heavy computational time requirements; high dimensionality explosion of inputs; sensitivity to missing data; concept drift; modality alignment task. |
| Feature-Level Fusion | Performs fusion on latent features produced by modalities (e.g., picture embeddings + clinical) prior to the last prediction levels. | Highest performance in most COVID-19 studies. Other models, such as transformer-based designs with concatenated image + clinical features, are superior. | Balances complexity that can be handled and the richness of information. Allows each mode encoder to become specialized. Resistant to noise; resistant to non- homogeneous data (CT + lab data). | Mandates scrupulous architectural design of fusion layers to align the dimensions and semantics of features. |
| Decision-Level Fusion | The modules of each modality are separated; the outputs (labels or probability scores) of each model are brought together through either voting, averaging, or meta-learners. | Performs well on small datasets or in heterogeneous modalities. Provides robustness at the cost of usually not being as accurate as feature-level fusion when abundant data are available | Practical to apply; independent modes minimized cross-modality interference; tolerant to failure in one of the modes. | Overlooks the interaction of deep features across modalities; has a lower performance limit than feature fusion. |
| Source of Non-Uniformity | Description | Examples from COVID-19 Datasets |
|---|---|---|
| Device and Scanner Variability | Variability across X-ray or CT machines (brand, model, imaging resolution, and imaging protocol). | - The COVIDx dataset contains scans from several hospitals, generated using different machines. - COVID-QU-Ex combines images from various equipment, which have various image qualities. |
| Acquisition Protocol Differences | Variation in patient positioning (AP vs. PA view in X-rays), slice thickness in CT, exposure time, and contrast use. | - Other datasets are a combination of AP and PA chest X-rays, which have not been standardized. - CT data differ in slice thickness (1 mm vs. 5 mm), which affects the level of detail observed in the lung images. |
| Patient Demographics | Differences in age, sex, ethnicity, geography, and comorbidities. | - COVID-QU-Ex has a worldwide distribution, but it lacks representation of some ethnicities. - The HUST-19 CT dataset has less diversity, as it primarily contains data from individuals from Wuhan. |
| Annotation Inconsistency | Lip antenna variation in labels because of divergent standards of diagnosis, manual vs. automated labeling, or mistakes. | - COVIDx contains labels derived from text reports as opposed to radiologist consensus. - Other CT datasets have slices, which are ambiguous, instead of patient labels. |
| Image Preprocessing Differences | Differences in normalization, resizing, windowing (particularly of CT), and cropping of images. | - Differing methods of normalizing CT scans. Some datasets normalize to [−1000, 400 HU], while others normalize to [−1024, 3071 HU]. - In certain datasets, chest X-rays can be cropped to lung areas, while there is no such cropping in other datasets. |
| Class Imbalance and Selection Bias | Artificially high proportions of COVID-19-positive cases or other conditions; selection within particular hospital groups. | - COVID-19 data are biased towards severe (hospitalized patients) vs. mild/asymptomatic cases. - It is possible that some datasets either excluded normal cases or included more pneumonia controls in populations. |
| Source | Impact |
|---|---|
| Device Variability | There is a risk that models are scanner-dependent and modules learn scanner patterns (gridlines and noise patterns) instead of pathology. Inefficient when used on various scanners in the hospital. |
| Acquisition Protocol Differences | Inconsistent characteristics on account of angles of projection (e.g., AP vs. PA), which results in a loss of accuracy when dependencies between deployment data are not the same in the methods of acquisition. Example: Models based on the frontal view may misdiagnose lateral images. |
| Patient Demographics | Poor generalization to unobservable populations (e.g., age groups and ethnicities). A model trained on a majority of elderly patient cases may not work on pediatric patients or young adults. |
| Annotation Inconsistency | Labeling errors are a source of noise in training data, resulting in an inflated level of false positives/negatives. Disadvantages: It decreases the maximum accuracy that can be attained and compromises reliability. |
| Preprocessing Variability | Models trained to perform on a specific preprocessing pipeline (e.g., cropped lungs) fail when test images have gone through a different preprocessing pipeline. It is particularly useful in the context of transfer learning and deployment. |
| Class Imbalance | Prejudice against overrepresented classes (e.g., overrepresentation of COVID-19-positive samples in a dataset where COVID-19 cases represent 80 percent of the data, whereas in the real world, it is only ~10 percent) causes inaccuracy in real-life screening. |
| Challenge Type | Description | Example Confounders |
|---|---|---|
| Image Artifacts | The non-biological characteristics that mask or resemble an image. | Motion blur, metal implants, ECG leads, portable X-ray artifacts, and under/overexposure. |
| Comorbidities | Other systemic or pulmonary diseases that can change the imaging findings. | COPD, pulmonary fibrosis, lung cancer, and heart failure (causing pulmonary edema). |
| Other Respiratory Infections | Non-COVID-19 pneumonias or viral infections that share imaging features. | Bacterial pneumonia, influenza, SARS, MERS, and tuberculosis. |
| Factor | Resilience Level | Observations from the Literature |
|---|---|---|
| Image Artifacts | Moderate to poor | -CNNs are highly sensitive to artifacts. Studies show that models often misclassify based on scanner noise, image borders, or embedded texts rather than lung pathology. -Vision transformers (ViTs) exhibit better resilience due to global context awareness but still degrade with severe motion blur or low-dose CT noise. -Data augmentation with synthetic artifacts improves robustness. |
| Comorbidities | Low to moderate | -AI models trained on COVID-19 datasets often fail to generalize to patients with overlapping conditions, such as heart failure or COPD. -Studies (e.g., Cohen et al., 2020 [89]) have found a high false-positive rate in elderly patients with comorbid pulmonary edema, as fluid accumulation mimics COVID-19 consolidations. -Feature-level fusion with clinical data improves performance (e.g., distinguishing COVID-19 from heart failure when oxygen saturation and BNP levels are included). |
| Other Respiratory Infections | Variable—Poor if not explicitly trained | -Differentiating between COVID-19 and non-COVID-19 pneumonia is the most challenging aspect in imaging-based diagnosis, since patterns such as ground-glass opacities are not COVID-19-specific. -Models trained without diverse non-COVID-19 pneumonia samples often show inflated accuracy, failing real-world deployment. -Well-curated datasets with balanced pneumonia classes (e.g., COVID-QU-Ex and COVIDx) improve resilience, but accuracy drops by 10–15% compared with COVID-19 vs. healthy tasks. |
| Rank | Authors | Population | Technique | Model | Imaging Type | Key Results |
|---|---|---|---|---|---|---|
| 1 | Loey et al. [37] | 306 | DL | GoogleNet | X-ray | Acc 100% |
| 2 | Ko et al. [106] | 3993 | DL | ResNet-50 (FCONet) | CT Scan | Acc 99.87%, Sens 99.58%, Spec 100%, and |
| 3 | Hasan et al. [107] | 321 | TL | LSTM Classifier | CT Scan | Acc 99.68% |
| 4 | Ardakani et al. [30] | 194 | DL | AlexNet, VGG-16, VGG-19, GoogleNet, and SqueezeNet | CT Scan | Acc 99.51%, Sen 100%, and Spec 99.02% |
| 5 | Apostolopoulos and Mpesiana [10,108] | 455 | CoroNet (DL-based) | MobileNetV2 | X-ray | Acc 99.18%, Sens 97.36%, and Spec 99.42% |
| 6 | Waheed et al. [108] | 1124 | GAN (CovidGAN) | ACGAN3 and VGG-16 | X-ray | Acc 95%, Sens 90%, and Spec 97% |
| 7 | Rahimzadeh and Attar [39] | 11,302 | DL | ResNet50V2 + Xception | X-ray | Acc 95.5%, and overall 91.4% |
| 8 | Saiz and Barandiaran | 1500 | CNN + TL | VGG-16 (Single-Depth Dilation) | X-ray | Acc 94.92%, Sens 94.92%, Spec 92%, and F1-score 97 |
| 9 | Wang et al. [62] | 181 | DL | VGG-19 | X-ray | Acc 96.3% |
| 10 | Brunese et al. [109] | 6523 | CoroNet (DL-based) | VGG-16 | X-ray | Acc 96.3% |
| 11 | Abbas et al. [110] | 6523 | CoroNet (DL-based) | VGG-16 | X-ray | Acc 97% |
| 12 | Panwar et al. [111] | 337 | DL | VGG-16 | X-ray | Acc 88.10%, Sens 97.62%, and Spec 85.7% |
| 13 | Ni et al. [112] | 14,531 | DL | 3D U-Net + MVPNet | CT Scan | Sens 100% and lobe lesion score 0.96 (no Acc mentioned) |
| 14 | Pathak et al. [113] | 852 | TL | ResNet-50 | CT Scan | Acc 93% |
| 15 | Yang et al. [19,114] | 295 | DL | DenseNet | CT Scan | Acc 92%, Sens 97%, and Spec 7% (very low specificity) |
| 16 | Pereira et al. [115] | 1144 | CNN | Inception-V3 | X-ray | F1-score: 89 (no Acc reported) |
| 17 | Sethy et al. [60] | 381 | CNN + SVM | ResNet-50 | X-ray | Sens 95.33% (no Acc mentioned) |
| 18 | Wang et al. [17] | 5372 | DL | DenseNet121-FPN | CT Scan | Acc 87–88% and Sens 76.3–81.1% |
| 19 | Wu et al. [22] | 495 | CoroNet (DL-based) | VGG-19 | CT Scan | Acc 76%, Sens 81.1%, and Spec 61.15% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caliman Sturdza, O.A.; Filip, F.; Terteliu Baitan, M.; Dimian, M. Deep Learning Network Selection and Optimized Information Fusion for Enhanced COVID-19 Detection: A Literature Review. Diagnostics 2025, 15, 1830. https://doi.org/10.3390/diagnostics15141830
Caliman Sturdza OA, Filip F, Terteliu Baitan M, Dimian M. Deep Learning Network Selection and Optimized Information Fusion for Enhanced COVID-19 Detection: A Literature Review. Diagnostics. 2025; 15(14):1830. https://doi.org/10.3390/diagnostics15141830
Chicago/Turabian StyleCaliman Sturdza, Olga Adriana, Florin Filip, Monica Terteliu Baitan, and Mihai Dimian. 2025. "Deep Learning Network Selection and Optimized Information Fusion for Enhanced COVID-19 Detection: A Literature Review" Diagnostics 15, no. 14: 1830. https://doi.org/10.3390/diagnostics15141830
APA StyleCaliman Sturdza, O. A., Filip, F., Terteliu Baitan, M., & Dimian, M. (2025). Deep Learning Network Selection and Optimized Information Fusion for Enhanced COVID-19 Detection: A Literature Review. Diagnostics, 15(14), 1830. https://doi.org/10.3390/diagnostics15141830







